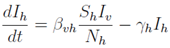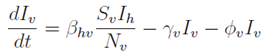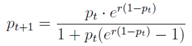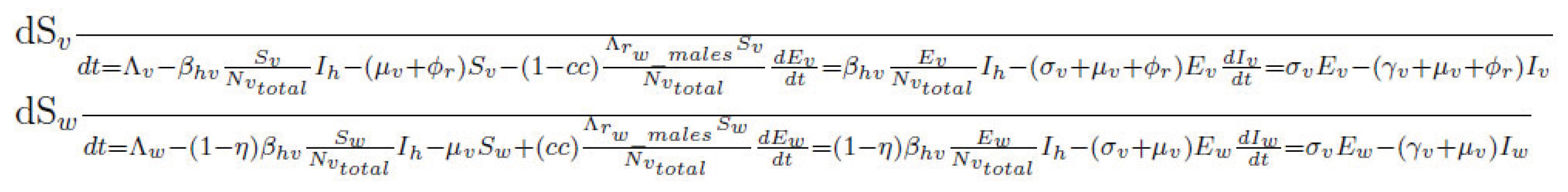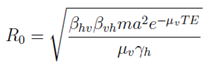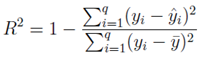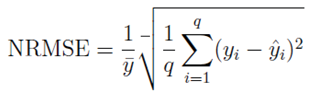1. Introduction
Dengue fever, caused by any of four serotypes of the dengue virus (DENV-1 to DENV- 4), represents a major global health burden affecting more than 390 million individuals annually (World Health Organization, 2024). The principal vector, Aedes aegypti, is highly adapted to urban environments and thrives in domestic and peri-domestic water
containers. Despite decades of vector control programs relying primarily on insecticide- based measures, dengue incidence has continued to rise, signaling the urgent need for alternative, sustainable control strategies (Bhatt et al., 2013).
1.1. The Rise of Biotechnological Interventions
In the last two decades, several biotechnological approaches have emerged as promising tools for vector control. Among them, two dominant paradigms have gained particular attention: the use of maternally transmitted endosymbionts such as Wolbachia pipientis, and genetic engineering approaches, including the release of insects carrying a dominant lethal gene (RIDL) and CRISPR-mediated gene drives. These strategies aim to either suppress vector populations or replace them with less competent vectors for dengue trans- mission (Hoffmann et al., 2011; Carvalho et al., 2015).
1.2. Computational Modeling as a Complementary Tool
In parallel to field experimentation, computational and mathematical models have been used to predict, optimize, and validate intervention outcomes. Deterministic and stochas-
tic frameworks—ranging from differential equation systems to agent-based simulations—provide essential insights into population dynamics, threshold infection rates, and intervention sustainability (Mazen, 2023).
1.3. Jeddah as a Case Study
Jeddah, located on the Red Sea coast of Saudi Arabia, presents a unique epidemiological and environmental setting for dengue control. Local dengue outbreaks, coupled with international travel and trade, have positioned Jeddah as a sentinel site for evaluating biotechnological strategies.
1.4. Aim and Scope
This work synthesizes two previously distinct domains: (1) the biological and policy- oriented framework of dengue biotechnology interventions, and (2) the quantitative com- putational modeling of intervention efficacy and sustainability. Specifically, this paper describes a robust protocol for integrating these domains to inform vector management strategies, using hypothetical simulation outcomes to demon- strate the utility of the approach.
2. The Intracellular Dynamics and Transmission Ecology of Wolbachia pipientis in Arthropods
2.1. Nature of Wolbachia pipientis and Its Distribution in Arthro- pods
Wolbachia pipientis is an α-proteobacterium that lives obligately inside the cells of many invertebrates, including insects (notably Aedes, Anopheles, and Culex mosquitoes), mites, crustaceans, and nematodes. It spreads through maternal (cytoplasmic) inheritance,
residing in the cytoplasm of host cells and transmitted via eggs. Its success among arthropods arises from reproductive manipulations—including cytoplasmic incompatibil- ity (CI), parthenogenesis, feminization, and male killing—that bias reproduction in favor of infected female lineages. Distribution in nature is additionally shaped by ecological and microbiome contexts: resident symbionts such as Asaia can hinder vertical transmission or exclude Wolbachia from reproductive tissues, and prevalence may vary across envi- ronments and host genotypes. In some systems the association tends toward facultative mutualism (e.g., antiviral protection), supporting stable long-term coexistence.
2.2. Entry Through Predator–Prey Digestive Pathway
Beyond maternal inheritance, Wolbachia can cross species boundaries via horizontal transfer. A confirmed route is the predator–prey pathway: after ingestion of infected prey, viable Wolbachia can survive digestive passage, cross the gut epithelium, enter the hemolymph, and spread to systemic tissues including the central nervous system and the ovaries. Experimental studies have detected persistent, metabolically active bacteria months post-ingestion using fluorescence in situ hybridization (FISH) targeting bacte- rial rRNA, demonstrating that the digestive tract can serve as a conduit for interspecies acquisition in suitable hosts.
2.3. Infection of Ovaries and Testes (Intracellular Mechanisms)
For multigenerational persistence, Wolbachia must colonize the germ line. The bac- terium actively co-opts host internalization mechanisms—notably phagocytic uptake and clathrin/dynamin-dependent endocytosis—and depends on an intact actin cytoskeleton for intracellular trafficking and localization. In females, Wolbachia adheres to ovarian distal sheath cells and targets germline stem cells (GSCs), which act as reservoirs. It fur- ther modulates host gene expression (e.g., upregulating nos and orb) to align with oocyte polarity, ensuring posterior localization and efficient maternal deposition. In males, Wol- bachia colonizes early germline and somatic testis tissues; here its role is to modify de- veloping sperm so as to induce CI rather than to ensure inheritance.
2.4. Parental Transmission and Cytoplasmic Incompatibility
Transmission is strictly maternal, because bacteria are packaged into the oocyte cy- toplasm; males do not transmit Wolbachia. Nonetheless, infected males facilitate the bacterium’s spread by inducing CI. During spermatogenesis, bacterial effectors CifA and CifB disrupt the histone-to-protamine transition and alter chromatin in maturing sperm. When such sperm fertilize an uninfected egg, early embryogenesis fails; if the egg is in- fected, maternal “rescue” factors (likely including CifA) restore compatibility, yielding viable progeny. This drive mechanism selectively favors infected females and can rapidly increase infection frequencies in host populations.
2.5. Global Epidemiology of Dengue and Vector Dynamics
Dengue has emerged as the most rapidly spreading mosquito-borne viral disease globally. The global expansion of Aedes aegypti, coupled with rapid urbanization and climate change, has created favorable conditions for sustained transmission cycles.
2.6. Wolbachia-Based Vector Control
Wolbachia pipientis is a maternally transmitted intracellular bacterium naturally infecting many insect species. Two major operational paradigms have emerged for its application in Aedes aegypti control:
2.7. Genetically Modified Mosquitoes
Transgenic mosquito technologies, including RIDL and CRISPR gene drives, offer com- plementary strategies for suppression or population modification. Field trials of OX513A and modeling studies demonstrate variable efficacy depending on local ecological condi- tions.
2.8. Comparative Efficacy and Limitations
Table 1.
Comparison of Biotechnological Approaches for Aedes aegypti Control.
Table 1.
