1. Introduction
Dengue virus (DENV) has seen a concerning rise in cases over recent decades, posing a major threat to global public health. It affects people of all ages, including infants, children, and adults, placing a heavy burden on healthcare systems. The World Health Organization reports that there are approximately 390 million dengue infections each year, with 96 million presenting clinical symptoms. Although dengue is endemic in over 100 countries, primarily in the Americas, Southeast Asia, and the Western Pacific, its presence has also spread to unexpected regions in Europe [
1,
2].
DENV, which belongs to the Flaviviridae family, is considered the most prevalent arbovirus worldwide. Its impact on public health is unquestionable, and there are still many challenges in controlling it, particularly in tropical and subtropical regions. [
3].
There are four distinct but antigenically related serotypes of dengue virus, known as DENV-1, DENV-2, DENV-3, and DENV-4. Similar to other flaviviruses, these serotypes possess a single-stranded RNA genome encased within an icosahedral structure and surrounded by a lipid envelope [
4]. DENV is transmitted to humans by the bite of an infected female mosquito of the Aedes genus [
5], however, the widespread distribution and high adaptive capacity of Aedes albopictus suggest that this vector could also become an active transmitter of dengue in endemic regions [
6]. During the usual five-day period of viremia, the mosquitoes become infected when they feed on viremic humans. The virus is transferred from the intestinal tract of the mosquito to the salivary glands after an extrinsic incubation period, a process that takes about 10 days and is most rapid when the ambient temperature is high [
7].
The lack of specific treatments for dengue and the need for early detection highlight the importance of developing and enhancing molecular diagnostic tools. Traditional methods, such as indirect immunofluorescence and serological tests, have significant limitations, including the requirement for a second serum sample to confirm results and the persistence of detectable antibodies for a lifetime. These issues are further complicated by cross-reactivity in diagnostic tests, as reported by the Centers for Disease Control and Prevention [
1]. In response to these limitations, molecular techniques like RT-PCR and qRT-PCR have emerged as promising alternatives for dengue diagnosis, offering greater sensitivity and speed. However, the complexity and high costs associated with these methods present challenges for large-scale accessibility and application. In recent years, significant progress has been made in the development and optimization of isothermal-based methodologies for the molecular diagnosis of dengue virus and other arboviruses, such as Zika and chikungunya. Among these emerging methodologies, the RT-LAMP test stands out for its speed, sensitivity, specificity, low cost, and suitability for use in primary healthcare centers with limited infrastructure [
8].
In this study, we evaluated the usefulness of the RT-LAMP test for dengue virus diagnosis in Colombia and demonstrated that this combined approach, referred to here as COMB-RT-LAMP, is highly sensitive, making it a good alternative for implementation as a point-of-care test in areas with high dengue endemicity in Colombia. This research is not only aligned with the objectives of the Comprehensive Health Care Model (MIAS), which places people and their wellbeing at the center of care, but also seeks to optimize and standardize the diagnosis of dengue from educational, social and political perspectives. The standardization of these techniques not only improves the training of health professionals, but also has a positive impact on patient care and facilitates decision-making based on accurate epidemiological data.
2. Materials and Methods
A cross-sectional study was conducted to evaluate an RT-LAMP assay for the detection of DENV in three phases: a) standardization, b) laboratory validation and c) field validation. q-RT-PCR was used as the reference standard in the study.
A total of 158 serological samples were collected from suspected DENV cases at the Clínica General del Norte in Barranquilla city and sent to the Life Science Research Center of the Simon Bolivar University in Barranquilla for further processing and molecular detection of DENV.
All 158 samples were subjected to RNA extraction from 200 μl of serum using the Quick-RNA Viral Kit™ extraction kit (ZYMO Research), which employs silica magnetic beads for total RNA purification, following the manufacturer's recommendations. In brief, 400 μl of viral RNA buffer was added to each 200 μl of sample (2:1) and mixed gently. Subsequently, successive washes were performed to finally elute the RNA in 15 μl of DNAase/RNase-free water.
To perform RT-PCR, the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase™ kit (Invitrogen, catalog: 1257401) was used to perform RT-PCR. 8 μl of 2X reaction buffer containing 0.4 mM of each dNTP, 3.2 mM MgSO4, 0.3 μl of reverse transcriptase (SuperScript III RT/Platinum Taq Mix), 200 nM of each primer directed against the E gene DENV and 100 nM of CAL Red 610 fluorophore conjugated TaqMan probe were used. In addition, an endogenous control was introduced to the reaction (human RNase P gene), 100 nM of each primer and 100 nM of TaqMan-Hex probe, for amplification. RT-qPCR was performed with the CFX96™ Bio-Rad kit, under the following amplification conditions: 50 °C for 20 min, followed by 95 °C for 3 min, and ending with 45 cycles of 95 °C for 15 s and 58 °C for 30 s. The protocols used was the molecular diagnosis of Denv reported by Nunes [
9].
The RT-LAMP isothermal amplification assay was performed using the Warmstart Colorimetric LAMP 2X Master Mix (DNA & RNA) kit from New England Biolabs (NEB), following the manufacturer's instructions. In this study, primers reported by Dauner, et. al. [
10] targeting the 5´ UTR conserved region of DENV and primers reported by by Teoh, et. al. [
11] targeting the 3'UTR of the DENV were validated in a single reaction [
10,
11]. The primers sequences are shown in the
Table 1.
Before each LAMP reaction, a 10X mixture of single use of primers was prepared and a short incubation at 95 °C for 5 min was performed to eliminate any formation of secondary structures that could affect the color change of the reaction [
12]. (
Table 2).
The LAMP reaction was prepared using 10 μl of Warmstart colorimetric reagent LAMP 2X Master Mix (DNA & RNA), 5 μl of water, 3 μl of the previously extracted RNA and 2 μl of the 10X master mix of the respective primer set that had previously been incubated at 95 °C. The 10X primer mixture was always added in the lid of the reaction tube, to avoid unspecific amplifications that could start before the optimal amplification temperature. The reaction was started by a fast spinning of the tubes and an incubation at 65 °C for 30 minutes in a thermal cycler (SimpliAmp Thermal Cycler™, Thermo Fisher Scientific). After the incubation time, the result was visualized by colorimetry. The color change from pink to yellow was indicative of a positive reaction.
