Submitted:
06 November 2025
Posted:
07 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
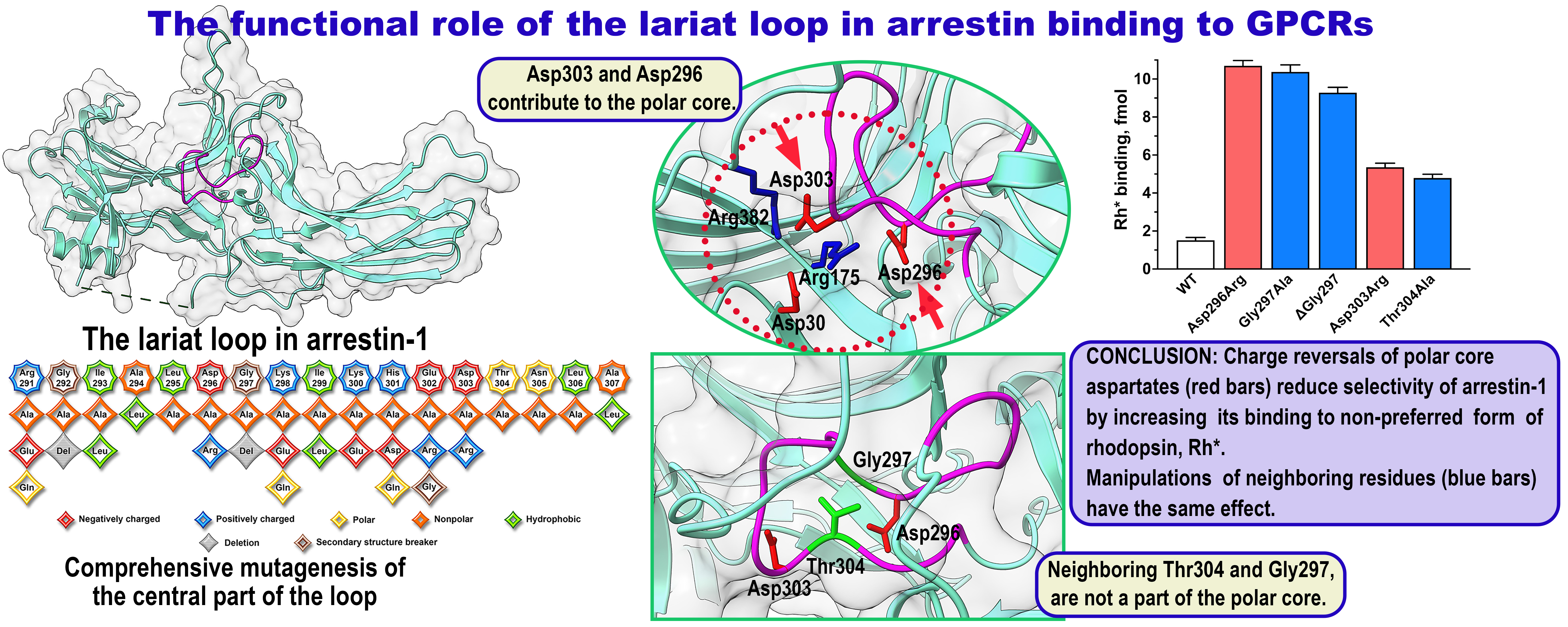
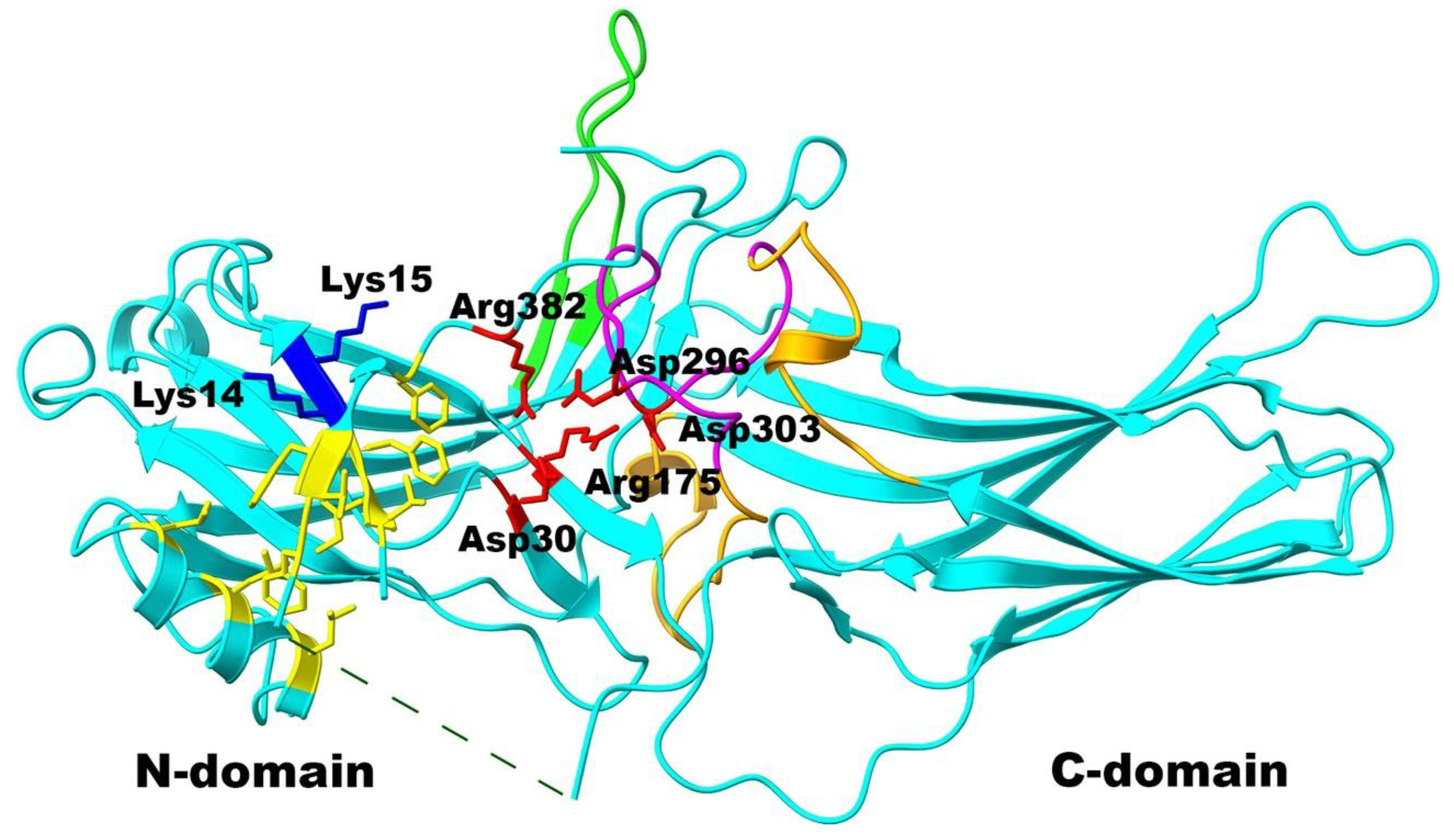
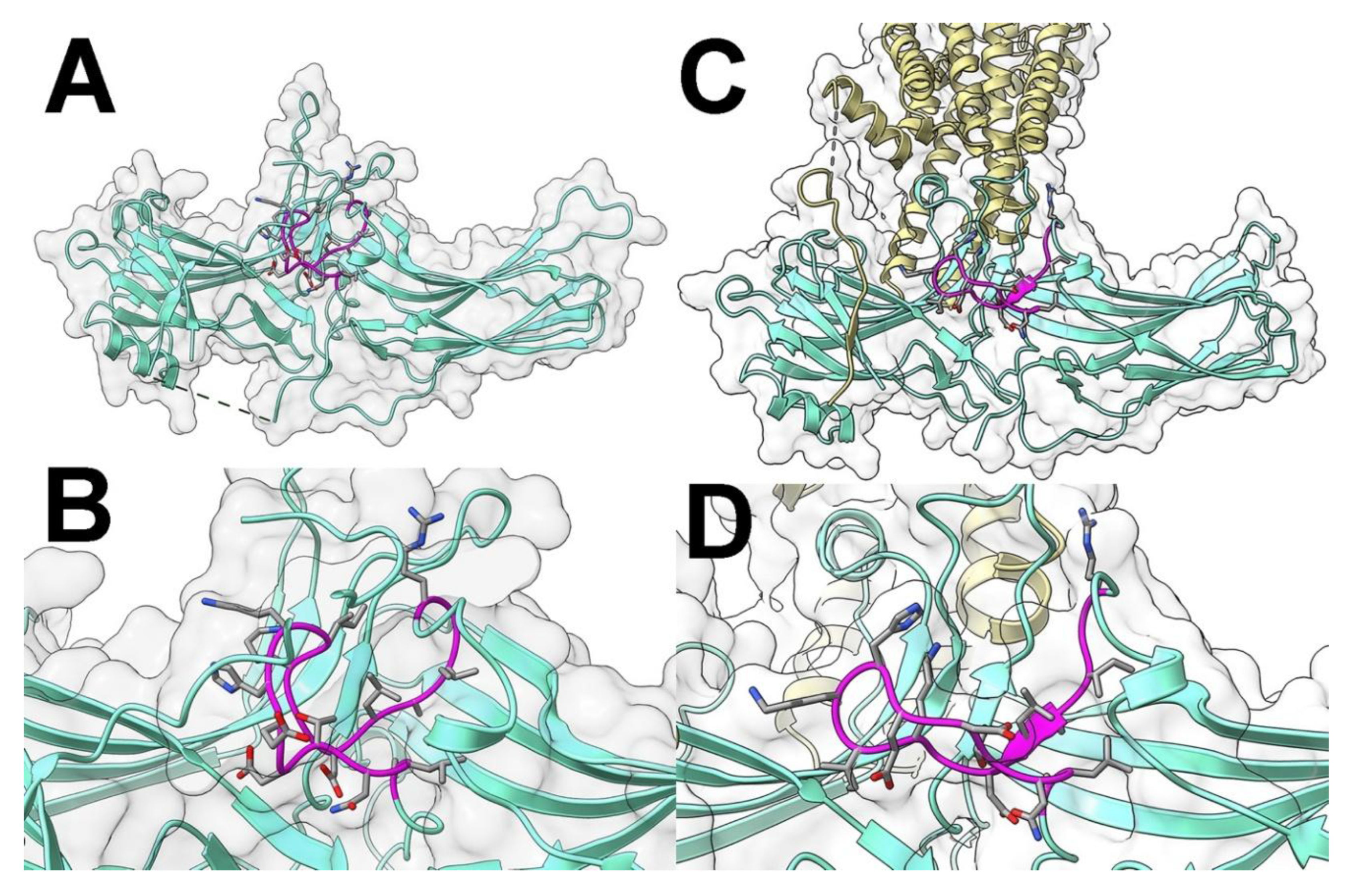
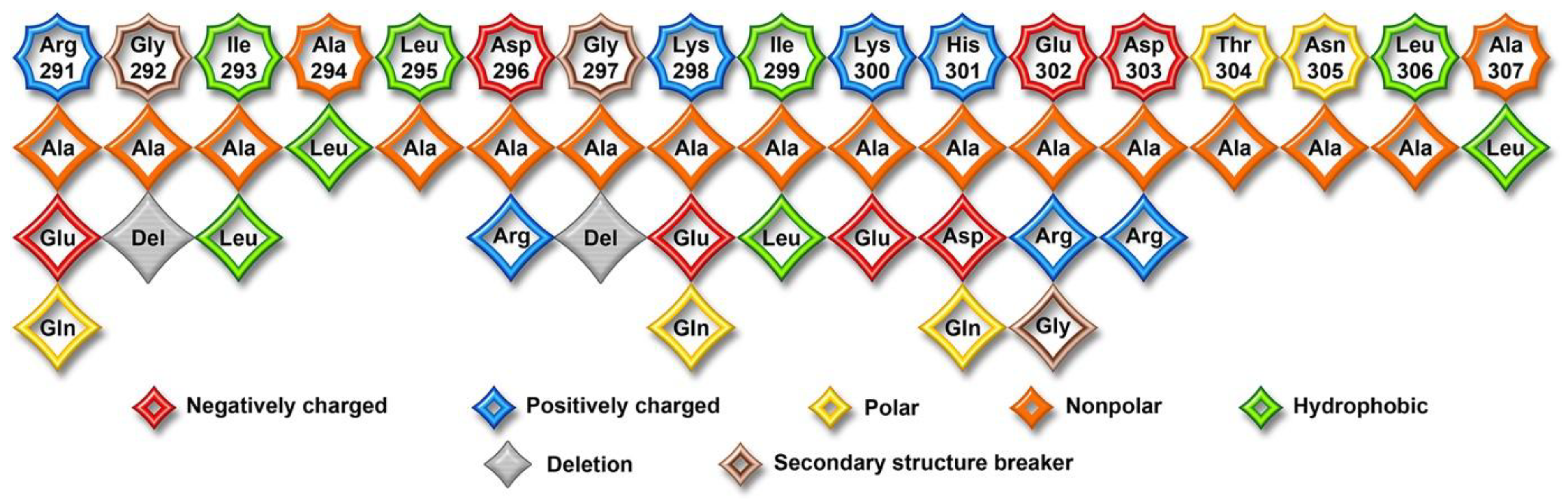
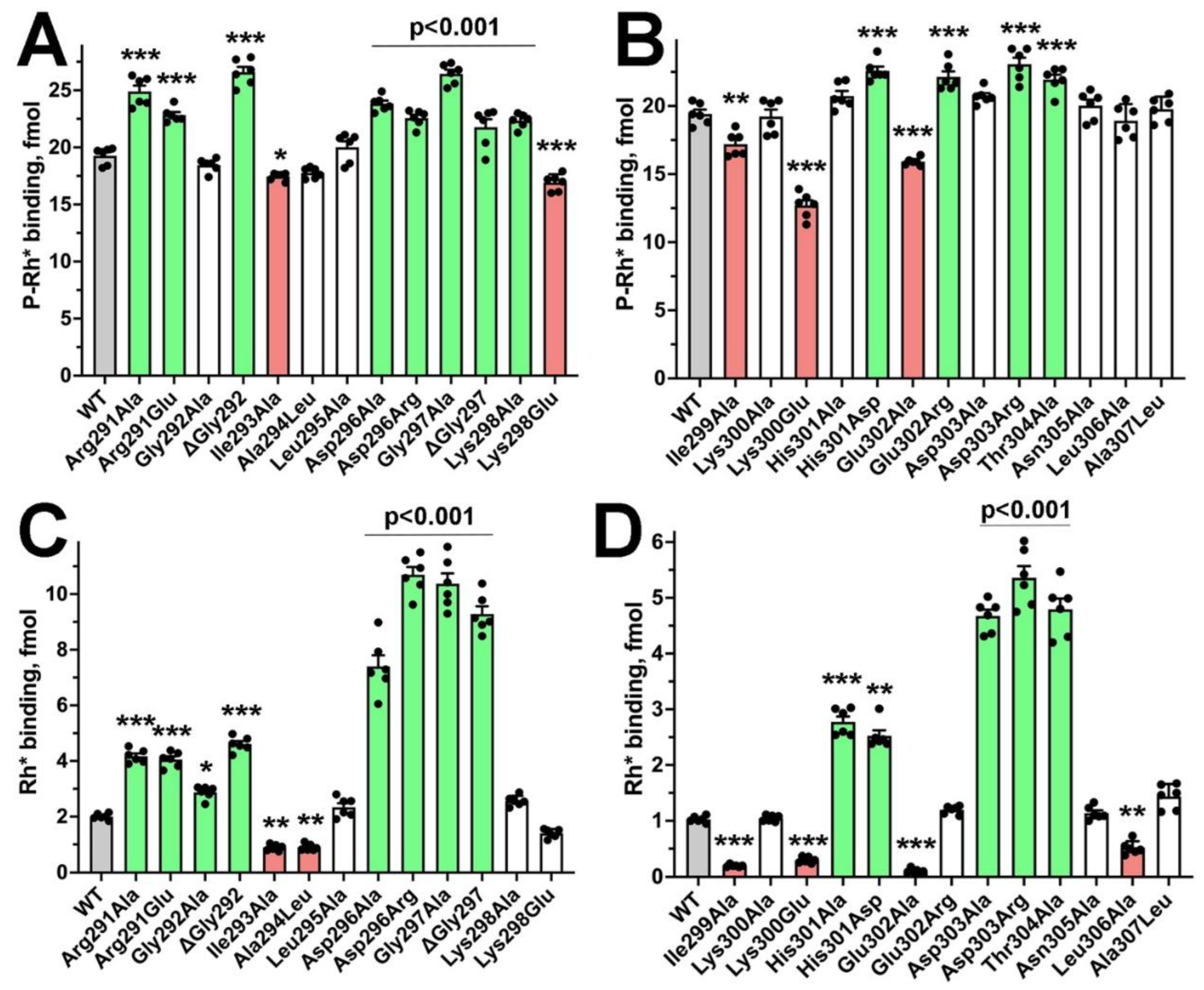
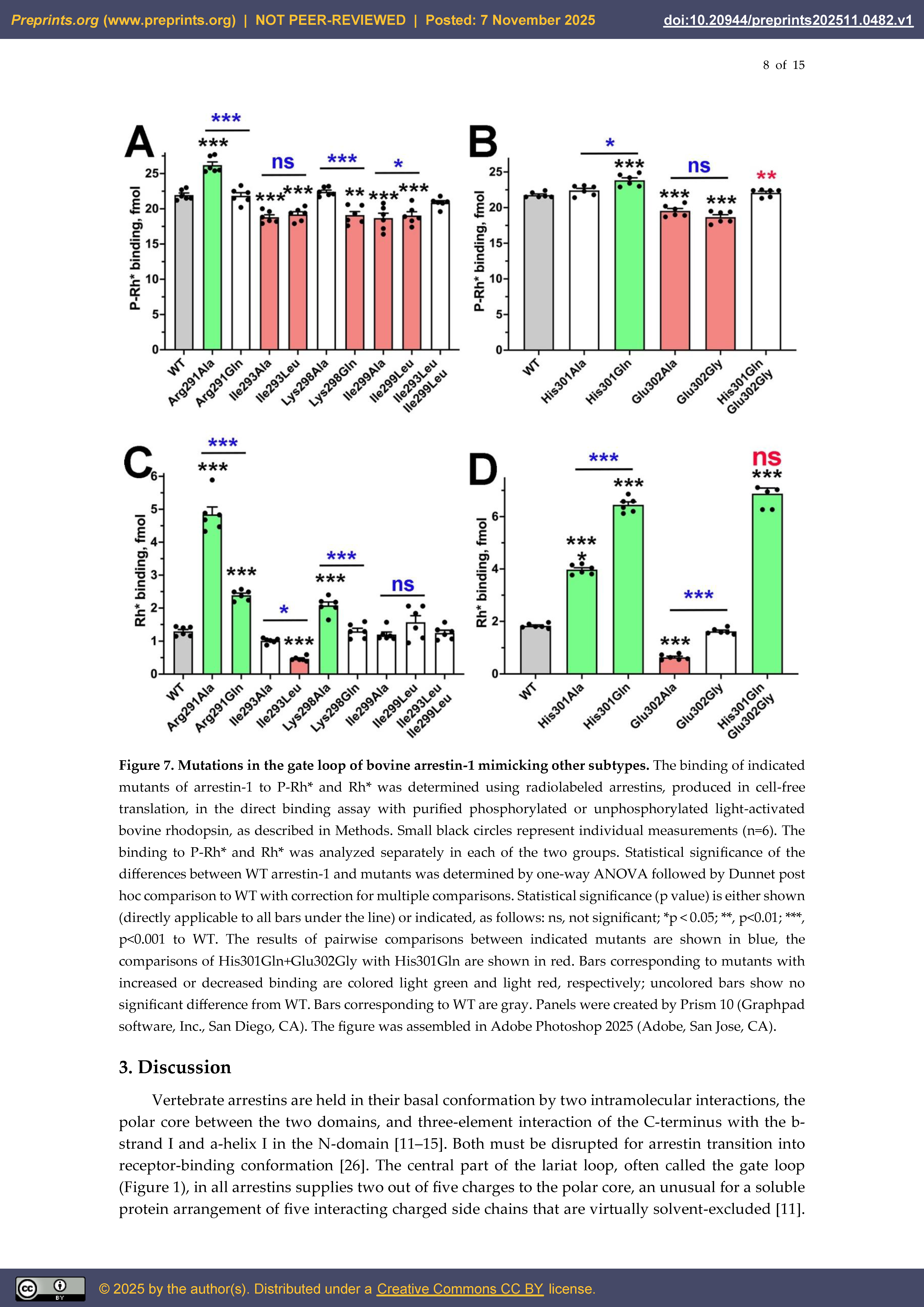
2. Results
3. Discussion
4. Materials and Methods
Footnote
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Indrischek, H.; Prohaska, S.J.; Gurevich, V.V.; Gurevich, E.V.; Stadler, P.F. Uncovering missing pieces: duplication and deletion history of arrestins in deuterostomes. BMC Evol Biol 2017, 17, 163. [Google Scholar] [CrossRef]
- Hofmann, K.P.; Scheerer, P.; Hildebrand, P.W.; Choe, H.W.; Park, J.H.; Heck, M.; Ernst, O.P. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci 2009, 34, 540–552. [Google Scholar] [CrossRef]
- Carman, C.V.; Benovic, J.L. G-protein-coupled receptors: turn-ons and turn-offs. Curr Opin Neurobiol 1998, 8, 335–344. [Google Scholar] [CrossRef]
- Kuhn, H. Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry 1978, 17, 4389–4395. [Google Scholar] [CrossRef]
- Shinohara, T.; Dietzschold, B.; Craft, C.M.; Wistow, G.; Early, J.J.; Donoso, L.A.; Horwitz, J.; Tao, R. Primary and secondary structure of bovine retinal S antigen (48 kDa protein). Proceedings of the National Academy of Sciences 1987, 84, 6975–6979. [Google Scholar] [CrossRef]
- Song, X.; Vishnivetskiy, S.A.; Seo, J.; Chen, J.; Gurevich, E.V.; Gurevich, V.V. Arrestin-1 expression in rods: balancing functional performance and photoreceptor health. Neuroscience 2011, 174, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Hanson, S.M.; Gurevich, E.V.; Vishnivetskiy, S.A.; Ahmed, M.R.; Song, X.; Gurevich, V.V. Each rhodopsin molecule binds its own arrestin. Proc Nat Acad Sci USA 2007, 104, 3125–3128. [Google Scholar] [CrossRef] [PubMed]
- Nikonov, S.S.; Brown, B.M.; Davis, J.A.; Zuniga, F.I.; Bragin, A.; Pugh, E.N., Jr.; Craft, C.M. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron 2008, 59, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V. Arrestins: A Small Family of Multi-Functional Proteins. Int J Mol Sci 2024, 25, 6284. [Google Scholar] [CrossRef]
