Introduction
Dengue, a viral infection caused by Dengue virus (DENV), is a wide-spreading mosquito-borne infection with more than 14 million cases and nearly 10,000 deaths in 2024 [
1]
. The disease is now endemic in over 100 countries across the tropics and subtropics, placing nearly half of the world’s population at risk. Transmission occurs mainly through
Aedes aegypti and
Aedes albopictus mosquitoes, whose expanding geographic range has fueled the spread of dengue to new regions. Nepal reflects the DENV global trajectory. Once confined to the Terai plains, dengue has expanded into mid-hill and even high-altitude districts, including Kathmandu Valley and Solukhumbu at an altitude of 2,500 meters [
2]
. The country has experienced successive large outbreaks, with over 54,000 cases in 2022, 51,000 in 2023, and 40,000 in 2024 [
3,
4].
Dengue virus is a positive-sense RNA virus of the Flaviviridae family that cycles between humans and Aedes mosquitoes. There are four antigenically distinct serotypes (DENV-1 to DENV-4), which share significant sequence divergence and are further divided into genotypes, contributing to variations in disease severity and immune response [
5]
. Studies conducted in several districts in Nepal showed the circulation of three serotypes DENV-1, DENV-2, and DENV-3 during the 2022 and 2023 outbreak [
6,
7,
8]. Clinically, dengue manifests in three phases: febrile, critical, and recovery, ranging from mild febrile illness to severe disease with plasma leakage, bleeding, or organ involvement [
9]. Disease outcomes are shaped by serotype, host immune status, and the timing of infection. Secondary infection with a heterologous serotype is a major risk factor for severe disease due to antibody-dependent enhancement (ADE) and original antigenic sin (OAS), where cross-reactive but suboptimal immune responses intensify viral replication and immunopathology [
10]
.
Currently available diagnostic tools for dengue are essential but have limitations [
11]. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR) provides high accuracy during the first week of fever, yet sensitivity declines as viremia wanes. NS1 antigen assays perform well in primary infections but lose reliability in secondary cases, while routine IgM/IgG serology faces challenges from cross-reactivity with other flaviviruses and variable antibody kinetics [
12]. These shortcomings underscore the difficulty of accurately staging disease and predicting progression using conventional methods alone. With the expanding outbreaks, changing serotype dynamics, and increasing secondary infections, the search for reliable biomarkers to guide diagnosis and prognosis is more pressing than ever.
Severe dengue is largely driven by host immune dysregulation rather than direct viral injury; hence, there is growing interest in host-derived biomarkers for early prognostication. Traditional clinical markers such as platelet count and aspartate aminotransferase (AST) show limited predictive value during the critical febrile phase. Given these limitations, circulating microRNAs offer a promising alternative: they are stable, reflect real-time host–pathogen interactions, and may capture molecular signatures of disease stage and severity.
MicroRNAs (miRNAs) are short, non-coding RNAs containing 18–25 nucleotides that regulate gene expression by binding to target mRNAs and controlling their stability or translation. Beyond their fundamental role in cellular processes, they are key modulators of host–virus interactions, either by directly binding viral RNA to influence replication or by shaping host immune pathways such as Toll-like receptor and interferon signaling [
13]. Viruses, in turn, can exploit host miRNA machinery to favor persistence and immune evasion, as seen in dengue virus (DENV) infections, where dysregulated miRNAs contribute to cytokine storms and disease severity [
14]. Among these, miR-1246 stands out due to its non-canonical biogenesis, exceptional stability in circulation, and enrichment in exosomes, making it an attractive biomarker [
15].
This study aims to evaluate the potential of miR-1246 as a host-derived biomarker for dengue infection in Nepal, where current diagnostic tools remain insufficient for predicting disease progression. This research quantifies serum miR-1246 levels in dengue-positive patients and healthy controls, examines its variation across different serological groups (IgM, Weakly positive IgM, and IgG), and explores its association with conventional clinical parameters. By addressing the lack of Nepal-specific evidence, the study aims to determine whether miR-1246 can complement or improve upon existing diagnostic approaches, thereby contributing to earlier risk stratification and better clinical decision-making in resource-limited settings.
Materials and Methods
This study had obtained ethical approval from the Nepal Health Research Council (NHRC) (Reference number: 1760). All patients’ data were totally anonymous and were requested from clinicians involved in this study. This retrospective observational study included a total of 41 serum samples collected from Peoples Diagnostics Laboratory & Clinic in Kathmandu or K.B. Hospital Pvt. Ltd. or Scheer Memorial Adventist Hospital in Banepa. The inclusion criteria included dengue-positive cases confirmed by IgM and/or IgG positivity, while the dengue-negative individuals were included as healthy controls. The final sample size was 41 which consisted of 21 dengue positive-cases selected on the basis of serological results and 20 healthy dengue-negative individuals used as control group.
