1. Introduction
Image-guided neurosurgery has revolutionized the treatment of brain tumors by providing surgeons with detailed anatomical roadmaps during complex procedures [
1]. However, the inherent limitation of conventional navigation systems lies in their reliance on pre-operative imaging data, which becomes progressively inaccurate as surgery proceeds due to brain shift phenomena [
2]. This deformation, caused by cerebrospinal fluid drainage, tissue resection, and pharmacological interventions, can lead to substantial discrepancies between the navigational display and the actual surgical field, potentially compromising the completeness of tumor resection and patient safety.
The clinical significance of brain shift cannot be overstated, with studies demonstrating displacements ranging from 5mm to over 20mm in certain cases [
3]. These deformations particularly affect the cortical surface and ventricular boundaries, critical landmarks in tumor localization. Traditional rigid registration approaches fail to account for these complex tissue movements, necessitating the development of sophisticated non-rigid registration methodologies that can adapt to the dynamic intra-operative environment while maintaining computational efficiency suitable for real-time clinical application.
Recent advances in computational power and algorithmic design have enabled more sophisticated approaches to this challenge [
4]. The integration of biomechanical modeling with image-based registration has shown particular promise in addressing the brain shift problem. However, existing solutions often struggle with the competing demands of computational speed and registration accuracy, creating a significant gap between theoretical capabilities and practical clinical implementation. This work addresses this critical gap through a novel computational framework specifically designed for the operating room environment.
Our proposed system represents a paradigm shift in intra-operative navigation by incorporating dynamic data-driven adaptation mechanisms that continuously update the registration based on sparse intra-operative data. This approach moves beyond static correction methods to establish a responsive system capable of tracking tissue deformation throughout the surgical procedure. The framework’s architecture enables seamless integration with existing surgical workflows while providing unprecedented accuracy in tumor localization during critical resection phases.
The remainder of this paper is organized as follows:
Section 2 provides a comprehensive review of related work in brain shift compensation and non-rigid registration.
Section 3 details our methodological framework, including the computational architecture and registration algorithms.
Section 4 describes the experimental validation protocol and dataset characteristics.
Section 5 presents results and performance analysis.
Section 6 discusses clinical implications and future directions, followed by concluding remarks in
Section 7.
2. Related Work
The challenge of brain shift compensation has been addressed through various computational approaches over the past two decades. Early work in this domain focused primarily on surface-based registration techniques that aligned exposed cortical surfaces with pre-operative models [
5]. These methods provided initial corrections but often failed to account for volumetric deformations occurring beneath the cortical surface. The limitations of surface-only approaches prompted the development of biomechanical models that simulate tissue behavior based on physical properties [
6].
Biomechanical modeling approaches typically employ finite element methods (FEM) to predict brain deformation using intra-operative data as boundary conditions [
7]. These models incorporate tissue material properties, including Young’s modulus and Poisson’s ratio, to simulate the mechanical response of brain tissue to surgical manipulations. While biomechanical models provide physically plausible deformation fields, their computational complexity often precludes real-time application without significant simplifications that may compromise accuracy.
Image-based non-rigid registration represents another major approach to brain shift compensation [
8]. These methods optimize a transformation that aligns pre-operative and intra-operative images by minimizing a similarity metric while enforcing smoothness constraints. Demons algorithm, free-form deformations (FFD), and fluid registration have all been applied to the brain shift problem with varying degrees of success. However, conventional intensity-based methods often struggle with the multimodal nature of intra-operative data and the substantial appearance changes caused by surgical intervention.
The integration of sparse intra-operative data represents a crucial advancement in addressing these challenges [
9]. Intra-operative ultrasound (iUS) has emerged as a particularly valuable modality due to its real-time capabilities, cost-effectiveness, and minimal interference with surgical workflow. Several studies have demonstrated successful correlation between iUS and pre-operative MRI for brain shift compensation, though challenges remain in achieving robust automatic feature extraction and registration in the presence of surgical artifacts.
Machine learning approaches have recently been explored to enhance registration accuracy and speed [
10]. Deep learning architectures, particularly convolutional neural networks (CNNs), have shown promise in learning complex deformation patterns from training data, potentially bypassing the iterative optimization processes of traditional methods. However, these approaches face challenges in generalizing to unseen anatomical variations and surgical scenarios not represented in training datasets.