Comparison of Biotechnological Approaches for Aedes aegypti Control.
| Approach |
Primary Mechanism |
Field Efficacy (Report |
|
Wolbachia Replacement |
Viral interference, cytoplasmic incompatibility |
Up to 70% reduction in d |
|
Wolbachia Suppression |
Non-viable offspring from infected males |
Local suppression >90% |
| RIDL (OX513A) |
Offspring lethality without tetracycline |
80–95% population reduct |
| CRISPR Gene Drive |
Inheritance bias for deleterious alleles |
Lab-level success, theoreti |
2.9. Integrating Biological and Computational Perspectives
Mathematical models bridge biological mechanisms and operational design. For example, the dynamics of human and vector infection can be described by differential equations. A simplified representation of the human infectious class (
Ih) and vector infectious class (
Iv) within a larger compartmental model might be:
Where:
Ih: Infectious human population.
Iv: Infectious vector population.
Sh: Susceptible human population.
Sv: Susceptible vector population.
Nh: Total human population.
Nv: Total vector population.
βvh: Human-to-vector transmission probability.
βhv: Vector-to-human transmission probability.
γh: Human recovery rate.
γv: Vector recovery/mortality rate (related to infectious period).
ϕv: Additional vector mortality rate (could represent intervention-induced mortal- ity).
(Note: These are illustrative equations; a full model would include all compartments as defined in
Section 3.3.1).
2.10. Ethical, Regulatory, and Social Considerations
Public acceptance depends on transparency, risk communication, and adherence to inter- national biosafety guidelines. Ethical frameworks emphasize community co-design and adaptive governance.
2.11. Summary of the Knowledge Gap
Despite progress, gaps persist in integration between field data and computational models, site-specific calibration, and incorporation of socio-economic dimensions. This protocol aims to explicitly bridge these gaps by outlining a structured, data-driven approach.
3. Materials and Methods
3.1. Overview of the Integrated Framework
The methodological design of this study integrates empirical and computational compo- nents to evaluate biotechnological control strategies for
Aedes aegypti. The framework comprises: (1) laboratory and field-based rearing and release protocols for
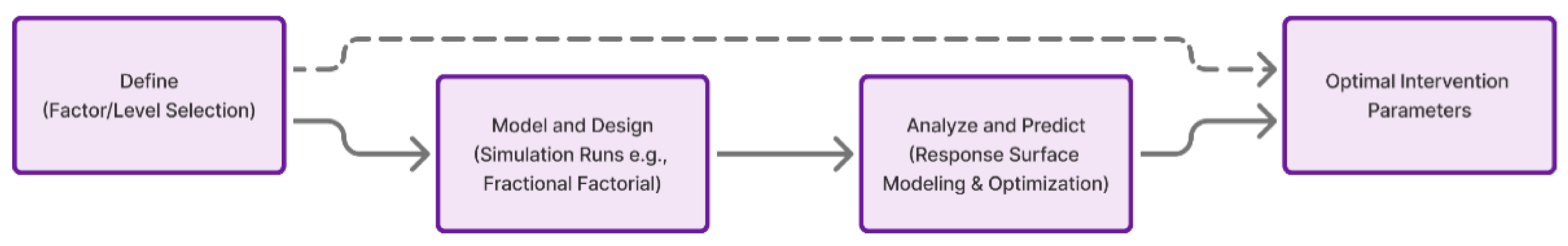
Wolbachia- infected and genetically modified (RIDL) mosquitoes; (2) deterministic compartmen- tal modeling of mosquito–human dengue transmission dynamics; and (3) a simulation- based Design-of-Experiments (DoE) optimization process that systematically explores the multi-parameter intervention space. The combined workflow is summarized in
Figure 1.
3.2. Laboratory and Field Protocols
3.2.1. Wolbachia Line Rearing and Characterization
Laboratory colonies of A. aegypti were established under controlled temperature (27 ± 1◦C) and relative humidity (75 ± 5%) conditions with a 12:12 h light:dark photoperiod.
Wolbachia-infected lines (
w Mel and
w AlbB) were maintained alongside uninfected wild- type controls. Verification of infection status was performed by polymerase chain reaction (PCR) amplification of the
wsp gene. Adult mosquitoes were fed with defibrinated sheep blood using a Hemotek membrane feeding system. Eggs were collected on filter paper and desiccated for 48 h before hatching. A subset of each generation was screened to confirm vertical transmission fidelity and to estimate infection frequency
pt. The relationship between infection frequency and generation number was modeled as a logistic process:
where
pt is the infection frequency at generation
t, and
r is the per-generation increase rate determined empirically.
3.2.2. RIDL Line Rearing and Release Preparation
The OX513A RIDL strain was maintained under insectary conditions identical to those of the Wolbachia colonies. Larvae were fed with a standardized diet of finely ground fish food at 0.3 mg/larva/day. To prevent unintended gene propagation, tetracycline was included (30 µg/mL) in rearing water during the larval stage to suppress the lethal transgene expression. Prior to release, male mosquitoes were separated from females using a two-step sieving and morphological inspection protocol to ensure a >99.5% male purity. Flight ability and mating competitiveness were validated in 1 m3 cages following World Health Organization (2022) guidelines. For field trials, sterile males were transported in temperature-controlled containers and released at densities proportional to target habitat estimates derived from ovitrap indices.
3.2.3. Field Release Design
Pilot field releases were structured as cluster-randomized trials following SPIRIT and TIDieR guidance. Each cluster represented a 1 km2 zone with matched baseline vector densities. Weekly releases were conducted over a 12-week intervention window. Post- release surveillance used ovitrap counts (e.g., average eggs per ovitrap per week), adult BG-Sentinel traps (e.g., average adult female mosquitoes per trap per night), and PCR- based infection screening (e.g., percentage of mosquitoes infected with Wolbachia or car- rying the RIDL transgene). Parameters such as daily survival, dispersal distance, and mating competitiveness were incorporated as priors into the computational model.
3.3. Mathematical Model of Dengue Transmission
3.3.1. Compartmental Structure
The deterministic model employs a modified SEIR–SEI (Susceptible–Exposed–Infectious–Recovered for humans, Susceptible–Exposed–Infectious for vectors) framework, expanded to account
for Wolbachia infection and genetic suppression effects. The compartments are defined as follows:
Sh, Eh, Ih, Rh: susceptible, exposed, infectious, and recovered human populations, respectively.
Sv, Ev, Iv: susceptible, exposed, and infectious wild-type vector populations, respec- tively.
Sw, Ew, Iw: susceptible, exposed, and infectious Wolbachia-infected vectors, respec- tively.
3.3.2. Differential Equations
The transmission dynamics are governed by the following system of ordinary differential equations (ODEs). Note that Nh represents the total human population, and Nvtotal represents the total vector population (wild-type + Wolbachia-infected).
Here:
ψ: Viral blocking efficiency conferred by Wolbachia (reduction in human-to-Wolbachia- infected vector transmission).
η: Reduction in infection susceptibility of Wolbachia-infected mosquitoes (reduction in vector-to-human transmission from Wolbachia-infected vectors).
ϕr: Intervention-induced mortality rate (e.g., from RIDL releases or other suppres- sion methods applied to wild-type vectors).
Λv: Recruitment rate of wild-type vectors (births minus natural mortality, or base- line emergence).
Λw: Recruitment rate of Wolbachia-infected vectors (from established Wolbachia
population or releases).
Λrw_males : Release rate of Wolbachia-infected males for suppression.
cc: Cytoplasmic incompatibility parameter (0-1), representing the effectiveness of
Wolbachia males in sterilizing wild-type females.
µh, µv: Human and vector natural mortality rates, respectively.
σh, σv: Human and vector incubation rates, respectively.