Total RNAs were extracted from serum samples using the Direct-zol RNA Miniprep Plus Kit supplied with 50 ml TRI Reagent (Zymo Research, Cat # R2071) following the manufacturer’s instructions. cDNA was synthesized from total extracted RNA using the miRNA first-strand cDNA Synthesis Kit (stem-loop method, Vazyme, Cat# MR101) following the manufacturer’s protocol. For first-strand cDNA synthesis, stem-loop primers specific for RNU6 (reference) and miR-1246 (target) were used (
Table 1). Genomic DNA contamination was removed from extracted RNA by incubating a 10 μL mixture of RNA, 5× gDNA Wiper Mix, and RNase-free water at 42°C for 2 minutes. The reverse transcription reaction contained 10 μL of previous mixture, 1 μL of stem-loop primer (2 μM), 2 μL 10× RT Mix, 2 μL HiScript II Enzyme Mix, and RNase-free water up to a final volume of 20 μL. The reaction was incubated with the following thermal profile: primer annealing at 25°C for 5 minutes, reverse transcription at 50°C for 15 minutes, and enzyme inactivation at 85°C for 5 minutes. The cDNA thus formed was stored at −20°C.
Quantification of miR-1246 was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, Cat# 1725124) on the Bio-Rad CFX96 C1000 real-time PCR system. Each 10 μL reaction contained 5 μL of 2× SYBR Green Supermix, 0.5 μL of miR-1246-specific forward primer (500 nM final concentration), 0.5 μL of universal reverse primer (500 nM final