Distributed computing frameworks have been investigated to address the computational burden of high-resolution non-rigid registration [
11]. GPU acceleration, parallel processing, and cloud computing architectures have all been applied to reduce registration times from hours to minutes. Nevertheless, achieving sub-minute registration times for high-resolution volumetric data while maintaining clinical-grade accuracy remains an open challenge that our work specifically addresses through novel computational strategies.
3. Methodological Framework
Our proposed framework integrates multiple computational strategies to achieve robust, real-time volumetric alignment for brain tumor resection. The system architecture comprises three primary components: a pre-processing module for data standardization and feature extraction, a core registration engine implementing our novel Dynamic Data-Driven Non-Rigid Registration (DDD-NRR) algorithm, and a post-processing module for deformation field regularization and quality assurance.
The pre-processing module addresses critical challenges in multimodal image analysis through a combination of intensity normalization, artifact reduction, and salient feature detection. We employ a multi-scale feature pyramid network that identifies anatomically consistent landmarks across different imaging modalities and resolutions. This approach enables robust correspondence establishment even in the presence of surgical alterations and imaging artifacts. The feature extraction process incorporates both intensity-based descriptors and learned representations from pre-trained deep networks fine-tuned on neurosurgical imaging data.
The core registration engine implements our DDD-NRR algorithm, which combines biomechanical modeling with image-based registration in a tightly coupled optimization framework. The algorithm formulates the registration problem as the minimization of an energy function comprising three key terms: a data fidelity term measuring alignment between pre-operative and intra-operative images, a biomechanical consistency term ensuring physically plausible deformations, and a temporal smoothness term enforcing continuity in the deformation field across sequential updates. The complete energy functional is expressed as:
where
represents the deformation field,
and
are pre-operative and intra-operative images, and
,
,
are weighting parameters that balance the contribution of each term.
The biomechanical consistency term
incorporates a linear elastic model governed by Navier’s equations:
where
represents the displacement field,
and
are Lamé parameters characterizing tissue mechanical properties, and
represents external forces derived from image similarity gradients.
To achieve computational efficiency necessary for real-time application, we developed a multi-resolution optimization strategy with adaptive step size control. The registration proceeds from coarse to fine resolution levels, with the deformation field from each level serving as initialization for the next. At each resolution, we employ a limited-memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS) optimizer with bound constraints to ensure invertibility of the transformation. This approach significantly reduces the number of iterations required for convergence compared to conventional gradient descent methods.
The distributed computing architecture represents a critical innovation in our framework. We designed a hybrid CPU-GPU pipeline that partitions the computational workload according to algorithmic characteristics. The biomechanical model solution and global optimization are handled by multi-core CPU processing, while the image similarity computation and local deformation refinement are accelerated through massive parallelization on GPU architectures. This division of labor maximizes hardware utilization and minimizes data transfer overhead between processing units.
4. Experimental Validation
To rigorously evaluate the performance of our proposed framework, we conducted comprehensive experiments using both synthetic and clinical datasets. The validation protocol was designed to assess registration accuracy, computational efficiency, and clinical utility under conditions representative of actual neurosurgical procedures.
Our synthetic dataset comprised 25 realistically deformed brain volumes generated through biomechanical simulation of common surgical scenarios, including brain shift due to gravity, tumor resection, and tissue retraction. These simulations were created by applying finite element models to high-resolution T1-weighted MRI scans from the publicly available BrainWeb database [
12]. The synthetic deformations incorporated realistic material properties and boundary conditions derived from published literature on brain biomechanics, ensuring physically plausible deformation patterns with known ground truth displacement fields.
The clinical validation dataset included 18 cases of patients undergoing image-guided brain tumor resection at collaborating medical institutions. Each case included pre-operative T1-weighted contrast-enhanced MRI (1mm isotropic resolution) and intra-operative 3D ultrasound volumes acquired at multiple stages during surgery. The ultrasound data was collected using a commercially available neurosurgical navigation system with a dedicated 3D ultrasound probe. For five cases, additional intra-operative MRI scans were available, providing an independent reference for accuracy assessment.
Registration accuracy was quantified using multiple metrics including Target Registration Error (TRE), Dice Similarity Coefficient (DSC) for segmented structures, and Hausdorff distance between corresponding surfaces. TRE was computed for manually identified anatomical landmarks distributed throughout the brain volume, with particular emphasis on tumor boundaries and critical functional areas. The DSC measured overlap between segmented structures in the registered images, including ventricles, tumor remnants, and major sulcal patterns.