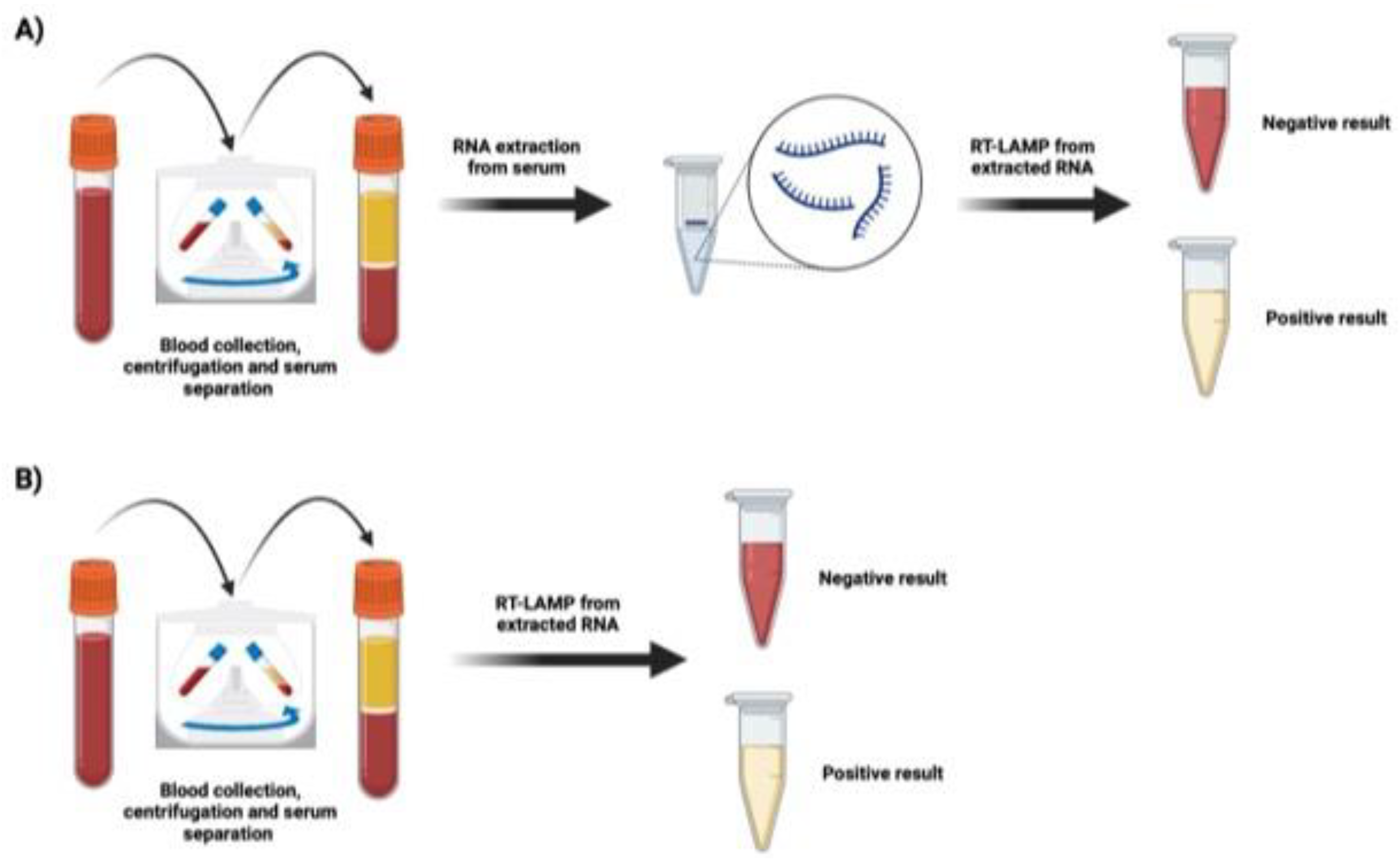
Figure 2 shows pink and yellow samples, pink samples correspond to negative samples and yellow samples correspond to positive results for DENV (figure 2A).
We apply a simple human blood lysis procedure for the LAMP reaction to simplify the DNA/RNA preparation process. A total of 100-200 µl of human blood in a heparin tube was transferred to a tube containing 900 µl of blood lysis buffer (0.1% TritonX-100 in DDW), vortexed for 1 minute, and the sample was allowed to stand for 30 minutes. Then 10 µl of lysed blood was used for the LAMP reaction which was carried out as usual.
Figure 2b.
The statistical analysis of the results was performed with the statistical software Rstudio version (1.2.50033). The specificity of the RT-LAMP test was calculated from the fraction of RT-qPCR negative samples that were also negative in the RT-LAMP assays. Sensitivity was given as the fraction of samples that were positive for RT-qPCR that were also positive for RT-LAMP. Positive predictive and negative predictive value and kappa index were calculated to estimate the concordance of RT-LAMP results with RT-qPCR.
3. Results
In this study, we aimed to explore the utility of two previously published RT-LAMP protocols for the rapid diagnosis of dengue virus in samples from febrile patients with a presumptive serological or antigen diagnosis of dengue [
10,
11]. When we evaluated both protocols separately, we observed sensitivity levels that did not exceed 40%, even in samples with a high viral load (data no shown). We wondered if combining both protocols using the primer sets reported in each study could improve the sensitivity of the method in samples from Colombian patients.