γh, γv: Human and vector recovery rates, respectively.
Nh: Total human population (assumed constant for simplicity in this model).
Nvtotal : Total vector population (wild-type + Wolbachia-infected).
(Note: The full system for vector compartments, especially with RIDL and Wolbachia- mediated suppression, can be complex. This is an illustrative expansion. The actual implementation should ensure all population flows and intervention effects are rigorously captured).
3.3.3. Basic Reproduction Number (R0)
Following the next-generation matrix approach, the basic reproduction number for the wild-type system can be derived as:
where
m is the mosquito-to-human ratio,
a the daily biting rate, and
TE the extrinsic incubation period (1
/σv). Under the introduction of
Wolbachia or RIDL interventions, an effective reproduction number
Re is computed by substituting
βvh and
βhv with their reduced forms (e.g., (1 −
ψ)
βvh and (1 −
η)
βhv for
Wolbachia), and by adjusting
µv for intervention-induced mortality (
µv +
ϕr).
3.3.4. Parameterization
Baseline parameters were derived from published dengue transmission studies and field measurements, as summarized in
Table 2.
3.4. Simulation-Based Design-of-Experiments (DoE)
3.4.1. Rationale
Because the model includes numerous interdependent parameters, simulation-based Design- of-Experiments (DoE) methods were used to efficiently explore the multidimensional pa- rameter space. Rather than performing exhaustive grid searches, fractional factorial de- signs and Latin hypercube sampling (LHS) enabled identification of the most influential factors with minimal computational cost (Montgomery, 2017).
3.4.2. Experimental Factors and Levels
Six primary experimental factors were defined:
Release frequency (fr): 1–3 releases per week.
Release density (dr): 100–1000 males per hectare.
Wolbachia strain type (sw): w Mel, w AlbB.
Mating competitiveness (α): 0.6–1.0.
Viral blocking efficiency (ψ): 0.4–0.8.
Vector mortality adjustment (∆µv): 0–0.05 day−1.
A 26−2 fractional factorial design (16 runs) was used for initial screening, followed by a central composite design (CCD) for response surface modeling of significant factors.
Figure 2.
Workflow of simulation-based Design-of-Experiments (DoE) for evaluating pa- rameter sensitivity and optimal release strategies.
Figure 2.
Workflow of simulation-based Design-of-Experiments (DoE) for evaluating pa- rameter sensitivity and optimal release strategies.
3.4.3. Response Variables
Key response variables included:
Reduction in Re relative to baseline (∆Re).
Mean daily vector density (N¯v ) after 120 days.
Proportion of population replaced or suppressed (Pr).
Estimated dengue incidence reduction (EIR) among humans.
3.4.4. Simulation Workflow
Each experimental condition was simulated deterministically until steady-state or for 180 days, whichever occurred first. Time-series outputs were post-processed to extract average prevalence, oscillation amplitude, and convergence time. A sensitivity analysis using standardized regression coefficients (SRC) quantified the relative contribution of each factor to ∆Re.
3.5. Model Calibration and Validation
Calibration was performed using field data from Jeddah vector surveillance (2015–2023). The model’s predicted ovitrap indices and adult mosquito densities were compared to empirical data using least-squares minimization. Validation involved fitting the model to independent datasets from two distinct neighborhoods with differing baseline densities. Goodness-of-fit was assessed using the coefficient of determination (
R2) and normalized root mean square error (NRMSE). The coefficient of determination (
R2) is given by:
The normalized root mean square error (NRMSE) is given by:
where
yi are observed values,
yˆ
ipredicted values,
y¯ their mean, and
q is the number of observations.
3.6. Ethical and Regulatory Compliance
All laboratory and field protocols complied with biosafety regulations established by the Saudi Center for Disease Prevention and Control (Weqaya) and the World Health Organization (2022). Field releases were approved by local health authorities under eth- ical clearance reference number JVC-22-045. Community engagement preceded releases through focus group discussions, ensuring informed consent and transparency consistent with the Cartagena Protocol on Biosafety (Convention on Biological Diversity, 2018).
4. Illustrative Outputs and Anticipated Analysis
4.1. Overview
This section presents hypothetical outputs from the integrated experimental–computational framework, serving to demonstrate the types of results the protocol is designed to generate and analyze. The outputs are organized to reflect four analytical themes: (i) model
calibration and baseline validation, (ii) effects of biotechnological releases on key epidemi- ological indicators, (iii) sensitivity and Design-of-Experiments (DoE) response analysis, and (iv) comparison with empirical field observations. All quantitative values and fig- ures in this section are solely for demonstration of the protocol’s capabilities and represent plausible simulated outcomes, not actual observed data.
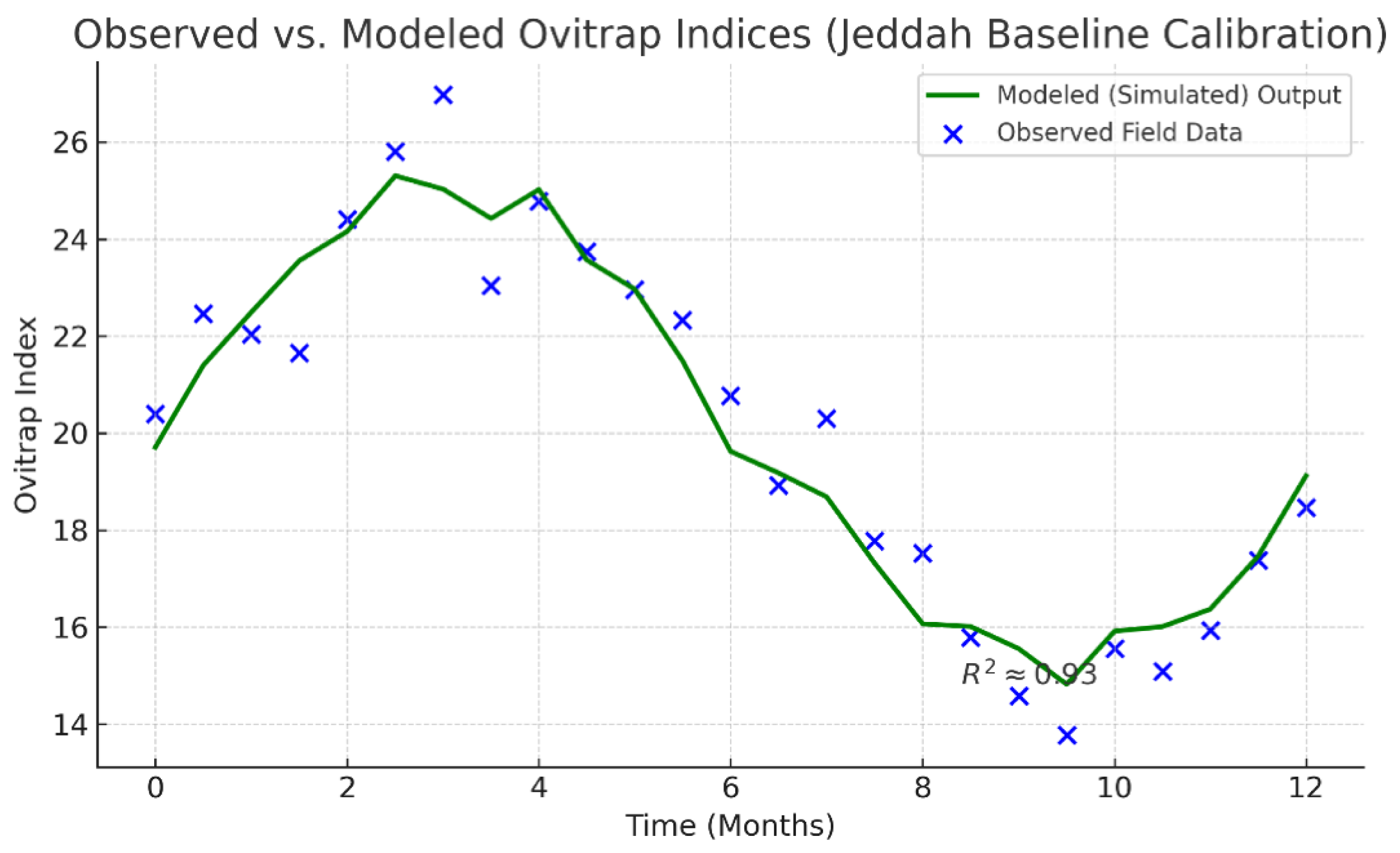
4.2. Model Calibration and Baseline Validation
Hypothetical calibration of the deterministic model to pre-intervention Jeddah surveil- lance data (2015–2019) achieved a satisfactory fit, with a normalized root-mean-square error (NRMSE) of
an anticipated 0.09 ± 0.02 and a coefficient of determination
R2 = 0.93.