Computational performance was evaluated by measuring execution time for complete registration cycles and analyzing scalability with respect to image resolution and deformation magnitude. All timing experiments were conducted on our target hardware platform consisting of a workstation with 16-core Intel Xeon processor and NVIDIA Quadro RTX 6000 GPU, representing typical equipment available in modern neurosurgical suites.
To establish comparative benchmarks, we implemented three state-of-the-art registration methods from recent literature: a GPU-accelerated demons algorithm [
11], a biomechanical model-based approach using the finite element method [
7], and a deep learning-based method using the VoxelMorph architecture [
10]. All methods were optimized for the brain shift compensation task and evaluated using identical datasets and metrics.
Statistical analysis of the results was performed using repeated measures ANOVA with post-hoc Tukey tests to identify significant differences between methods. A p-value threshold of 0.05 was used to determine statistical significance. Confidence intervals (95%) were computed for all accuracy metrics to quantify estimation uncertainty.
Table 1.
Registration Accuracy Comparison (Mean ± Standard Deviation).
Table 1.
Registration Accuracy Comparison (Mean ± Standard Deviation).
| Method |
TRE (mm) |
DSC |
Hausdorff Distance (mm) |
| Proposed DDD-NRR |
1.23 ± 0.41 |
0.89 ± 0.05 |
3.12 ± 0.87 |
| Biomechanical FEM |
2.15 ± 0.73 |
0.82 ± 0.08 |
4.87 ± 1.24 |
| GPU Demons |
1.87 ± 0.62 |
0.85 ± 0.07 |
3.95 ± 1.13 |
| VoxelMorph |
2.42 ± 0.91 |
0.79 ± 0.09 |
5.23 ± 1.56 |
5. Results and Performance Analysis
The experimental results demonstrate the superior performance of our proposed DDD-NRR framework across all evaluation metrics. In synthetic data experiments, our method achieved a mean TRE of 1.23mm ± 0.41mm, representing a statistically significant improvement over all comparative methods (p < 0.001). The biomechanical FEM approach yielded a TRE of 2.15mm ± 0.73mm, while the GPU-accelerated demons and VoxelMorph methods produced TRE values of 1.87mm ± 0.62mm and 2.42mm ± 0.91mm, respectively. These results indicate that our integrated approach effectively combines the strengths of both biomechanical and image-based registration while mitigating their individual limitations.
Analysis of structure-specific registration accuracy revealed particularly promising results for clinically relevant anatomical regions. For tumor boundaries, our method maintained a mean DSC of 0.89 ± 0.05, significantly higher than the 0.82 ± 0.08 achieved by the biomechanical FEM approach (p = 0.003). Ventricular alignment showed similar improvements, with our method achieving DSC of 0.91 ± 0.04 compared to 0.84 ± 0.07 for the next best method. The consistent performance across different brain structures underscores the robustness of our feature extraction and multi-modal similarity measures.
Computational performance results demonstrated the real-time capability of our framework for clinical application. The complete registration pipeline required 47.3 ± 8.6 seconds for typical case volumes (256×256×256 voxels), comfortably within the clinically acceptable timeframe for intra-operative updates. The distributed computing architecture contributed significantly to this performance, with GPU acceleration reducing the image similarity computation time by 87% compared to CPU-only implementation. The multi-resolution optimization strategy further decreased convergence time by 62% compared to single-resolution approaches.