concentration), 1 μL of cDNA template, and 3 μL nuclease-free water. The mixture was thoroughly mixed and aliquoted into qPCR tubes. The quantitative PCR was performed with initial polymerase activation and DNA denaturation at 95°C for 30 seconds, followed by 40 amplification cycles of 95°C for 5 seconds and 60°C for 30 seconds. The melt curve analysis was performed from 65°C to 95°C with 0.5°C increments, held for 2 seconds at each step, to verify primer specificity. The end products of PCR were visualized in 2% (w/v) agarose gel to confirm that no non-specific products or primer dimers were formed.
All statistical analyses were performed using R statistical software (R Core Team, 2024). The analysis workflow included three main steps: group comparison, diagnostic evaluation, and subgroup analysis. To compare ΔCt values between healthy controls and dengue patients, the non-parametric Wilcoxon rank-sum test (Mann–Whitney U test) was used due to non-normal data distribution and unequal group sizes. The diagnostic potential of miR-1246 expression was assessed using Receiver Operating Characteristic (ROC) curve analysis implemented via the pROC package in R. The Area Under the Curve (AUC) was calculated with 95% confidence intervals, where values nearer to 1.0 indicate the best discriminative ability. For subgroup comparisons among dengue patients and healthy individuals, the Kruskal–Wallis test