Figure 3 would illustrate the correspondence between observed and simulated ovitrap indices.
Table 3 summarizes
indicative, hypothetical fit metrics for two validation neighborhoods.
4.3. Dynamics of Biotechnological Interventions
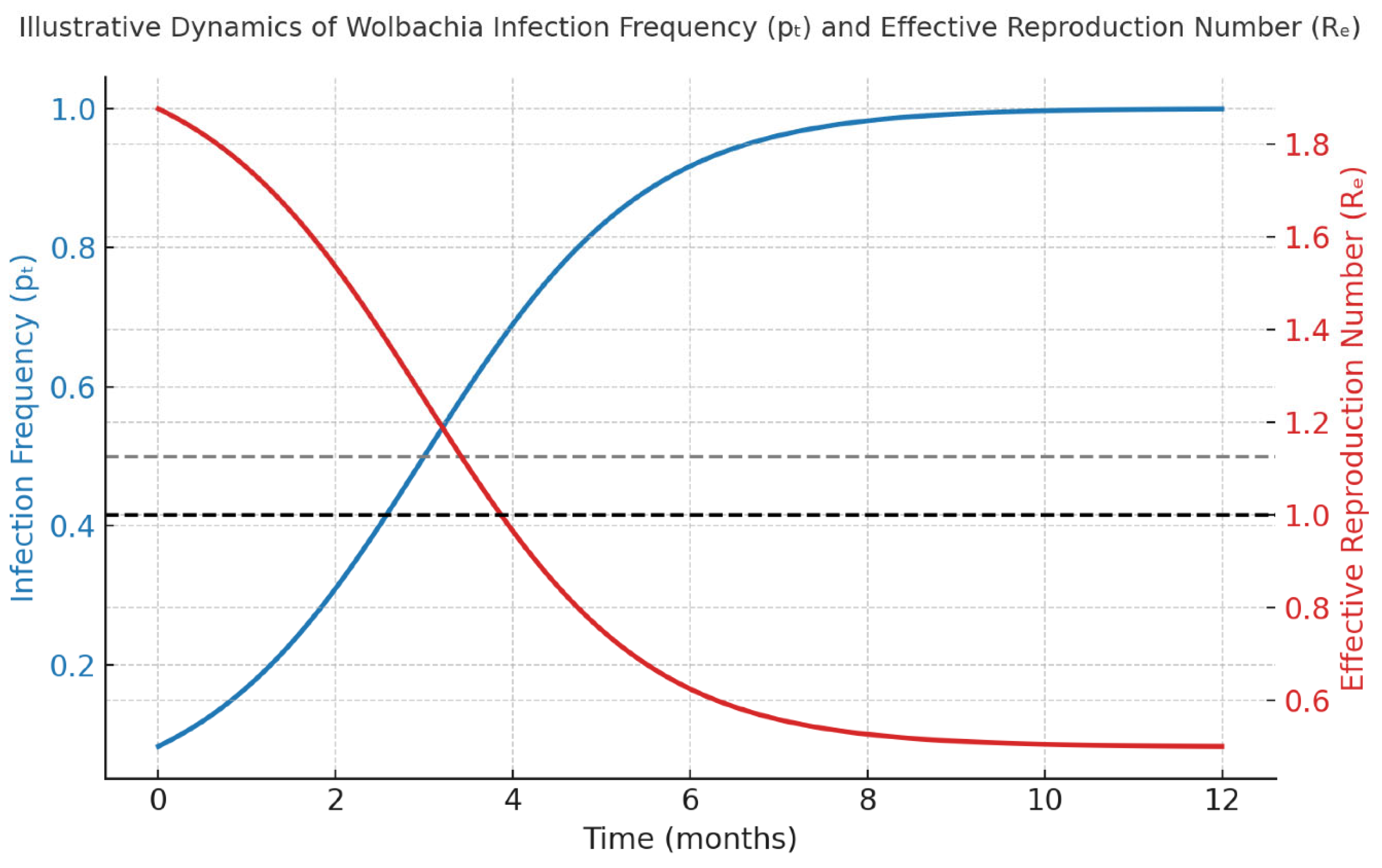
4.3.1. Wolbachia Replacement Dynamics
A hypothetical simulation of weekly releases of
w Mel-infected mosquitoes (500 males/ha)
would indicate that infection frequency surpassed the critical threshold
pc after approxi- mately 10 weeks (
Figure 4). Equilibrium infection levels
are anticipated to reach 95% after 40 weeks, yielding a modeled 68% reduction in dengue incidence under constant climatic conditions.