We evaluated the performance of the COMB-RT-LAMP protocol in 158 samples previously diagnosed as DENV-positive by IgM or NS1 antigen test. RNA was extracted from these samples, and they were confirmed as DENV-positive by qRT-PCR using the CDC protocol with primers targeting the E gene [
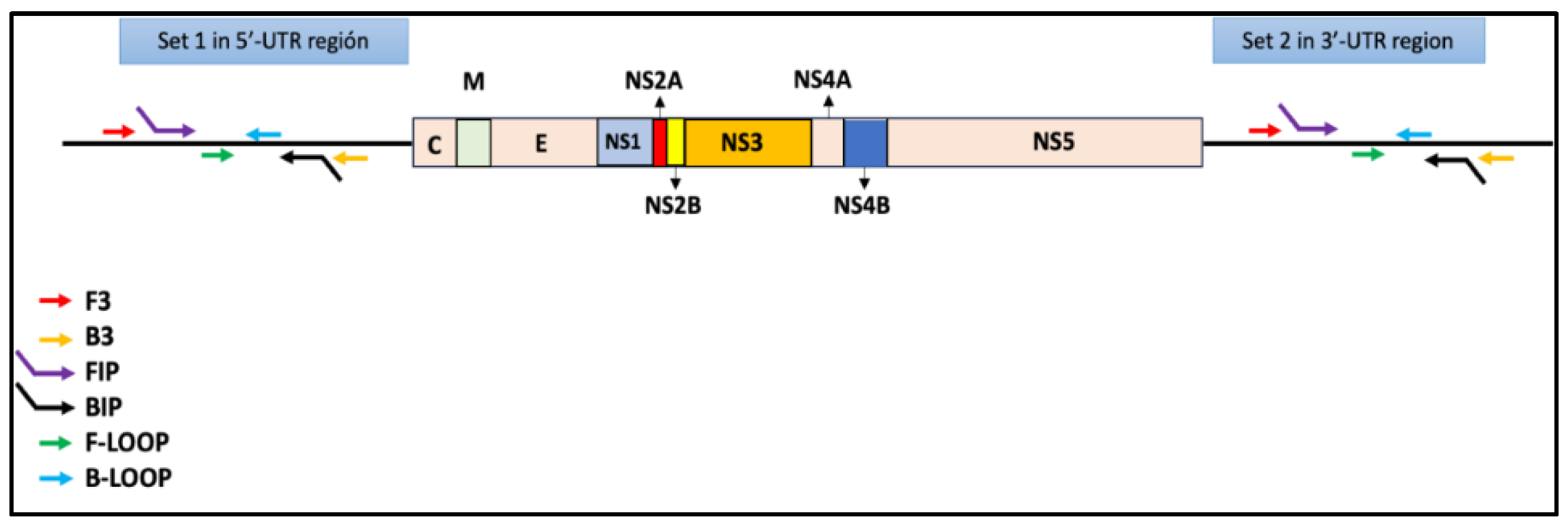
9]. In parallel, the samples were evaluated using the COMB-RT-LAMP method, which amplifies two conserved sequences in the 5' and 3'-UTR regions of the DENV genome (
Figure 1). Initially, we evaluated a conventional RT-LAMP protocol using extracted RNA from serum samples of febrile patients as starting material (
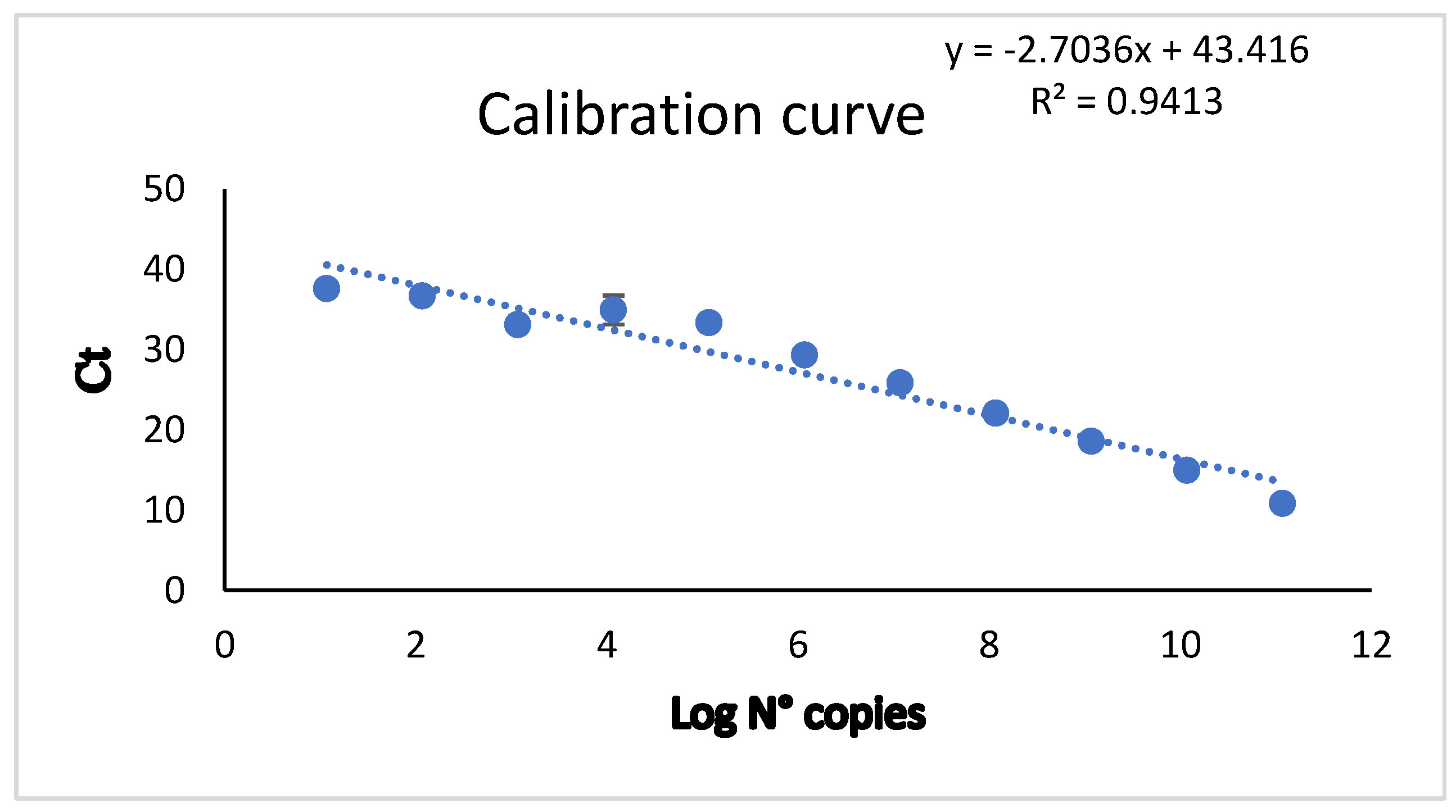
Figure 2A). We generated a calibration curve to determine the copy number in relation to Ct values. To achieve this, we used a DENV2 control and performed serial dilutions. The formula and curve were prepared according to the methodology described by Hurtado et al. [
12], as shown in
Table 3.