was applied. Significant results were further explored with Dunn’s post-hoc pairwise comparisons and adjusting p-values using the Bonferroni correction to control for multiple testing. Statistical significance was set at p < 0.05 with a confidence interval of 95%.
Results
The study comprised 21 dengue patients and 20 healthy controls. Dengue-positive samples were categorized into three subgroups: IgM positive, Weakly positive IgM, and IgG positive. The distribution of dengue serological results of the patients is presented in
Table 2.
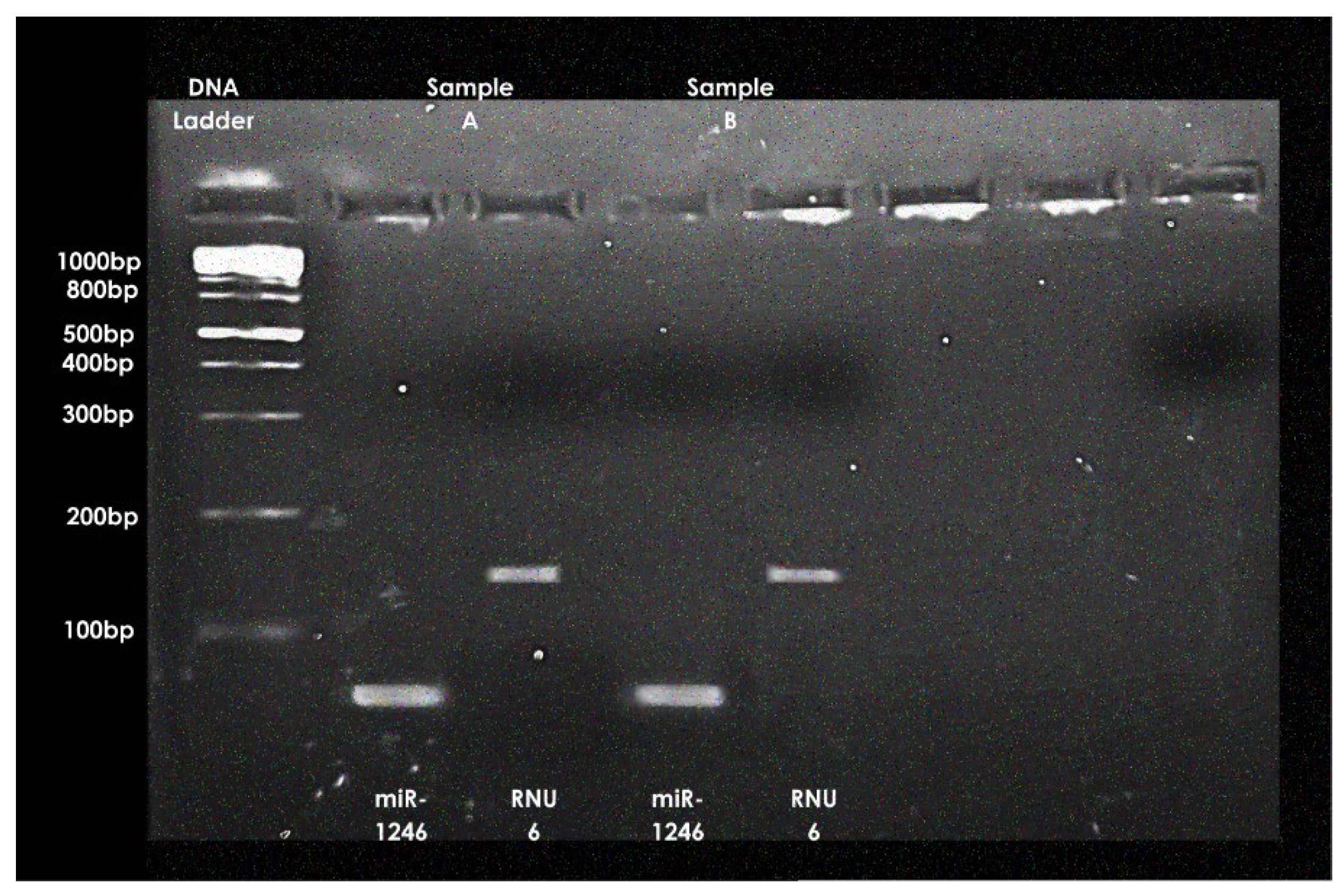
Specific amplification of miR-1246 (~80 bp) and RNU6 (~120 bp) was confirmed by end-point PCR and agarose gel electrophoresis (
Figure 1).
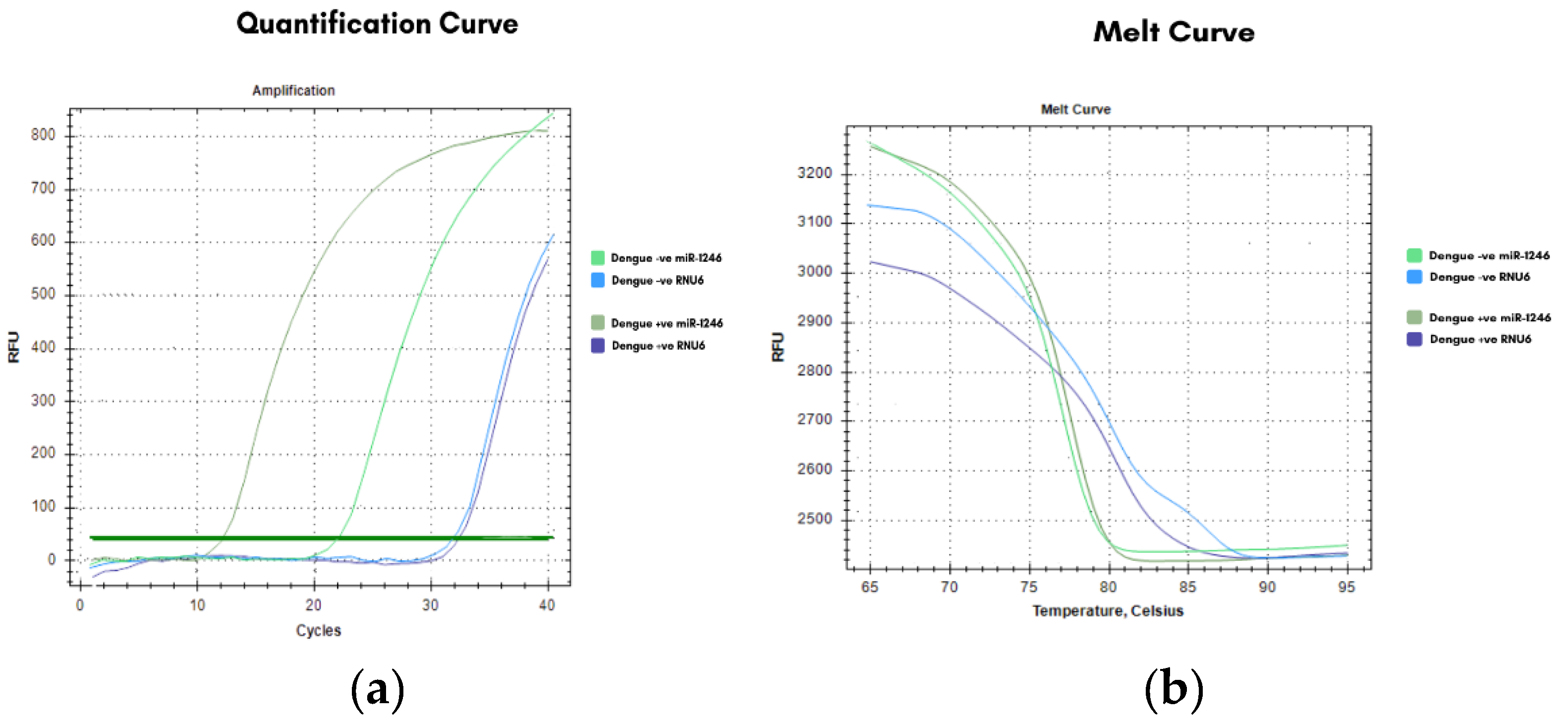
Real-time PCR analysis showed that the miR-1246 amplification curves (
Figure 2a) in dengue-positive samples appeared earlier than in dengue-negative controls, indicating higher expression, whereas RNU6 curves remained nearly identical across groups, confirming its stability as a reference gene. Melt curve and peak analyses (
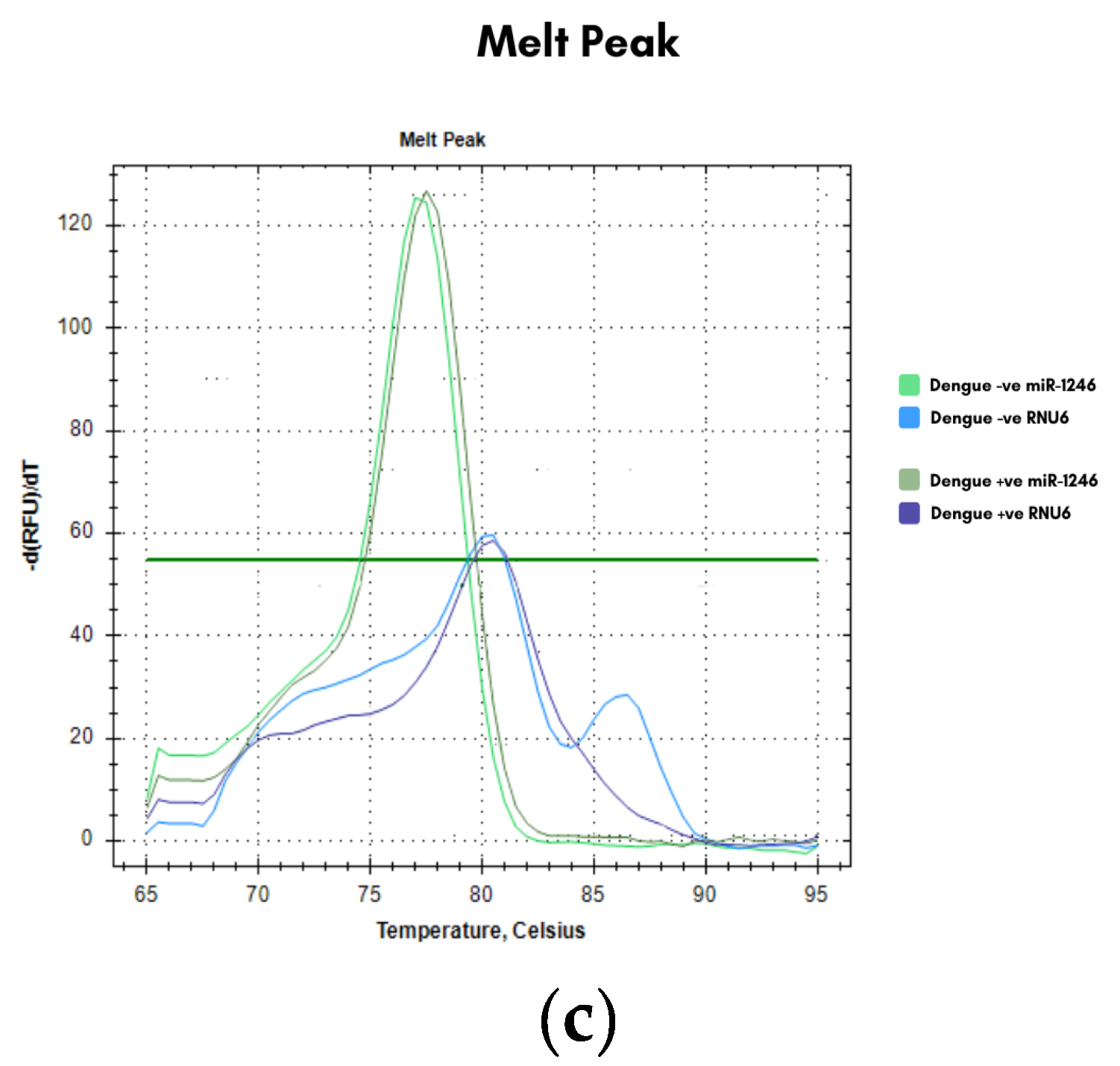
Figure 2b and 2c) demonstrated single, specific products without primer-dimer formation.
The average Ct for miR-1246 was significantly lower in dengue-positive patients (15.23 ± 2.46) compared to controls (18.97 ± 3.86), while RNU6 values remained stable (32.08 ± 3.08 vs. 32.39 ± 2.68).