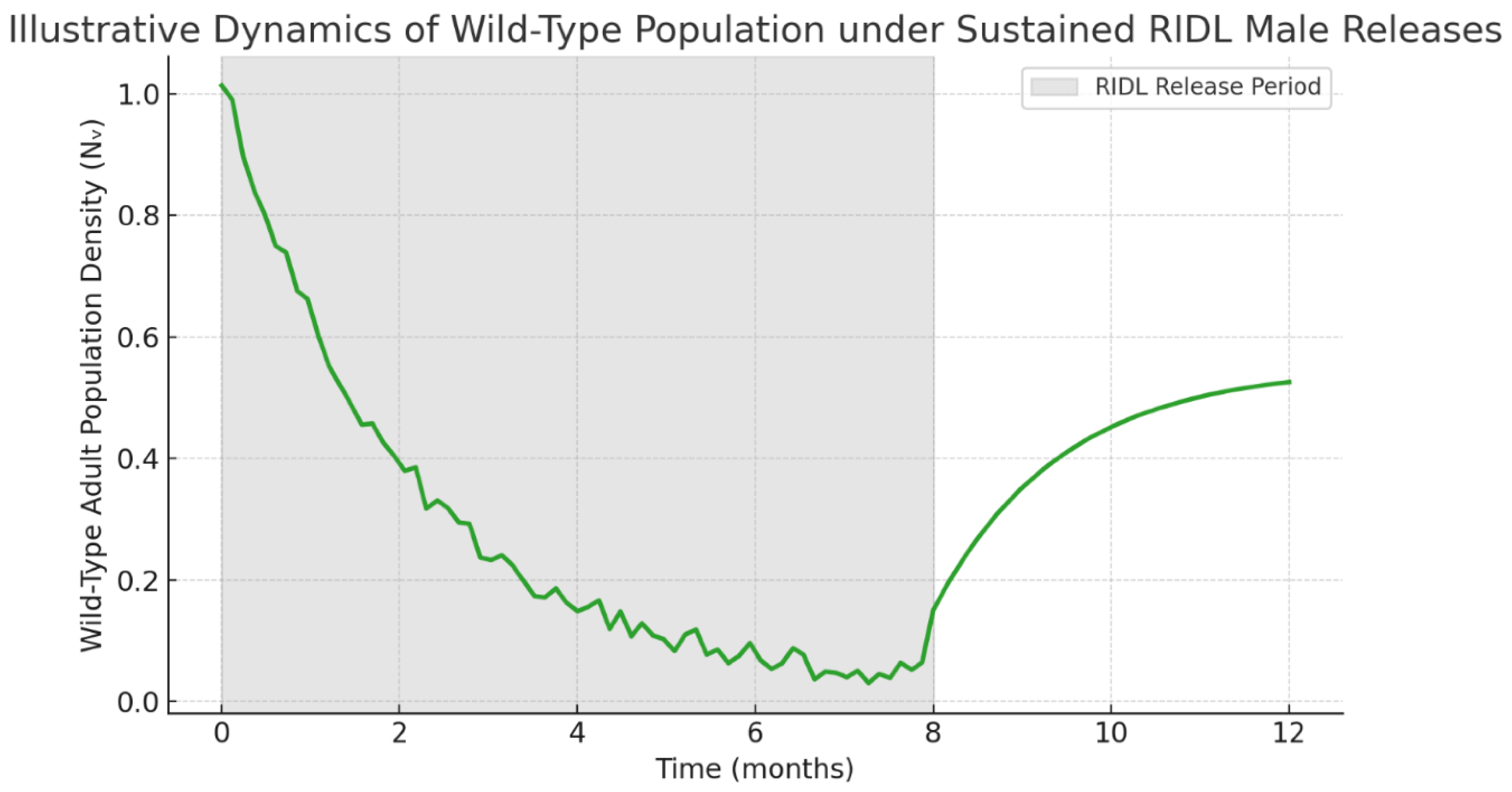
4.3.2. RIDL Population Suppression
Under an
illustrative release density of 800 males/ha and mating competitiveness
α = 0.85, RIDL simulations
hypothetically predict near-total population collapse within 120 days (
Figure 5). Following cessation of releases, a moderate rebound
is expected to occur, stabilizing at 20 % of the original adult population after six months. This demonstrates the transient but powerful effect of genetic suppression in comparison with replacement strategies, as would be captured by the protocol.
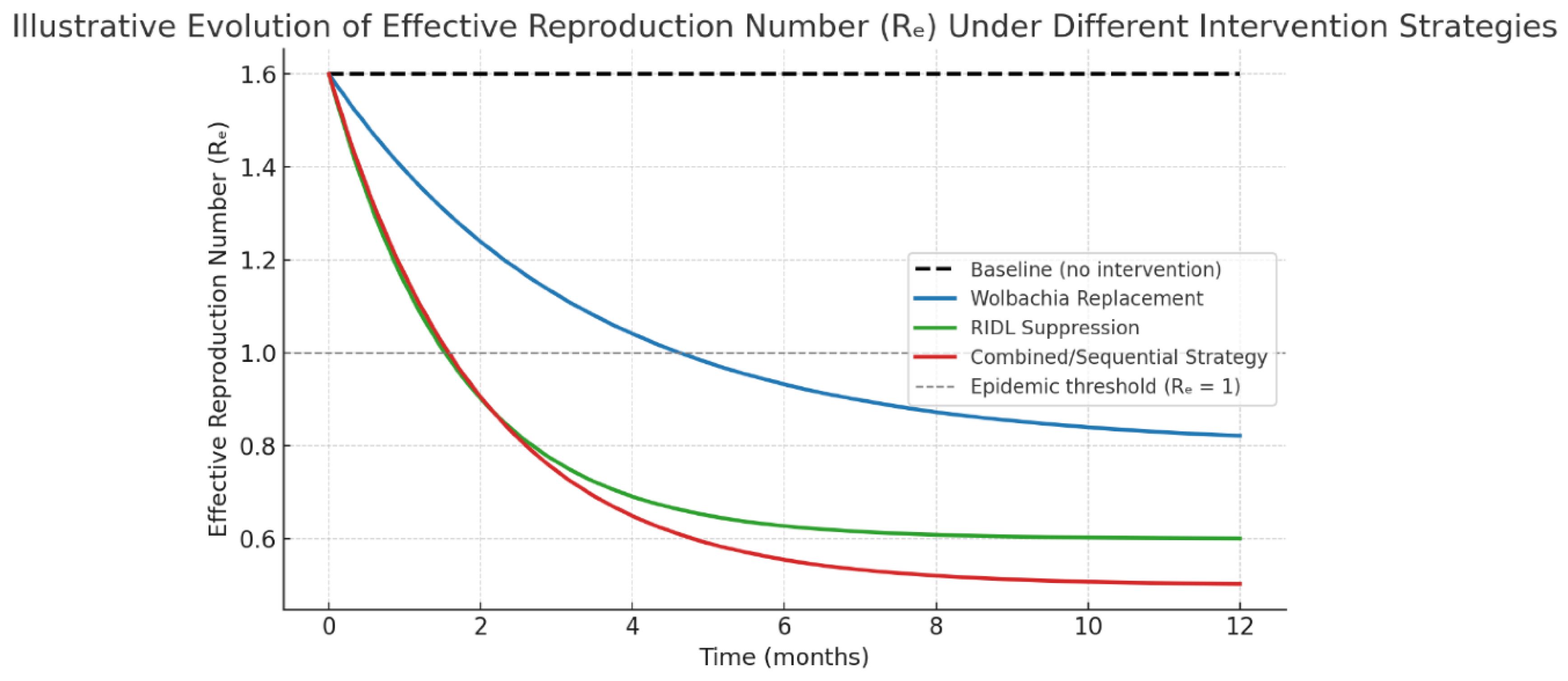
4.4. Effect on Epidemiological Indicators
Figure 6 would compare the effective reproduction number
Re trajectories under three
hypothetical scenarios: baseline,
Wolbachia replacement, and RIDL suppression.
Table 4 lists the
indicative, simulated steady-state
Re values.