- Hirsch, J.A.; Schubert, C.; Gurevich, V.V.; Sigler, P.B. The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell 1999, 97, 257–269. [Google Scholar] [CrossRef]
- Han, M.; Gurevich, V.V.; Vishnivetskiy, S.A.; Sigler, P.B.; Schubert, C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure 2001, 9, 869–880. [Google Scholar] [CrossRef]
- Milano, S.K.; Pace, H.C.; Kim, Y.M.; Brenner, C.; Benovic, J.L. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry 2002, 41, 3321–3328. [Google Scholar] [CrossRef]
- Sutton, R.B.; Vishnivetskiy, S.A.; Robert, J.; Hanson, S.M.; Raman, D.; Knox, B.E.; Kono, M.; Navarro, J.; Gurevich, V.V. Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J Mol Biol 2005, 354, 1069–1080. [Google Scholar] [CrossRef]
- Zhan, X.; Gimenez, L.E.; Gurevich, V.V.; Spiller, B.W. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J Mol Biol 2011, 406, 467–478. [Google Scholar] [CrossRef]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin determined by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef]
- Zhou, X.E.; He, Y.; de Waal, P.W.; Gao, X.; Kang, Y.; Van Eps, N.; Yin, Y.; Pal, K.; Goswami, D.; White, T.A.; Barty, A.; Latorraca, N.R.; Chapman, H.N.; Hubbell, W.L.; Dror, R.O.; Stevens, R.C.; Cherezov, V.; Gurevich, V.V.; Griffin, P.R.; Ernst, O.P.; Melcher, K.; Xu, H.E. Identification of Phosphorylation Codes for Arrestin Recruitment by G protein-Coupled Receptors. Cell 2017, 170, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Li, Z.; Jin, M.; Yin, Y.L.; de Waal, P.W.; Pal, K.; Yin, Y.; Gao, X.; He, Y.; Gao, J.; Wang, X.; Zhang, Y.; Zhou, H.; Melcher, K.; Jiang, Y.; Cong, Y.; Zhou, X.E.; Yu, X.; Xu, H.E. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res 2019, 29, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Staus, D.P.; Hu, H.; Robertson, M.J.; Kleinhenz, A.L.W.; Wingler, L.M.; Capel, W.D.; Latorraca, N.R.; Lefkowitz, R.J.; Skiniotis, G. Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Nature 2020, 579, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Warne, T.; Nehmé, R.; Pandey, S.; Dwivedi-Agnihotri, H.; Chaturvedi, M.; Edwards, P.C.; García-Nafría, J.; Leslie, A.G.W.; Shukla, A.K.; Tate, C.G. Molecular basis of β-arrestin coupling to formoterol-bound β(1)-adrenoceptor. Nature 2020, 583, 862–866. [Google Scholar] [CrossRef]