The miR-1246 expression was found highest (147.97 ± 30.58) in IgM patients, intermediate (52.33 ± 16.02) in weakly positive IgM, and lowest in IgG patients (3.50 ± 3.31), as shown in
Table 3.
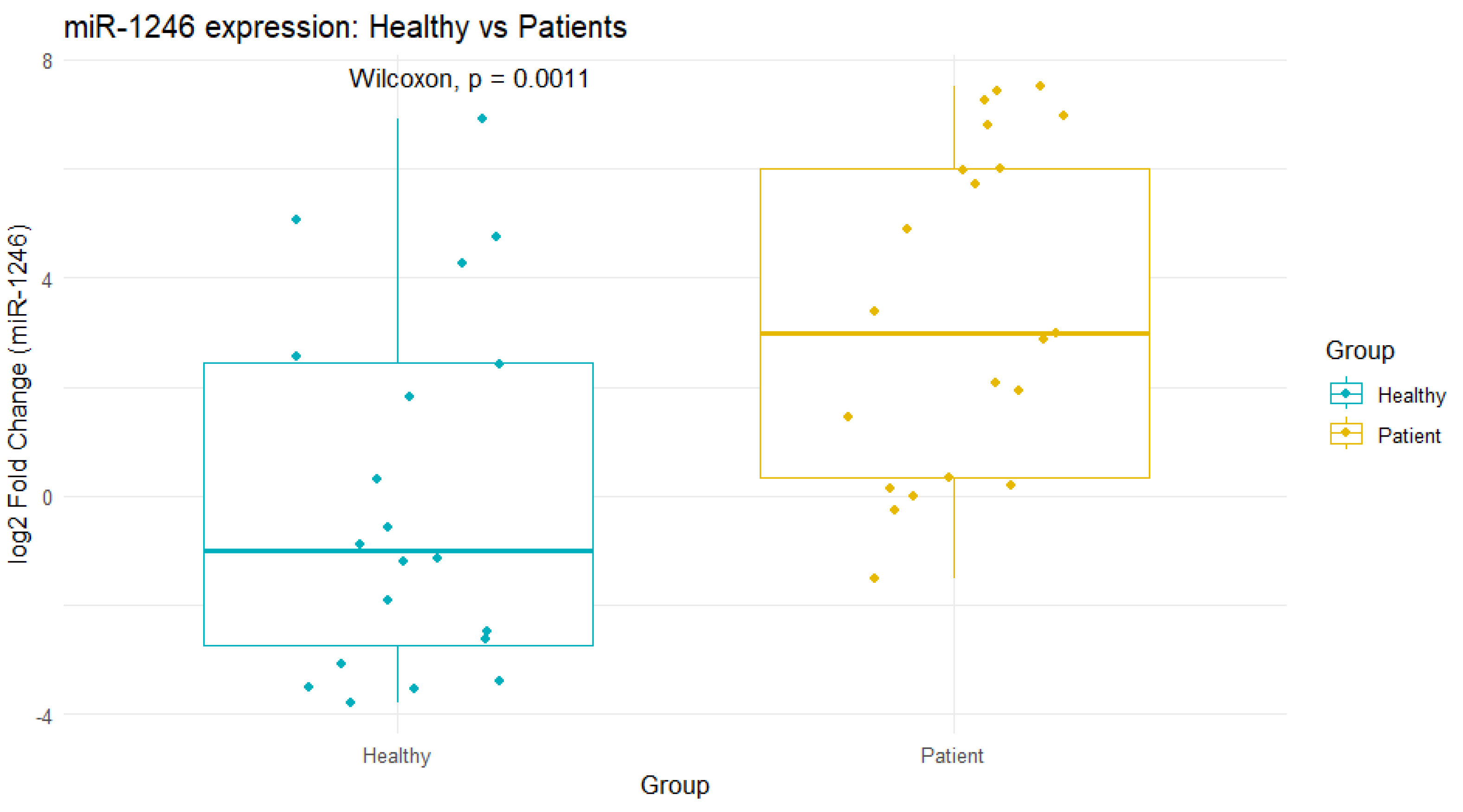
The mean fold change of miR-1246 in dengue-positive patients was 47.2 ± 62.6 compared to healthy individuals. This indicated a substantial upregulation of miR-1246 in dengue-positive serum. A Wilcoxon rank-sum test comparing healthy individuals (n = 20) versus dengue patients (n = 21) revealed near-complete separation between the two groups [p = 0.001, Cliff’s Δ = 1.00 (95% CI )] (
Figure 3)
The Kruskal–Wallis H test showed a statistically significant difference in miR-1246 expression across four groups (Healthy, IgM positive, Weakly positive IgM, and IgG positive) (p = 0.0001). Further, Dunn's post-hoc test was performed to identify specific group pairs with significant differences (
Table 4).