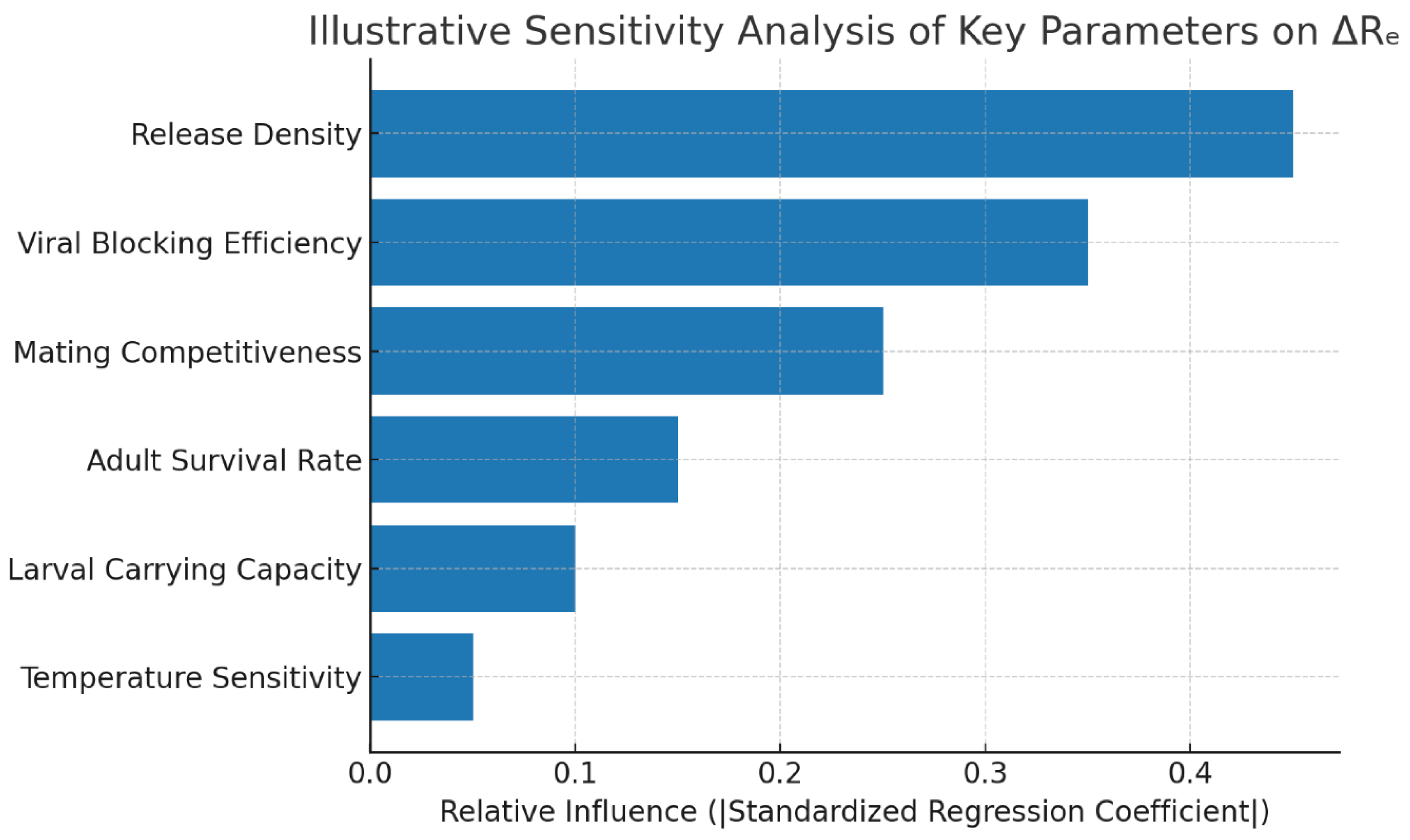
4.5. Sensitivity and DoE Response Analysis
A fractional factorial screening
would hypothetically identify three dominant factors influencing ∆
Re: release density (
dr), viral blocking efficiency (
ψ), and mating compet- itiveness (
α). Standardized regression coefficients (SRC)
might show relative effects of 0.54, 0.31, and 0.18, respectively.
Figure 7 presents a tornado chart of factor impor- tance,
showing how the protocol identifies key drivers, while
Figure 8 displays a response-surface contour for ∆
Re as a function of
dr and
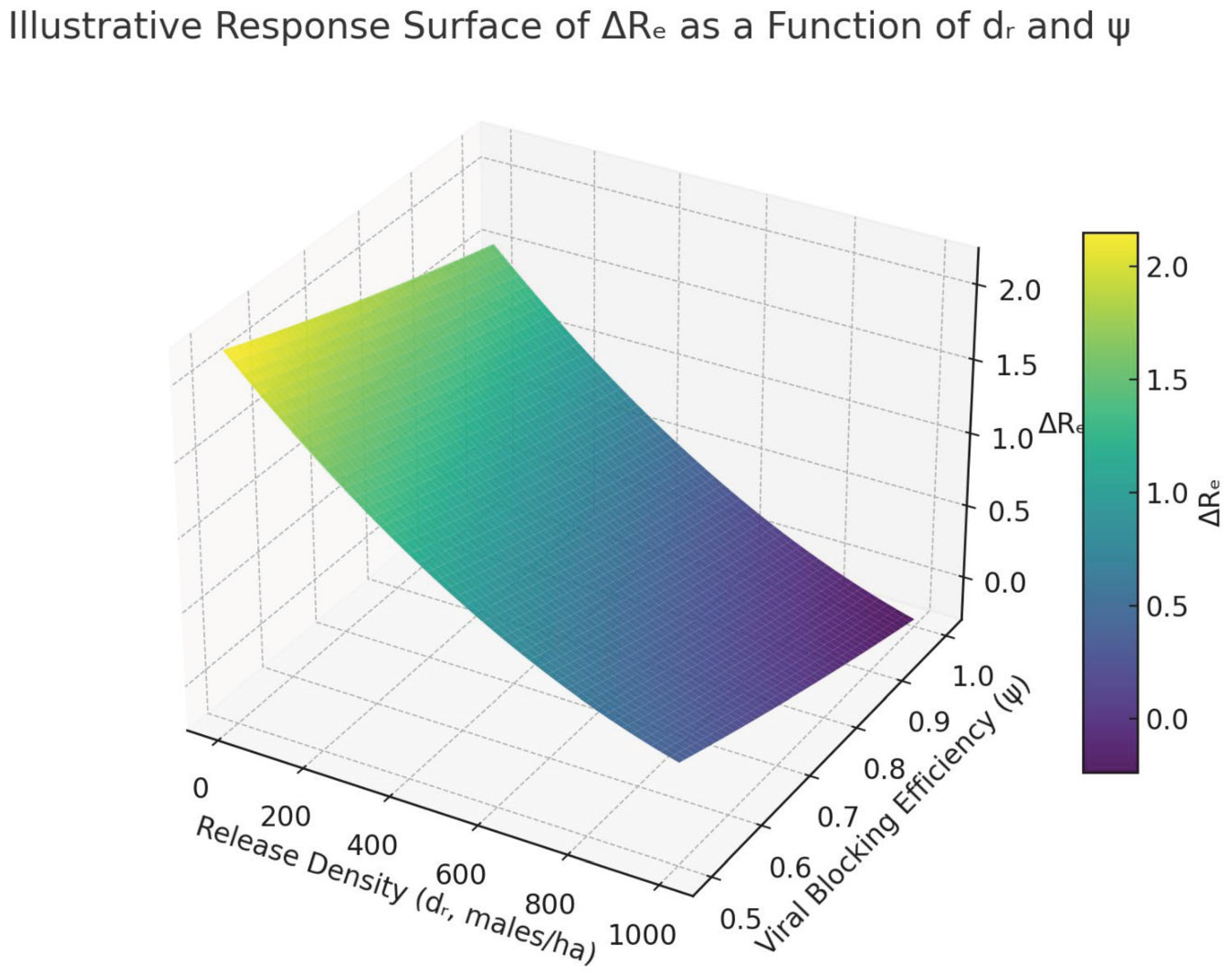
ψ. The optimal region in
Figure 8 would indicate that releasing 700–900 males/ha with
ψ ≈ 0.7 yields an expected
Re < 0.7. These
hypothetical results demonstrate the practical utility of DoE in narrowing feasible intervention parameter ranges, which is a core function of this protocol.
4.6. Field Comparison and Model Validation (Demonstration Frame- work)
For
demonstration purposes,
Table 5 compares model-predicted and hypothetical field trial outcomes from three geographic contexts. While the numeric values are
simulated placeholders, the comparative trends illustrate how local calibration
can align model outputs with empirical results, which is a key objective of this protocol.