- Liao, Y.Y.; Zhang, H.; Shen, Q.; Cai, C.; Ding, Y.; Shen, D.D.; Guo, J.; Qin, J.; Dong, Y.; Zhang, Y.; Li, X.M. Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism. Cell 2023, 186, 5784–5797. [Google Scholar] [CrossRef]
- Huang, W.; Masureel, M.; Qianhui, Q.; Janetzko, J.; Inoue, A.; Kato, H.E.; Robertson, M.J.; Nguyen, K.C.; Glenn, J.S.; Skiniotis, G.; Kobilka, B.K. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 2020, 579, 303–308. [Google Scholar] [CrossRef]
- Bous, J.; Fouillen, A.; Orcel, H.; Trapani, S.; Cong, X.; Fontanel, S.; Saint-Paul, J.; Lai-Kee-Him, J.; Urbach, S.; Sibille, N.; Sounier, R.; Granier, S.; Mouillac, B.; Bron, P. Structure of the vasopressin hormone-V2 receptor-β-arrestin1 ternary complex. Sci Adv 2022, 8, eabo7761. [Google Scholar] [CrossRef]
- Cao, C.; Barros-Álvarez, X.; Zhang, S.; Kim, K.; Dämgen, M.A.; Panova, O.; Suomivuori, C.M.; Fay, J.F.; Zhong, X.; Krumm, B.E.; Gumpper, R.H.; Seven, A.B.; Robertson, M.J.; Krogan, N.J.; Hüttenhain, R.; Nichols, D.E.; Dror, R.O.; Skiniotis, G.; Roth, B.L. Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD. Neuron 2022, 110, 3154–3167. [Google Scholar] [CrossRef]
- Chen, Q.; Schafer, C.T.; Mukherjee, S.; Wang, K.; Gustavsson, M.; Fuller, J.R.; Tepper, K.; Lamme, T.D.; Aydin, Y.; Agrawal, P.; Terashi, G.; Yao, X.Q.; Kihara, D.; Kossiakoff, A.A.; Handel, T.M.; Tesmer, J.J.G. Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor. Nature 2025, 643, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Sente, A.; Peer, R.; Srivastava, A.; Baidya, M.; Lesk, A.M.; Balaji, S.; Shukla, A.K.; Babu, M.M.; Flock, T. Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat Struct Mol Biol 2018, 25, 538–545. [Google Scholar] [CrossRef]