Analysis of clinical parameters across serological groups showed that miR-1246 expression was highest in IgM positive patients, who also exhibited mild to moderate thrombocytopenia (110–209 ×10³/µL), intermediate in the weakly positive IgM group with variable platelet recovery, and lowest in IgG positive patients with largely normalized counts. Leukocyte levels varied across individuals, with the lowest values observed in some weakly positive IgM patients (as low as 2.1 ×10³/µL), but no consistent phase-dependent trend was identified. Red cell indices [hemoglobin (Hb), red blood cells (RBC), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), and mean corpuscular hemoglobin concentration(MCHC)] remained within normal or slightly altered ranges across groups, with transient decreases during the transitional phase.
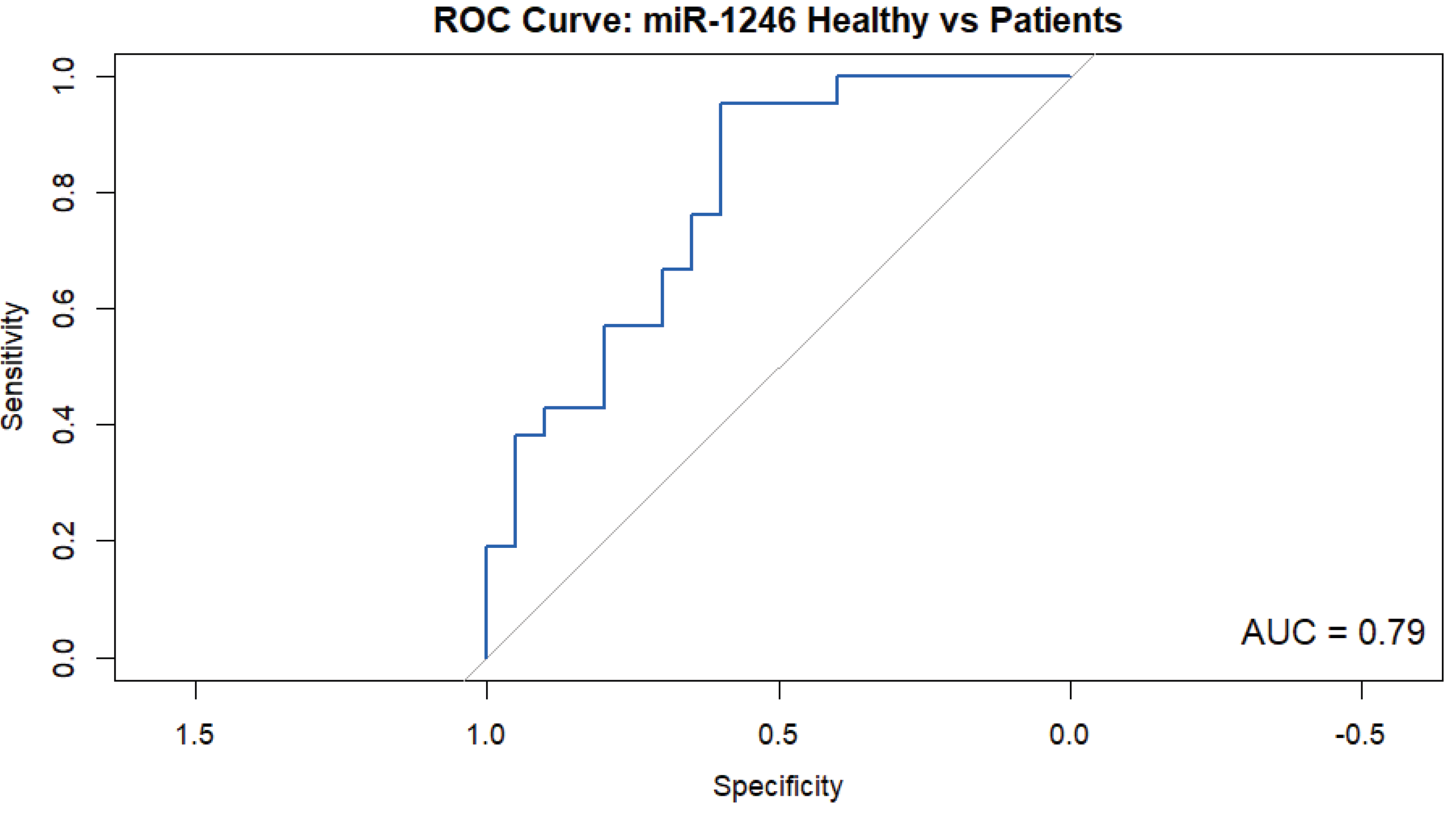
ROC analysis revealed an AUC of 0.79 (95% CI: 0.65–0.93), with a sensitivity of 95.24% and a specificity of 60.00%, showing the promising diagnostic capability of miR-1246 as a biomarker for dengue infection (
Figure 4).
Discussion
Dengue is a significant health challenge in Nepal, as current diagnostic techniques have inadequate predictive capacity regarding disease progression. Our research underscores the potential of miR-1246 as a molecular biomarker that may enhance conventional assays and act as a diagnostic and prognostic tool [
16,
17].