Figure 1.
Diagram depicting the DENV genome, showing the 5'-UTR and 3'-UTR regions recognized by set 1 and set 2, respectively.
Figure 1.
Diagram depicting the DENV genome, showing the 5'-UTR and 3'-UTR regions recognized by set 1 and set 2, respectively.
Figure 2.
COMB-RT-LAMP colorimetric reaction for DENV diagnosis. Figure generated in Biorender.
Figure 2.
COMB-RT-LAMP colorimetric reaction for DENV diagnosis. Figure generated in Biorender.
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
The standard curves were generated through linear regression analysis of the Ct values obtained from real-time PCR for each amplification reaction against the log
10 RNA copy number (
Figure 3). This allowed us to determine the copy number, as shown in
Table 3 and
Table S1 table (
Supplementary Data). Fifty-one samples were positive for RT-qPCR and 107 negatives, while 47 were positive for RT-LAMP and 111 negatives. There were 152 concordant samples and 6 discordant samples.
The results of the COMB-RT-LAMP test for DENV are shown below. The reaction was performed using the colorimetric reagent Warmstart (NEB), the 2 sets of primers mentioned in
Table 1 and 2, and samples with suspected DENV, water was included as a negative control and a DENV isolate from infected cell cultures was included as a positive control.
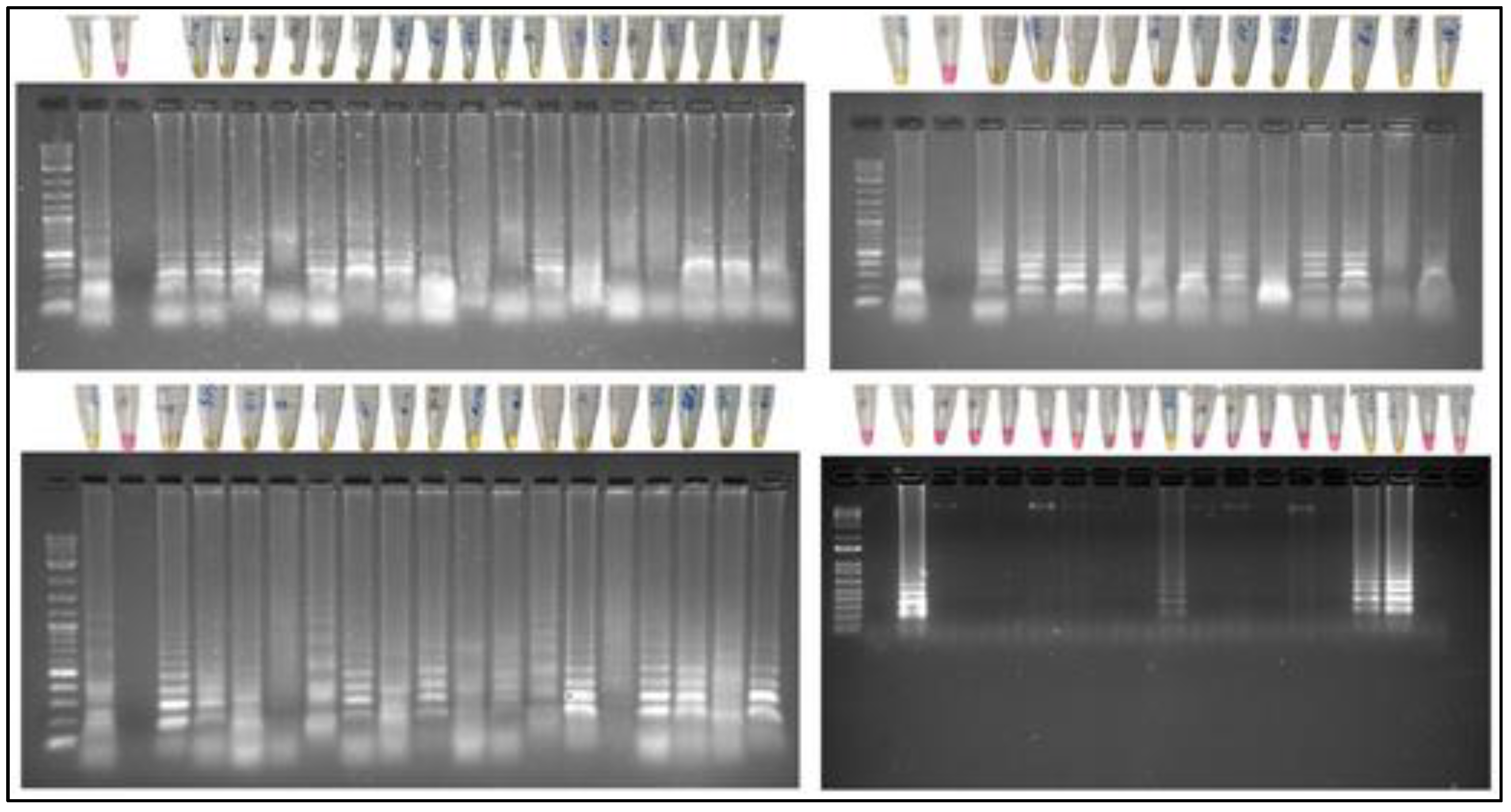
Figure 4 shows pink and yellow samples, pink samples correspond to negative samples and yellow samples correspond to positive results for DENV.