- Sander, C.L.; Luu, J.; Kim, K.; Furkert, D.; Jang, K.; Reichenwallner, J.; Kang, M.; Lee, H.J.; Eger, B.T.; Choe, H.W.; Fiedler, D.; Ernst, O.P.; Kim, Y.J.; Palczewski, K.; Kiser, P.D. Structural evidence for visual arrestin priming via complexation of phosphoinositols. Structure 2022, 30, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Van Eps, N.; Eger, B.T.; Rauscher, S.; Yedidi, R.S.; Moroni, T.; West, G.M.; Robinson, K.A.; Griffin, P.R.; Mitchell, J.; Ernst, O.P. A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin. J Mol Biol 2018, 430, 4102–4118. [Google Scholar] [CrossRef]
- Granzin, J.; Wilden, U.; Choe, H.W.; Labahn, J.; Krafft, B.; Buldt, G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature 1998, 391, 918–921. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Paz, C.L.; Schubert, C.; Hirsch, J.A.; Sigler, P.B.; Gurevich, V.V. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem 1999, 274, 11451–11454. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskiy, S.A.; Huh, E.K.; Gurevich, E.V.; Gurevich, V.V. The finger loop as an activation sensor in arrestin. J Neurochem 2021, 157, 1138–1152. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Zheng, C.; May, M.B.; Karnam, P.C.; Gurevich, E.V.; Gurevich, V.V. Lysine in the lariat loop of arrestins does not serve as phosphate sensor. J Neurochem 2021, 156, 435–444. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Schubert, C.; Climaco, G.C.; Gurevich, Y.V.; Velez, M.-G.; Gurevich, V.V. An additional phosphate-binding element in arrestin molecule: implications for the mechanism of arrestin activation. J. Biol. Chem. 2000, 275, 41049–41057. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem 1998, 273, 15501–15506. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskiy, S.A.; Francis, D.J.; Van Eps, N.; Kim, M.; Hanson, S.M.; Klug, C.S.; Hubbell, W.L.; Gurevich, V.V. The role of arrestin alpha-helix I in receptor binding. J. Mol. Biol. 2010, 395, 42–54. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Kim, M.; Hanson, S.M.; Vishnivetskiy, S.A.; Song, X.; Cleghorn, W.M.; Hubbell, W.L.; Gurevich, V.V. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry 2011, 50, 2235–2242. [Google Scholar] [CrossRef]