The RT-qPCR analysis confirmed that the level of miR-1246 was significantly higher in dengue-positive patients compared to the healthy individuals, as evidenced by consistently lower Ct values. The integrity of these results was reinforced by clean amplification and melt curves, as well as negative no-template controls. Importantly, RNU6 remained stable across groups, confirming its reliability as an endogenous control. Dengue patients exhibited an approximate 47-fold upregulation compared to controls, with results statistically significant (p = 0.001). Based on the ROC curve analysis, miR-1246 showed good diagnostic performance with an AUC of 0.79 and a sensitivity of 95.24%, suggesting a high value for screening. However, specificity was moderate at 60%, highlighting the need for miR-1246 to be integrated with additional biomarkers for improved diagnostic accuracy. These findings parallel earlier reports, such as those of Limothai et al. [
18], although slight differences in diagnostic performance may be attributable to cohort size, timing of sample collection, and technical variability.
The robustness of our assay was analyzed by amplification efficiency analysis, which was found to be 95.6% for miR-1246 and 89.4% for RNU6, both within the acceptable range for accurate relative quantification. Standard curves showed strong linearity and precision, with efficiency differences between target and reference genes under 7%, further confirming the robustness of the assay. Furthermore, to confirm the suitability of the assay endpoint, gel electrophoresis of both miR-1246 and RNU6, which resulted in the appearance of sharp, specific bands without evidence of non-specific amplification, supported the integrity of our PCR conditions.
We further analyzed miR-1246 expression across different serological phases to better understand its relationship with disease progression. Notably, expression peaked (about 148-fold) in IgM-positive patients representing the acute phase, and declined progressively (about 52-fold) through the transitional IgM/IgG group to the IgG-only convalescent group, where levels approached baseline (about 4-fold). This expression trajectory mirrors clinical parameters: acute-phase patients demonstrated severe thrombocytopenia [
19], a hallmark of active infection[
20], whereas convalescent patients showed recovery of blood counts [
21]. These findings suggest that miR-1246 tracks systemic immune activation and may serve as a proxy for disease phase. Statistical analysis using the Kruskal–Wallis H test confirmed these intergroup differences as significant (p < 0.05).
The correlation of these findings with clinical manifestations further confirmed our hypothesis. Thrombocytopenia was most prominent in the acute IgM-positive group, corresponding with the highest miR-1246 expression and consistent with dengue pathogenesis. In contrast, leukopenia did not show a consistent correlation with miR-1246 in our cohort, with the most severe case in transitional-phase patients rather than in the acute group, a discrepancy likely reflecting sample size limitations and phase misclassification. The miR-1246 concentration was also positively correlated with clinical parameters such as Hb, RBC, MCV, MCH, and MCHC level but negatively correlated with platelet counts, showing phase-specific changes[
21]. Taken together, these findings demonstrate that miR-1246 is significantly upregulated in dengue patients compared to controls, peaks during the acute phase of infection, and gradually returns to baseline during recovery.
This study has some limitations. The relatively small sample size and lack of comprehensive longitudinal sampling restricted our ability to fully capture expression changes across all stages of infection. Despite these limitations, the study provides important Nepal-specific evidence that supports the relevance of miR-1246 as a complementary biomarker.
Conclusions
Expression of miR-1246 increases by nearly 47 times during dengue (p = 0.001, Mann-Whitney U test), peaks during the acute phase around the time that patients exhibit thrombocytopenia, and then progressively decreases as patients recover. Although the miRNA's mediocre specificity of 60% suggests that some healthy people may have been flagged as false positives, its AUC of 0.79 and sensitivity of 95.24% demonstrate its effectiveness in identifying dengue cases. When combined, these results suggest that miR-1246 may be a promising biomarker for determining the severity of infection and the stage of the disease in dengue patients. Hence, although miR-1246 is not unique to dengue, within dengue, it can help us answer both who is infected and how far along they are, making it a valuable tool for risk stratification and an important foundation for future biomarker research.
References
- Haider, N.; Hasan, M.N.; Onyango, J.; Billah, M.; Khan, S.; Papakonstantinou, D.; Paudyal, P.; Asaduzzaman, M. Global Dengue Epidemic Worsens with Record 14 Million Cases and 9000 Deaths Reported in 2024. Int. J. Infect. Dis. 2025, 158, 107940. [Google Scholar] [CrossRef] [PubMed]
- Mathema, P. Climate Shifts and Urbanisation Drive Nepal Dengue Surge Available online:. Available online: https://www.rfi.fr/en/international-news/20241101-climate-shifts-and-urbanisation-drive-nepal-dengue-surge (accessed on 3 October 2025).
- 2025_04_searo_epi_bulletin.Pdf.