Electrophoresis was performed to corroborate the results by observing the electrophoretic run.
Figure 4 show that the LAMP-positive samples show a banding pattern characteristic of the positive samples, because the LAMP reaction generates fragments of different sizes, while the negative samples do not show any banding. Therefore, the colorimetric results correspond with the electrophoretic runs.
The sensitivity and specificity of the RT-LAMP technique were determined in comparison to RT-qPCR. A sensitivity of 0.96 was achieved, indicating that samples positive by RT-qPCR were also positive by RT-LAMP. A specificity of 0.96 was also obtained, meaning that the RT-LAMP technique correctly identified 96% of the RT-qPCR-negative samples. These results are summarized in the contingency table (
Table 5).
As shown in
Table 5, out of the 158 samples tested, 45 were true positives, 107 were true negatives, 4 were false positives, and 2 were false negatives.
The COMB-RT-LAMP technique for DENV diagnosis using the primer sets showed a sensitivity value of 0.96, suggesting that the technique was efficient in detecting DENV-positive samples. As shown below:
While the specificity found for the COMB-RT-LAMP technique was 0.96, i.e., it was specific for detecting negative samples.
While the specificity found for the COMB-RT-LAMP technique was 0.96, i.e., it was specific for detecting negative samples.
The positive predictive value (VPP) was 0.91, as shown below:
VPP= A/(A+B)
VPP= 45 / (45+4) =0.91.
The negative predictive value (NPV) was 0.98.
NPV= D/(C+D)
NPV= 107/ (2+107) =0.98.
R studio showed VPP of COMB-RT-LAMP DENV about 0.92 (0.80, 0.98) and NPV about 0.98 (0.94, 1.00).
The COMB-RT-LAMP technique for the detection of Dengue shows promising results since it is an easy-to-use diagnostic technique in less time and with characteristics of sensitivity and specificity that are competitive with the gold standard.
It was analyzed the concordance through kappa index (κ) for COMB-RT-LAMP DENV, the formula and interpretation of the Cohen kappa coefficient, defined as a function of the proportion of observed agreement and proportion of chance agreement (
Table 6).
The positive predictive value (VPP) was 0.91, as shown below:
VPP= A/(A+B)
VPP= 45 / (45+4) =0.91.
The negative predictive value (NPV) was 0.98.
NPV= D/(C+D)
NPV= 107/ (2+107) =0.98.
R studio showed VPP of COMB-RT-LAMP DENV about 0.92 (0.80, 0.98) and NPV about 0.98 (0.94, 1.00).
The COMB-RT-LAMP technique for the detection of Dengue shows promising results since it is an easy-to-use diagnostic technique in less time and with characteristics of sensitivity and specificity that are competitive with the gold standard.
It was analyzed the concordance through kappa index (κ) for COMB-RT-LAMP DENV, the formula and interpretation of the Cohen kappa coefficient, defined as a function of the proportion of observed agreement and proportion of chance agreement.
Table 6.
The Index kappa was 0.91, the COMB-RT-LAMP DENV results were almost perfect agreement in relation to PCR DENV results. The Po was 0.96 and Pe was 0.576. These values demonstrated it was possible to detect DENV using COMB-RT-LAMP and the technique had almost perfect agreement with the Gold Standard test. The COMB-RT-LAMP DENV show to be very sensible and specific test to diagnostic DENV. The
Table 7 resume the results of the COMB-RT-LAMP DENV test.
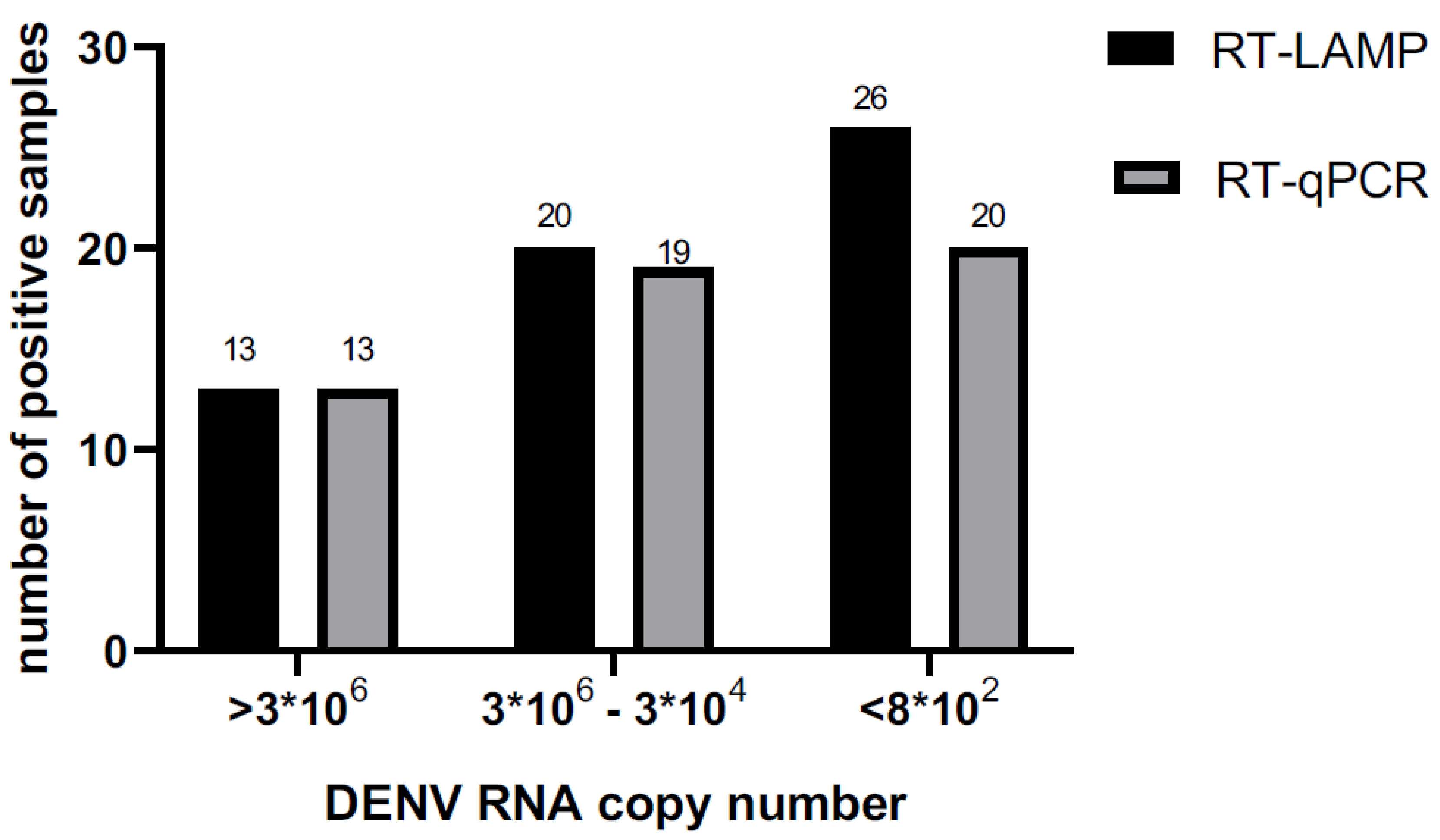
In relation to viral number copy we calculated LAMP sensitivity, so the sensitivity of LAMP in samples that have more than 3*106 copies/ µl was 1. Samples with copies number/ µl since 3*104 copies/ µl the sensitivity was 0.95. The LAMP test to detect DENV was very sensitive, it was sensible to detect samples with load viral lower than 8*102 copies number/µl.
Table 8.
According to the viral copy number, it was observed that samples with a viral load higher than 3 × 10^6 copies/μl showed concordance in both the RT-LAMP and RT-qPCR tests.