- Hanson, S.M.; Van Eps, N.; Francis, D.J.; Altenbach, C.; Vishnivetskiy, S.A.; Arshavsky, V.Y.; Klug, C.S.; Hubbell, W.L.; Gurevich, V.V. Structure and function of the visual arrestin oligomer. EMBO J 2007, 26, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Vishnivetskiy, S.A.; Gross, O.P.; Emelianoff, K.; Mendez, A.; Chen, J.; Gurevich, E.V.; Burns, M.E.; Gurevich, V.V. Enhanced arrestin facilitates recovery and protects rods lacking rhodopsin phosphorylation. Current biology: CB 2009, 19, 700–705. [Google Scholar] [CrossRef]
- Smith, W.C. A splice variant of arrestin from human retina. Exp Eye Res 1996, 62, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Syed, M.; Bugra, K.; Whelan, J.P.; McGinnis, J.F.; Shinohara, T. Structural analysis of mouse S-antigen. Gene 1988, 73, 11–20. [Google Scholar] [CrossRef]
- Sterne-Marr, R.; Gurevich, V.V.; Goldsmith, P.; Bodine, R.C.; Sanders, C.; Donoso, L.A.; Benovic, J.L. Polypeptide variants of beta-arrestin and arrestin3. J Biol Chem 1993, 268, 15640–15648. [Google Scholar] [CrossRef] [PubMed]
- Parruti, G.; Peracchia, F.; Sallese, M.; Ambrosini, G.; Masini, M.; Rotilio, D.; De Blasi, A. Molecular analysis of human beta-arrestin-1: cloning, tissue distribution, and regulation of expression. Identification of two isoforms generated by alternative splicing. J Biol Chem 1993, 268, 9753–9761. [Google Scholar] [CrossRef]
- Kingsmore, S.F.; Peppel, K.; Suh, D.; Caron, M.G.; Lefkowitz, R.J.; Seldin, M.F. Genetic mapping of the beta-arrestin 1 and 2 genes on mouse chromosomes 7 and 11 respectively. Mamm Genome 1995, 6, 306–307. [Google Scholar] [CrossRef]
- Rapoport, B.; Kaufman, K.D.; Chazenbalk, G.D. Cloning of a member of the arrestin family from a human thyroid cDNA library. Mol Cell Endocrinol 1992, 84, R39–R43. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Ohguro, H.; Sohma, H.; Kuroki, Y.; Wada, H.; Okisaka, S.; Murakami, A. Purification and characterization of bovine cone arrestin (cArr). FEBS Lett. 2000, 470, 336–340. [Google Scholar] [CrossRef]
- Murakami, A.; Yajima, T.; Sakuma, H.; McLaren, M.J.; Inana, G. X-arrestin: a new retinal arrestin mapping to the X chromosome. FEBS Lett. 1993, 334, 203–209. [Google Scholar] [CrossRef]
- Hyde, D.R.; Mecklenburg, K.L.; Pollock, J.A.; Vihtelic, T.S.; Benzer, S. Twenty Drosophila visual system cDNA clones: one is a homolog of human arrestin. Proc Natl Acad Sci U S A 1990, 87, 1008–1012. [Google Scholar] [CrossRef]