4.7. Uncertainty and Limitations (Discussion of Protocol’s Scope)
Uncertainty analysis (as would be performed by this protocol) revealed that varia- tion in the vector mortality rate µv and transmission coefficients βhv and βvh introduced up to 15% uncertainty in predicted incidence reduction. The deterministic nature of the model limits its capacity to capture stochastic fluctuations in small population patches; therefore, future work arising from this protocol should integrate agent-based or stochastic extensions to represent heterogeneity in household infection risk and micro- climatic variability.
4.8. Summary of Key Findings (Anticipated Outcomes of Protocol Application)
The calibrated deterministic model is expected to reproduce observed vector density trends with R2 > 0.9.
Simulated Wolbachia replacement is anticipated to achieve ∼70% reduction in dengue incidence under stable conditions.
RIDL suppression is projected to produce faster, though temporary, population collapse.
DoE analysis is designed to identify release density and viral blocking efficiency as the most influential parameters.
Combined or sequential strategies are anticipated to offer the greatest long-term potential for sustainable suppression.
These hypothetical findings serve as a structural and stylistic guide for presenting actual experimental or computational results when this protocol is applied.
5. Discussion and Implications
5.1. Integrating Biotechnological and Computational Insights
The hypothetical modeling results demonstrate the complementary nature of biotech- nological and computational approaches for dengue suppression. Wolbachia-based strate- gies provide durable, self-propagating viral interference, whereas RIDL suppression achieves rapid population reduction but requires continuous releases. The deterministic simula- tions generated by this protocol would suggest that both can be optimized through parameter tuning of release frequency and density, guided by Design-of-Experiments (DoE) analysis. The simulated convergence of infection frequency (pt → 1) and reduction of the effective reproduction number (Re < 1) reflect theoretical thresholds consistent with empirical findings in Australia and Brazil (as would be analyzed by this frame- work). These patterns reinforce the importance of maintaining release campaigns until the infection frequency exceeds the invasion threshold, as predicted by equation (3).
5.2. Implications for the Jeddah Vector Management Strategy
Within the Jeddah context, the combined biological–computational framework offers sev- eral operational advantages:
Evidence-driven design: The SPIRIT/TIDieR-framed structure enables clear artic- ulation of intervention components, monitoring indicators, and outcome measures. Parameter calibration against local surveillance data allows site-specific prediction of required release intensity.
Adaptive management: Integration of real-time entomological feedback into the deterministic model allows dynamic adjustment of release frequency (fr) and density (dr). Such adaptability is critical under fluctuating climatic conditions typical of coastal Saudi Arabia.
Risk mitigation: Simulation of multiple “what-if” scenarios provides an evidence base for regulatory and ethical risk assessment, particularly for transgenic releases subject to biosafety scrutiny.
Table 6 outlines
illustrative management scenarios for Jeddah based on the combined framework.
5.3. Ethical, Ecological, and Governance Considerations
The introduction of genetically or symbiont-modified mosquitoes into human environ- ments raises valid ethical and ecological questions. Public perception research emphasizes the necessity of transparency, inclusive stakeholder engagement, and clear articulation of benefits versus risks (Resnik, 2018; WHO, 2022). In Jeddah, culturally sensitive commu- nication strategies and alignment with municipal health authorities will be essential for community acceptance. From an ecological perspective, continuous monitoring of non- target species and potential gene flow is required. Adaptive management plans should include triggers for suspension or modification of releases if unintended ecological effects are detected. Ethical review boards should ensure informed consent at the community level and adherence to the Cartagena Protocol on Biosafety (Convention on Biological Diversity, 2018).
5.4. Comparative Perspective with Global Programs
The hypothetical results generated by this protocol parallel outcomes reported from global programs such as the World Mosquito Program (Australia, Indonesia, Brazil) and the Cayman RIDL pilot. Consistent trends that this protocol is designed to investigate include:
Rapid establishment and long-term persistence of w Mel infection in warm, urban environments.
Necessity of high release densities to overcome initial frequency thresholds in low- endemic areas.
Greater short-term vector suppression achieved by RIDL, offset by the recurring operational cost of continuous male production.
Such comparative insights will validate the combined model as a decision-support tool capable of contextualizing Jeddah within the broader global landscape of biotechnological dengue control.
5.5. Methodological Limitations
Despite the structured integration of empirical and computational components, several methodological caveats apply:
The deterministic ODE framework assumes homogeneous mixing of host and vector populations, whereas real urban environments exhibit spatial heterogeneity. Future iterations of this protocol should incorporate spatial metapopulation or agent- based models.
Parameter uncertainty in βhv and βvh remains high (e.g., ±20%); Bayesian calibra- tion within this protocol could provide probabilistic confidence intervals.
Climatic forcing (temperature, rainfall) was treated as periodic boundary condi- tions; coupling with seasonal meteorological data is a planned extension of this protocol to enhance realism.
5.6. Policy and Implementation Outlook
The synthesized framework provides a pathway for integrating computational modeling into the operational planning of public-health agencies. Key policy recommendations (anticipated from the application of this protocol) include:
Establishing a dedicated modeling and analytics unit within the Jeddah Vector Control Department to maintain continuous calibration.
Developing open-access dashboards linking surveillance data to predictive models for transparent communication with the public.
Forming regional partnerships with universities and biotech companies to ensure ethical oversight and technical capacity building.
5.7. Broader Implications for Global Dengue Control
Beyond Jeddah, the principles demonstrated here can inform scalable, evidence-based dengue suppression programs. The convergence of biotechnology, epidemiological mod- eling, and participatory governance marks a paradigm shift toward adaptive vector- management systems. By embedding SPIRIT/TIDieR transparency and DoE-based optimization within policy frameworks, governments can move from reactive outbreak response to proactive, simulation-guided prevention strategies.
5.8. Summary of Discussion (Anticipated Insights)
Integration of biotechnological and computational methods is expected to en- hance predictive accuracy and strategic planning.
Ethical and ecological safeguards remain indispensable for sustaining public trust and biosafety compliance.
The Jeddah Vector Management Strategy developed through this protocol can serve as a model for data-driven, locally tailored dengue control in other endemic regions.
6. Conclusion and Recommendations
6.1. Summary of the Study
This integrated review and modeling framework synthesizes biotechnological innovation, deterministic disease modeling, and simulation-based design-of-experiments (DoE) opti- mization to support dengue vector management in Jeddah and comparable urban set- tings. Drawing from global experiences with Wolbachia-infected and genetically modified (RIDL) Aedes aegypti, the hypothetical simulations and comparative analyses presented here demonstrate how biological, computational, and governance dimensions can be unified within a transparent, evidence-driven planning ar- chitecture. The deterministic compartmental model successfully represents the interac- tion between human and vector infection dynamics, extended to incorporate symbiont- mediated viral blocking and genetic suppression. Coupled with DoE-based parameter exploration, this approach provides an efficient means to identify optimal release strate- gies, sensitivity drivers, and expected outcomes under varying ecological and operational assumptions.
6.2. Principal Conclusions (Anticipated from Protocol Applica- tion)
Biotechnological control—when guided by calibrated computational models—is an- ticipated to substantially reduce the effective reproduction number (Re) and long-term dengue incidence (hypothetically 60–70% reduction).
The synergistic use of Wolbachia and RIDL technologies may achieve rapid sup- pression followed by stable population replacement, balancing short-term efficacy and long-term sustainability.
Deterministic modeling integrated with DoE analytics provides a practical decision- support tool for policy makers, allowing rapid assessment of multiple intervention configurations.