As illustrated in the figure 6, 13 samples were positive in both tests. In the viral load range between 3 × 10^6 and 3 × 10^4 copies/μl, high sensitivity was recorded; however, the results were not completely concordant between the two techniques. As the viral load decreased, the sensitivity of the LAMP test also reduced. Samples with less than 8 × 10^2 copies/μl were found to be inconsistent between the two techniques: 25 samples were RT-qPCR positive, while 19 were LAMP positive.
Figure 7.
It is important for these isothermal approaches to be feasible for use in primary care settings. Therefore, we decided to explore the possibility of performing our COMB-RT-LAMP approach directly on serum samples from four febrile patients without the need for RNA extraction. One hundred µl of human blood were lysed using lysis buffer (0.1% TritonX-100 in DDW), vortexed for 1 minute, and the sample was allowed to stand for 30 minutes. Then the RT-LAMP was carried at 65°C for 45 minutes, all the four samples were positive to DENV (
Supplementary Figure S1).
4. Discussion
Dengue is a serious global public health issue, exacerbated by climatic and migratory phenomena that have expanded the range of the Aedes aegypti vector. The World Health Organization, through the ASSURED criteria, has urged scientists to develop molecular diagnostic methods that overcome the limitations of PCR and real-time PCR [
13]. This call has led to the development of various molecular diagnostic methodologies for the dengue virus, with enormous potential for use in low-resource settings. Among these, LAMP has gained significant importance due to its simplicity, sensitivity, specificity, low cost, and suitability for use in rural areas with inadequate infrastructure. For the dengue virus, many protocols have been reported to date, each with its own unique features and efficiencies [
10,
11,
14,
15,
16,
17,
18,
19,
20].
However, existing protocols are often less reproducible when applied to different geographic regions, with less common serotypes, or even different viral genotypes within each serotype. Therefore, there is a need to optimize these protocols for each region to establish their utility as a reproducible molecular test on a global scale.
In this study, we sought to validate the efficacy of two previously reported RT-LAMP (in Peru and Malaysia) protocols for dengue diagnosis in an endemic area from Colombia [
10,
11]. Although both protocols had demonstrated high sensitivity and specificity in studies conducted in Peru and Malaysia, respectively, these protocols did not replicate this performance in our population. The sensitivity and specificity obtained by applying each protocol separately were unsatisfactory, which led us to explore the possibility of combining both approaches to improve diagnostic accuracy.
The hypothesis that combining the two protocols could improve diagnostic accuracy was confirmed in this study. By integrating the 20 primers from both protocols into a COMB-RT-LAMP assay, a significant number of positive samples were detected. In fact, the concordance between RT-LAMP and RT-qPCR results was high (152 out of 158 samples), with a sensitivity and specificity of 0.96, suggesting that the COMB-RT-LAMP has a potential as a diagnostic tool from dengue in Colombia
Out of the 158 suspected DENV cases tested, our RT-LAMP detected 47 positive samples, while RT-qPCR detected 51 positive samples. The accuracy between the two methods was high (152/158 samples), suggesting that the combined RT-LAMP may have diagnostic potential. The variations in performance observed between qRT-PCR and LAMP were mainly due to viral load, as it was noted that as RNA levels decrease, the sensitivity of LAMP also decreases (Figure 6 and
Table 6).
The 3'-UTR region of the dengue virus is a highly conserved sequence among the four circulating serotypes worldwide, which is why it is routinely used as a diagnostic target. However, serotype 4 presents some variations in this region, making it necessary to design a specific set of primers for this serotype. A probable cause for the low sensitivity of the Dauner and Teoh primer sets is that those studies did not analyze clinical samples from serotype 4, whereas in the clinical samples analyzed in this study, there were several serotype 4 samples.