- Yamada, T.; Takeuchi, Y.; Komori, N.; Kobayashi, H.; Sakai, Y.; Hotta, Y.; Matsumoto, H. A 49-kilodalton phosphoprotein in the Drosophila photoreceptor is an arrestin homolog. Science 1990, 246, 483–486. [Google Scholar] [CrossRef]
- Mayeenuddin, L.H.; Mitchell, J. Squid visual arrestin: cDNA cloning and calcium-dependent phosphorylation by rhodopsin kinase (SQRK). J Neurochem 2003, 85, 592–600. [Google Scholar] [CrossRef]
- Johnson, E.C.; Tift, F.W.; McCauley, A.; Liu, L.; Roman, G. Functional characterization of kurtz, a Drosophila non-visual arrestin, reveals conservation of GPCR desensitization mechanisms. Insect Biochem Mol Biol 2008, 38, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Palmitessa, A.; Hess, H.A.; Bany, I.A.; Kim, Y.M.; Koelle, M.R.; Benovic, J.L. Caenorhabditus elegans arrestin regulates neural G protein signaling and olfactory adaptation and recovery. J Biol Chem 2005, 280, 24649–24662. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Orii, H.; Yoshida, N.; Jojima, E.; Horie, T.; Yoshida, R.; Haga, T.; Tsuda, M. Ascidian arrestin (Ci-arr), the origin of the visual and nonvisual arrestins of vertebrate. Eur J Biochem 2002, 269, 5112–5118. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, C.; Lin, S.; Yan, X.; Cai, H.; Yi, C.; Ma, L.; Chu, X.; Liu, Y.; Zhu, Y.; Han, S.; Zhao, Q.; Wu, B. Tail engagement of arrestin at the glucagon receptor. Nature 2023, 620, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, X.; Yuan, Q.; Wang, Y.; Shi, P.; Zhang, H.; Wang, T.; Sun, W.; Ling, S.; Liu, Y.; Lai, J.; Xie, W.; Yin, W.; Liu, L.; Xu, H.E.; Tian, C. Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator. Cell Res 2025, 35, 284–295. [Google Scholar] [CrossRef]
- Chen, Q.; Perry, N.A.; Vishnivetskiy, S.A.; Berndt, S.; Gilbert, N.C.; Zhuo, Y.; Singh, P.K.; Tholen, J.; Ohi, M.D.; Gurevich, E.V.; Brautigam, C.A.; Klug, K.S.; Gurevich, V.V.; Iverson, T.M. Structural basis of arrestin-3 activation and signaling. Nat Commun 2017, 8, 1427. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Pals-Rylaarsdam, R.; Benovic, J.L.; Hosey, M.M.; Onorato, J.J. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem 1997, 272, 28849–28852. [Google Scholar] [CrossRef]