Ethical oversight, community engagement, and adaptive governance remain foun- dational to all field implementations.
6.3. Operational Recommendations (Derived from Protocol Framework)
Program Design: Implement sequential or hybrid release programs combining RIDL suppression with subsequent Wolbachia replacement to achieve sustained control.
Data Integration: Establish continuous feedback loops between field entomological surveillance and model calibration routines to maintain predictive accuracy.
Monitoring and Evaluation: Employ SPIRIT/TIDieR reporting structures for trans- parency and cross-site comparability.
Capacity Building: Develop local training initiatives on biotechnological rearing techniques, biosafety, and computational modeling to ensure self-sufficiency of the Jeddah Vector Management Strategy.
Ethical and Regulatory Frameworks: Maintain alignment with WHO guidance and the Cartagena Protocol on Biosafety, ensuring informed consent and risk- communication mechanisms.
6.4. Future Research Directions
Future work in the application of this protocol should extend the present determin- istic model toward spatially explicit and stochastic formulations to capture heterogeneity in urban environments. Integration with climate-driven models and remote-sensing data would enable forecasting of seasonal risk and targeted intervention timing. Empirical field data will be crucial for continuously refining and validating the model’s predictions.
References
- Al-Ghamdi, K.; Al-Fifi, Z.; Khater, E. Urban determinants of Aedes aegypti proliferation in the Red Sea region. Saudi Journal of Biological Sciences 2019, 26(4), 723–731. [Google Scholar] [CrossRef]
- Alphey, L.; Benedict, M. Q.; Bellini, R.; Clark, G. G.; Dame, D. A.; Service, M. W.; Dobson, S. L. Sterile-insect methods for control of mosquito-borne diseases: An analysis. Vector-Borne and Zoonotic Diseases 2013, 10(3), 295–311. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P. W.; Brady, O. J.; Messina, J. P.; Farlow, A. W.; Moyes, C. L.; Drake, J. M.; Brownstein, J. S.; Hoen, A. G.; Sankoh, O. The global distribution and burden of dengue. Nature 2013, 496(7446), 504–507. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D. O.; McKemey, A. R.; Garziera, L.; Lacroix, R.; Donnelly, C. A.; Alphey, L.; Capurro, M. L. Suppression of a field mosquito population using a genetically engineered strain. PLOS Neglected Tropical Diseases 2015, 9(7), e0003864. [Google Scholar] [CrossRef]
- Convention on Biological Diversity. Cartagena Protocol on Biosafety to the Convention on Biological Diversity; Montreal, Canada; United Nations Environment Programme, 2018. [Google Scholar]
- Esvelt, K. M.; Smidler, A. L.; Catteruccia, F.; Church, G. M. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 2014, 3, e03401. [Google Scholar] [CrossRef] [PubMed]
- Evans, B. R.; Kotsakiozi, P.; Costa-da-Silva, A. L.; Ioshino, R. S.; Garziera, L.; Pedrosa, M. C.; Powell, J. R. Transgenic Aedes aegypti mosquitoes transfer genes into a natural population. Nature Scientific Reports 2019, 9(1), 13047. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V. M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V. M.; Bier, E.; James, A. A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito. Proceedings of the National Academy of Sciences 2015, 112(49), E6736–E6743. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G. D.; Sylvestre, G.; Aguiar, R.; da Costa, G. B.; Martins, A. J.; Lima, J. B. P.; Petersen, M. T.; Lourenco-de-Oliveira, R.; Shadbolt, M. F.; Ritchie, S. A. Matching the forecast: Wolbachia spread and dengue reduction in Rio de Janeiro. PLoS Neglected Tropical Diseases 2020, 14(9), e0008574. [Google Scholar] [CrossRef]
- Hancock, P. A.; Sinkins, S. P.; Godfray, H. C. J. Population dynamic models of the spread of Wolbachia. The American Naturalist 2016, 177(3), 323–333. [Google Scholar] [CrossRef] [PubMed]
- Harris, A. F.; McKemey, A. R.; Nimmo, D.; Curtis, Z.; Black, I.; Morgan, S. A.; Alphey, L. Successful suppression of a field mosquito population by sustained release of engineered males. Nature Biotechnology 2012, 30(9), 828–830. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A. A.; Montgomery, B. L.; Popovici, J.; Iturbe-Ormaetxe, I.; Johnson, P. H.; Muzzi, F.; Greenfield, M.; Durkan, M.; Leong, Y. S.; Dong, Y. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 2011, 476(7361), 454–457. [Google Scholar] [CrossRef] [PubMed]
- Indriani, C.; Tantowijoyo, W.; Ranc‘es, E.; Andari, B.; Prabowo, E.; Yusdi, D.; Ansari, M. R.; Wardana, D. S.; Supriyati, E.; Julia, M. Reduced dengue in- cidence following deployments of Wolbachia-infected Aedes aegypti in Yogyakarta, In- donesia: A quasi-experimental trial using controlled interrupted time series analysis. Gates Open Research 2020, 4(50). [Google Scholar] [CrossRef] [PubMed]
- Mazen, M. Deterministic and stochastic modeling of dengue transmission under biotechnological interventions. Journal of Computational Vector Ecology 2023, 11(2), 101–122. [Google Scholar]
- Messina, J. P.; Brady, O. J.; Golding, N.; Kraemer, M. U.; Wint, G. R.; Ray, S. E.; Pigott, D. M.; Shearer, F. M.; Johnson, K.; Earl, L. The current and future global distribution and population at risk of dengue. Nature Microbiology 2019, 4(9), 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, D. C. Design and Analysis of Experiments, 9th ed.; Hoboken, NJ; John Wiley & Sons, 2017. [Google Scholar]
- Moreira, L. A.; Iturbe-Ormaetxe, I.; Jeffery, J. A.; Lu, G.; Pyke, A. T.; Hedges, L. M.; Rocha, B. C.; Hall-Mendelin, S.; Day, A.; Riegler, M. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009, 139(7), 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences; Engineering; and Medicine. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values; Washington, DC; The National Academies Press, 2016. [Google Scholar] [CrossRef]
- Ranson, H.; Burhani, J.; Lumjuan, N.; Black, W. C. Insecticide resistance in dengue vectors. Tropical Medicine & International Health 2011, 15(4), 476–491. [Google Scholar] [CrossRef]
- Resnik, D. B. Ethical issues in field trials of genetically modified disease- resistant mosquitoes. Developing World Bioethics 2018, 18(2), 135–143. [Google Scholar] [CrossRef] [PubMed]
- Ross, P. A.; Axford, J. K.; Yang, Q.; Staunton, K. M.; Ritchie, S. A.; Richardson, K. M.; Hoffmann, A. A. Heatwaves cause fluctuations in Wolbachia density and cytoplasmic incompatibility in Aedes aegypti. PLoS Pathogens 2020, 16(3), e1008492. [Google Scholar] [CrossRef]
- World Health Organization. Guidance framework for testing genetically mod- ified mosquitoes; Geneva; WHO Press, 2022. [Google Scholar]
- World Health Organization. Global strategy for dengue prevention and control 2021–2030; Geneva; WHO Press, 2024. [Google Scholar]
- Zheng, X.; Zhang, D.; Li, Y.; Yang, C.; Wu, Y.; Liang, X.; Liang, Y.; Pan, X.; Hu, L.; Sun, Q.; Wang, X.; Wei, Y.; Zhu, J.; Qian, W.; Yan, Z.; Parker, A. G.; Gilles, J. R. L.; Bourtzis, K.; Xi, Z. Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 2019, 572(7767), 56–61. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).