- Bhandari, S.; Blackburn, J.K.; Ryan, S.J. Spatial Patterns of Dengue Incidence in Nepal During Record Outbreaks in 2022 and 2023: Implications for Public Health Interventions. 2025. [Google Scholar] [CrossRef]
- Gutiérrez-Barbosa, H.; Castañeda, N.Y.; Castellanos, J.E. Differential Replicative Fitness of the Four Dengue Virus Serotypes Circulating in Colombia in Human Liver Huh7 Cells. Braz. J. Infect. Dis. 2020, 24, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Rimal, S.; Kharbuja, A.; Ray, M.K.; Shrestha, S.; Dulal, A.; Subedi, S.; Khadka, A.; Adhikari, N.; Dhimal, M.; et al. Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal. Pathogens 2025, 14, 639. [Google Scholar] [CrossRef] [PubMed]
- Rimal, S.; Shrestha, S.; Pandey, K.; Nguyen, T.V.; Bhandari, P.; Shah, Y.; Acharya, D.; Adhikari, N.; Rijal, K.R.; Ghimire, P.; et al. Co-Circulation of Dengue Virus Serotypes 1, 2, and 3 during the 2022 Dengue Outbreak in Nepal: A Cross-Sectional Study. Viruses 2023, 15, 507. [Google Scholar] [CrossRef] [PubMed]
- Rimal, S.; Shrestha, S.; Paudel, S.W.; Shah, Y.; Bhandari, G.; Pandey, K.; Kharbuja, A.; Kapandji, M.; Gautam, I.; Bhujel, R.; et al. Molecular and Entomological Characterization of 2023 Dengue Outbreak in Dhading District, Central Nepal. Viruses 2024, 16, 594. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control [CDC] Clinical Features of Dengue Available online:. Available online: https://www.cdc.gov/dengue/hcp/clinical-signs/index.html (accessed on 3 October 2025).
- An In-Depth Analysis of Original Antigenic Sin in Dengue Virus Infection | Journal of Virology Available online:. Available online: https://journals.asm.org/doi/10.1128/jvi.01826-10 (accessed on 6 October 2025).
- Gupta, B.P.; Haselbeck, A.; Kim, J.H.; Marks, F.; Saluja, T. The Dengue Virus in Nepal: Gaps in Diagnosis and Surveillance. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Hermann, L.L.; Thaisomboonsuk, B.; Poolpanichupatam, Y.; Jarman, R.G.; Kalayanarooj, S.; Nisalak, A.; Yoon, I.-K.; Fernandez, S. Evaluation of a Dengue NS1 Antigen Detection Assay Sensitivity and Specificity for the Diagnosis of Acute Dengue Virus Infection. PLoS Negl. Trop. Dis. 2014, 8, e3193. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Kothari, S.; Gohir, W.; Camargo, J.F.; Husain, S. MicroRNAs in Infectious Diseases: Potential Diagnostic Biomarkers and Therapeutic Targets. Clin. Microbiol. Rev. 2023, 36, e00015–23. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Lin, T.; Liu, C.; Cheng, C.; Han, X.; Jiang, X. microRNAs, the Link Between Dengue Virus and the Host Genome. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- St. John, A.L.; Rathore, A.P.S. Adaptive Immune Responses to Primary and Secondary Dengue Virus Infections. Nat. Rev. Immunol. 2019, 19, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Hoshino, I.; Mori, M.; Akutsu, Y.; Hanari, N.; Yoneyama, Y.; Ikeda, N.; Isozaki, Y.; Maruyama, T.; Akanuma, N.; et al. Serum microRNA Expression Profile: miR-1246 as a Novel Diagnostic and Prognostic Biomarker for Oesophageal Squamous Cell Carcinoma. Br. J. Cancer 2013, 108, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Samadian, M. A Review on the Role of miR-1246 in the Pathoetiology of Different Cancers. Front. Mol. Biosci. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Limothai, U.; Jantarangsi, N.; Suphavejkornkij, N.; Tachaboon, S.; Dinhuzen, J.; Chaisuriyong, W.; Trongkamolchai, S.; Wanpaisitkul, M.; Chulapornsiri, C.; Tiawilai, A.; et al. Discovery and Validation of Circulating miRNAs for the Clinical Prognosis of Severe Dengue. PLoS Negl. Trop. Dis. 2022, 16, e0010836. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Abreu, C.; Harris, M.; Shrader, J.; Sarvepalli, S. Severe Thrombocytopenia Associated with Dengue Fever: An Evidence-Based Approach to Management of Thrombocytopenia. Case Rep. Hematol. 2022, 2022, 3358325. [Google Scholar] [CrossRef] [PubMed]
- Nwe, K.M.; Ngwe Tun, M.M.; Myat, T.W.; Sheng Ng, C.F.; Htun, M.M.; Lin, H.; Hom, N.S.; Soe, A.M.; Elong Ngono, A.; Hamano, S.; et al. Acute-Phase Serum Cytokine Levels and Correlation with Clinical Outcomes in Children and Adults with Primary and Secondary Dengue Virus Infection in Myanmar between 2017 and 2019. Pathogens 2022, 11, 558. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, P.M.; Ranathunga, R.A.N.; Muthuthamby, M.M.; Bowatte, P.G.C.S.; Jeevathayaparan, S.; Perera, K.P.J. Study on Changes in a Subset of Full Blood Count Parameters of Patients with Dengue Fever and Dengue Haemorrhagic Fever Presented to Teaching Hospital – Kurunegala, Sri Lanka – a Descriptive Cross-Sectional Study. BMC Infect. Dis. 2025, 25, 1229. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).