- Celver, J.; Vishnivetskiy, S.A.; Chavkin, C.; Gurevich, V.V. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J. Biol. Chem. 2002, 277, 9043–9048. [Google Scholar] [CrossRef]
- Kovoor, A.; Celver, J.; Abdryashitov, R.I.; Chavkin, C.; Gurevich, V.V. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. J. Biol. Chem. 1999, 274, 6831–6834. [Google Scholar] [CrossRef]
- Ostermaier, M.K.; Peterhans, C.; Jaussi, R.; Deupi, X.; Standfuss, J. Functional map of arrestin-1 at single amino acid resolution. Proc Natl Acad Sci U S A 2014, 111, 1825–1830. [Google Scholar] [CrossRef]
- Peterhans, C.; Lally, C.C.; Ostermaier, M.K.; Sommer, M.E.; Standfuss, J. Functional map of arrestin binding to phosphorylated opsin, with and without agonist. Sci Rep 2016, 6, 28686. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Huh, E.K.; Karnam, P.C.; Oviedo, S.; Gurevich, E.V.; Gurevich, V.V. The Role of Arrestin-1 Middle Loop in Rhodopsin Binding. Int J Mol Sci 2022, 23, 13887. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskiy, S.A.; Weinstein, L.D.; Zheng, C.; Gurevich, E.V.; Gurevich, V.V. Functional Role of Arrestin-1 Residues Interacting with Unphosphorylated Rhodopsin Elements. Int J Mol Sci 2023, 24, 8903. [Google Scholar] [CrossRef]
- Gurevich, E.V.; Gurevich, V.V. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol 2006, 7, 236. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskiy, S.A.; Chen, Q.; Palazzo, M.C.; Brooks, E.K.; Altenbach, C.; Iverson, T.M.; Hubbell, W.L.; Gurevich, V.V. Engineering visual arrestin-1 with special functional characteristics. J Biol Chem 2013, 288, 11741–11750. [Google Scholar] [CrossRef]
- Alloway, P.G.; Dolph, P.J. A role for the light-dependent phosphorylation of visual arrestin. Proc Natl Acad Sci U S A 1999, 96, 6072–6077. [Google Scholar] [CrossRef]
- Bentrop, J.; Paulsen, R. Light-modulated ADP-ribosylation, protein phosphorylation and protein binding in isolated fly photoreceptor membranes. Eur J Biochem 1986, 161, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Satoh, A.K.; Xia, H.; Yan, L.; Liu, C.H.; Hardie, R.C.; Ready, D.F. Arrestin translocation is stoichiometric to rhodopsin isomerization and accelerated by phototransduction in Drosophila photoreceptors. Neuron 2010, 67, 997–1008. [Google Scholar] [CrossRef]