Our findings underscore the need for further exploration and optimization of molecular diagnostic methods for DENV in specific populations. The variability in the efficacy of RT-LAMP protocols according to geographic and demographic context highlights the importance of validating these techniques in local cohorts before widespread implementation in public health programs In addition, the integration of complementary methods and a more comprehensive approach for the detection of all DENV serotypes in populations with high genetic diversity and different levels of endemicity needs to be considered.
LAMP is a highly robust assay, allowing it to be performed even without extensive sample preparation in scenarios where false negatives due to low-copy-number samples are acceptable. It is estimated that individuals with acute dengue have varying viral loads, ranging from 10³ to 10¹² plaque-forming units per milliliter [
19], and these high viral titers can be detected by LAMP even without prior sample preparation. We tested COMB-RT-LAMP in samples extraction free, LAMP showed be sensitive to detect DENV without previous RNA extraction.
Figure S1 Supplementary Data.
It has been previously reported that asymptomatic (non-febrile) individuals with dengue exhibit viral loads like those of symptomatic individuals [
21]. Additionally, it has been observed that the decay rate of viral RNA in asymptomatic individuals is slower. This underscores the potential of the LAMP test to be used as a screening tool in dengue-endemic areas, serving as a strategy for active dengue surveillance.
The discrepancies observed between the RT-LAMP and RT-qPCR results can be attributed to various factors, including differences in viral load among samples, the efficiency of RNA extraction, and the specificity of the primers for the different DENV serotypes. Previous studies have demonstrated that using degenerate primers can help mitigate some of these challenges, enabling the detection of multiple serotypes in a single reaction. However, the variability in PAN-LAMP efficacy observed in our study indicates that further optimization of the protocol is necessary to account for the genetic diversity of DENV strains across different regions [
19].
We found that the limit of detection of PAN_LAMP was around 800 viral copies of DENV for all serotypes using the primers reported by Teoh [
11], however Zhou et al. using the same primers showed different limit of detection for the four serotypes, 74 for DEN-1, 252 for DENV-2, 78 copies for DENV-3 and 35 copies for DENV-4 [
22]. They used a high-fidelity polymerase DNA that had exonuclease activity 3′-5′, which improved the error no pared, so the Bst polymerase start extension. They used it to establish the novel mismatch-tolerant RT-LAMP system [
22].
Hu et al. using RT-LAMP assay detected 100% of clinical strain and 98.9% of DENV-infected patient samples, no false positives were found [
14]. The detection limit was about 10 copies template of DENV1-4, ten-fold more sensitive than RT-PCR or real-time PCR. They used different targets for all DENV serotypes: non-structural protein (NS) 2A of DENV1, NS4B of DENV2, NS4A of DENV3 and the 3′ untranslated region of the NS protein of DENV4 [
14]. They used 63 °C, as isothermal temperature for 45 min while we used 65°C for 30 minutes as isothermal temperature. Parida et al. used primers from the 3′ noncoding region (NCR) [
18]. They found a higher sensitivity was 100% to detect the viral RNA in patient serum samples and 93% specificity. The RT-LAMP were carried out at 63°C for 1 hour using Loopamp real-time turbidimeter. Lau et al. using primers target to NCR to detect DENV at real time, so they developed RT-LAMP real time using Loopamp real-time turbidimeter, it was positive sample when the turbidity reached 0.1 within 60 min at 650 nm, the isothermal temperature was 65°C, but the incubation time was 30 min for DENV 1, 2 and 3 serotypes and 45 min for DENV 4 serotype. It was the first study that included a HNB dye to RT-LAMP to detect DENV in real time. This assay was very sensitive and specific to detect DENV compared to ELISA and qRT-PCR. The detection limit was 10 copies template too [
15].
Li et al. developed a RT-LAMP assay for detection DENV1-4, Japanese encephalitis virus (JEV), and West Nile virus (WNV) simultaneously. They used the primers reported by Parida et al. and carried out at 63°C for 15-30 min. The detection limits were 100-fold higher than of RT-PCR, they used sera of 168 patients and 279 pools of field-caught blood sucked, mosquitoes, the concordance between the both test, RT-LAMP and RT-PCR was 1 [
16].
Development sequencing portable together with isothermal amplification is a novel method reported in 2017 in Indonesian patient samples, the use of both methodologies allowed amplify and sequence viral genome easier than the conventional techniques previously reported, with a high sensitivity and specificity. This would be further evidence that LAMP is a good, fast, reliable and sensitive method for genotyping [
20].
Finally, our results are consistent with findings from other studies that highlight the importance of assay characteristics such as robustness and ease of use in resource-limited settings. In such environments, methods like RT-LAMP can be especially valuable. The ability of RT-LAMP to operate effectively across a wide range of temperatures and without requiring extensive sample preparation makes it a practical option for use in rural areas or during outbreak situations where access to well-equipped laboratories is limited.
5. Conclusions
The COMB-RT-LAMP test proved to be a confident approach for the molecular diagnosis of dengue virus in Colombia. It was much faster than PCR and showed promising competitive results, which were confirmed by the results obtained with PCR.
The sensitivity and specificity of the COMB-RT-LAMP technique for molecular diagnosis of DENV were 0.96 for both parameters. The positive predictive value was 0.91 and the negative predictive value was 0.98. This indicates that the two sets of primers used to detect the 5'UTR and 3'UTR regions of the 4 DENV serotypes were specific for amplifying the region of interest and diagnosing DENV in the samples.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Author Contributions
Conceptualization, P.L.L.A., A.H.A. and D.O.Y.; methodology, D.P.A., P.L.L.C., E.L.K, H.G.L., R.J.S., S.L.N., B.S.C., G.J., B.L.Y.; validation, E.L.K, H.G.L., R.J.S., S.L.N., B.S.C., G.J., B.L.Y.; formal analysis, E.L.K., H.G.L., P.L.L.A.; investigation, E.L.K., H.G.L., P.L.L.A., A.H.A., D.O.Y.,; data curation, E.L.K., H.G.L., P.L.L.A.; writing—original draft preparation, E.L.K., H.G.L., P.L.L.A..; writing—review and editing, P.L.L.A.; supervision, P.L.L.A., A.H.A..; project administration, P.L.L.A., A.H.A.; funding acquisition, P.L.L.A., A.H.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science, Technology Innovation through Sistema General de Regalías, grant number BPIN: 2021000100270 and the APC was funded by BPIN: 2021000100270.