- Gurevich, V.V. Use of bacteriophage RNA polymerase in RNA synthesis. Methods Enzymol 1996, 275, 382–397. [Google Scholar]
- Vishnivetskiy, S.A.; Lee, R.J.; Zhou, X.E.; Franz, A.; Xu, Q.; Xu, H.E.; Gurevich, V.V. Functional role of the three conserved cysteines in the N domain of visual arrestin-1. J Biol Chem 2017, 292, 12496–12502. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Hosey, M.M.; Benovic, J.L.; Gurevich, V.V. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem 2004, 279, 1262–1268. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Benovic, J.L. Visual arrestin interaction with rhodopsin: Sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. J. Biol. Chem. 1993, 268, 11628–11638. [Google Scholar] [CrossRef]
- McDowell, J.H. Preparing Rod Outer Segment Membranes, Regenerating Rhodopsin, and Determining Rhodopsin Concentration. Methods in Neurosciences 1993, 15, 123–130. [Google Scholar]
- McDowell, J.H.; Nawrocki, J.P.; Hargrave, P.A. Phosphorylation sites in bovine rhodopsin. Biochemistry 1993, 32, 4968–4974. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Raman, D.; Wei, J.; Kennedy, M.J.; Hurley, J.B.; Gurevich, V.V. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem 2007, 282, 32075–32083. [Google Scholar] [CrossRef] [PubMed]
- Mendez, A.; Burns, M.E.; Roca, A.; Lem, J.; Wu, L.W.; Simon, M.I.; Baylor, D.A.; Chen, J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron 2000, 28, 153–164. [Google Scholar] [CrossRef]
- Wilden, U.; Kühn, H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry 1982, 21, 3014–3022. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V.; Benovic, J.L. Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. J Biol Chem 1992, 267, 21919–21923. [Google Scholar] [CrossRef]
- Vishnivetskiy, S.A.; Sullivan, L.S.; Bowne, S.J.; Daiger, S.P.; Gurevich, E.V.; Gurevich, V.V. Molecular Defects of the Disease-Causing Human Arrestin-1 C147F Mutant. Invest Ophthalmol Vis Sci 2018, 59, 13–20. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