Institutional Review Board Statement
This work was approved by the ethics committee of Simón Bolívar University (The project was formalized through Project Approval Record No. CIE-USB-CE-0382-00 on August 11, 2021), and all subjects signed an informed consent prior to sample collection.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
We are truly grateful to the microbiology unit of the Clinica de la Costa and Clínica General del Norte in Barranquilla City (Colombia) for the recruitment and sample collection of the patients enrolled in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- CDC, «Clinical Testing Guidance for Dengue», Dengue. Accedido: 18 de agosto de 2024. [En línea]. Disponible en: https://www.cdc.gov/dengue/hcp/diagnosis-testing/index.html.
- I. Gjenero-Margan et al., «Autochthonous dengue fever in Croatia, August-September 2010», Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull., vol. 16, n.o 9, p. 19805, mar. 2011.
- S. D. Sekaran y H. Artsob, «Molecular diagnostics for the detection of human flavivirus infections», Expert Opin. Med. Diagn., vol. 1, n.o 4, pp. 521-530, dic. 2007. [CrossRef]
- B. L. Ligon, «Dengue fever and dengue hemorrhagic fever: A review of the history, transmission, treatment, and prevention», Semin. Pediatr. Infect. Dis., vol. 16, n.o 1, pp. 60-65, ene. 2005. [CrossRef]
- M. G. Guzman et al., «Dengue: a continuing global threat», Nat. Rev. Microbiol., vol. 8, n.o S12, pp. S7-S16, dic. 2010. [CrossRef]
- C. Paupy, H. Delatte, L. Bagny, V. Corbel, y D. Fontenille, «Aedes albopictus, an arbovirus vector: From the darkness to the light», Microbes Infect., vol. 11, n.o 14-15, pp. 1177-1185, dic. 2009. [CrossRef]
- R. E. Whitmire, D. S. Burke, A. Nisalak, B. A. Harrison, y D. M. Watts, «Effect of Temperature on the Vector Efficiency of Aedes aegypti for Dengue 2 Virus», Am. J. Trop. Med. Hyg., vol. 36, n.o 1, pp. 143-152, ene. 1987. [CrossRef]
- T. Notomi, «Loop-mediated isothermal amplification of DNA», Nucleic Acids Res., vol. 28, n.o 12, pp. 63e-663, jun. 2000. [CrossRef]
- P. C. G. Nunes, M. R. Q. Lima, y F. B. Dos Santos, «Molecular Diagnosis of Dengue», en Dengue Virus, vol. 2409, R. Mohana-Borges, Ed., en Methods in Molecular Biology, vol. 2409. , New York, NY: Springer US, 2022, pp. 157-171. [CrossRef]
- A. L. Dauner et al., «Development of a pan-serotype reverse transcription loop-mediated isothermal amplification assay for the detection of dengue virus», Diagn. Microbiol. Infect. Dis., vol. 83, n.o 1, pp. 30-36, sep. 2015. [CrossRef]
- B.-T. Teoh et al., «Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification», BMC Infect. Dis., vol. 13, n.o 1, p. 387, dic. 2013. [CrossRef]
- L. Hurtado et al., «Validación clínica de la prueba RT-LAMP para el diagnóstico rápido del SARS-CoV-2», Biomédica, vol. 42, pp. 59-72, oct. 2022. [CrossRef]
- K. J. Land, D. I. Boeras, X.-S. Chen, A. R. Ramsay, y R. W. Peeling, «REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes», Nat. Microbiol., vol. 4, n.o 1, pp. 46-54, dic. 2018. [CrossRef]
- S. Hu et al., «Development of reverse-transcription loop-mediated isothermal amplification assay for rapid detection and differentiation of dengue virus serotypes 1–4», BMC Microbiol., vol. 15, p. 265, nov. 2015. [CrossRef]
- Y.-L. Lau et al., «Colorimetric Detection of Dengue by Single Tube Reverse-Transcription-Loop-Mediated Isothermal Amplification», PLOS ONE, vol. 10, n.o 9, p. e0138694, sep. 2015. [CrossRef]
- S. Li et al., «Simultaneous detection and differentiation of dengue virus serotypes 1-4, Japanese encephalitis virus, and West Nile virus by a combined reverse-transcription loop-mediated isothermal amplification assay», Virol. J., vol. 8, n.o 1, p. 360, dic. 2011. [CrossRef]
- B. Lopez-Jimena et al., «Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype», PLoS Negl. Trop. Dis., vol. 12, n.o 5, p. e0006381, may 2018. [CrossRef]
- M. Parida et al., «Rapid Detection and Differentiation of Dengue Virus Serotypes by a Real-Time Reverse Transcription-Loop-Mediated Isothermal Amplification Assay», J. Clin. Microbiol., vol. 43, n.o 6, pp. 2895-2903, jun. 2005. [CrossRef]
- D. W. Vaughn et al., «Dengue Viremia Titer, Antibody Response Pattern, and Virus Serotype Correlate with Disease Severity», J. Infect. Dis., vol. 181, n.o 1, pp. 2-9, ene. 2000. [CrossRef]
- J. Yamagishi et al., «Serotyping dengue virus with isothermal amplification and a portable sequencer», Sci. Rep., vol. 7, n.o 1, p. 3510, jun. 2017. [CrossRef]
- P. Matangkasombut et al., «Dengue viremia kinetics in asymptomatic and symptomatic infection», Int. J. Infect. Dis., vol. 101, pp. 90-97, dic. 2020. [CrossRef]
- Y. Zhou et al., «A Mismatch-Tolerant Reverse Transcription Loop-Mediated Isothermal Amplification Method and Its Application on Simultaneous Detection of All Four Serotype of Dengue Viruses», Front. Microbiol., vol. 10, p. 1056, may 2019. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).