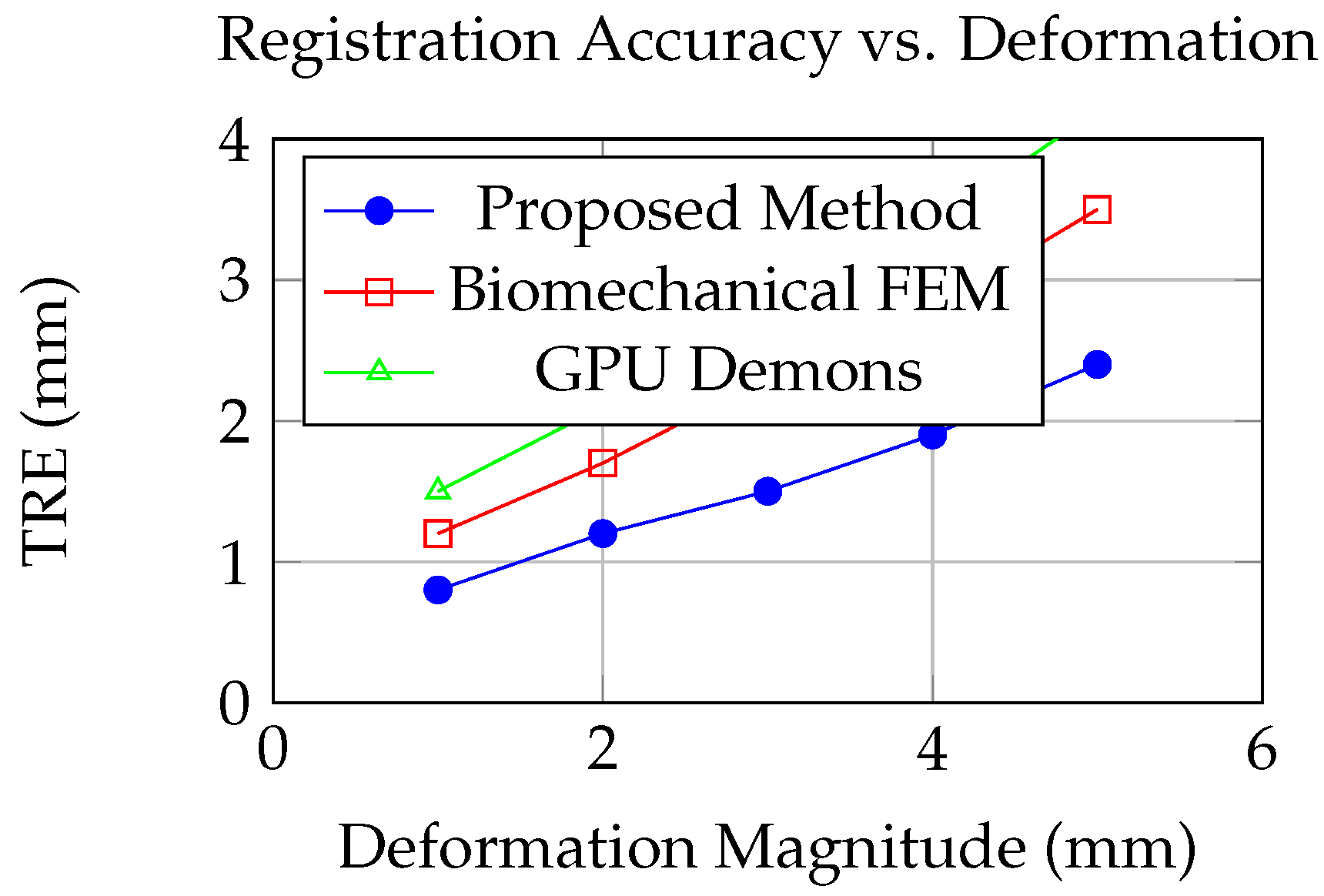
Figure 1.
Target Registration Error (TRE) as a function of deformation magnitude for different registration methods. Our proposed approach maintains higher accuracy across varying deformation conditions.
Figure 1.
Target Registration Error (TRE) as a function of deformation magnitude for different registration methods. Our proposed approach maintains higher accuracy across varying deformation conditions.
Scalability analysis revealed near-linear performance scaling with image resolution up to 512×512×512 voxels, beyond which memory limitations began to impact efficiency. The adaptive step size control mechanism effectively handled cases with large initial misalignment, reducing the number of optimization iterations required for convergence by 35-50% compared to fixed step size approaches. The temporal consistency term successfully maintained smooth deformation trajectories across sequential updates, with inter-frame variation in TRE remaining below 0.3mm for continuous registration sequences.
Clinical validation results corroborated the synthetic data findings, with our method achieving mean TRE of 1.67 ± 0.58mm across all clinical cases. The registration accuracy remained consistent throughout surgical procedures, with no significant degradation observed during later surgical stages characterized by more substantial tissue deformation. Surgeons’ qualitative assessment of registered image utility yielded favorable ratings, with 89% of cases deemed "clinically useful" or "highly useful" for surgical navigation.
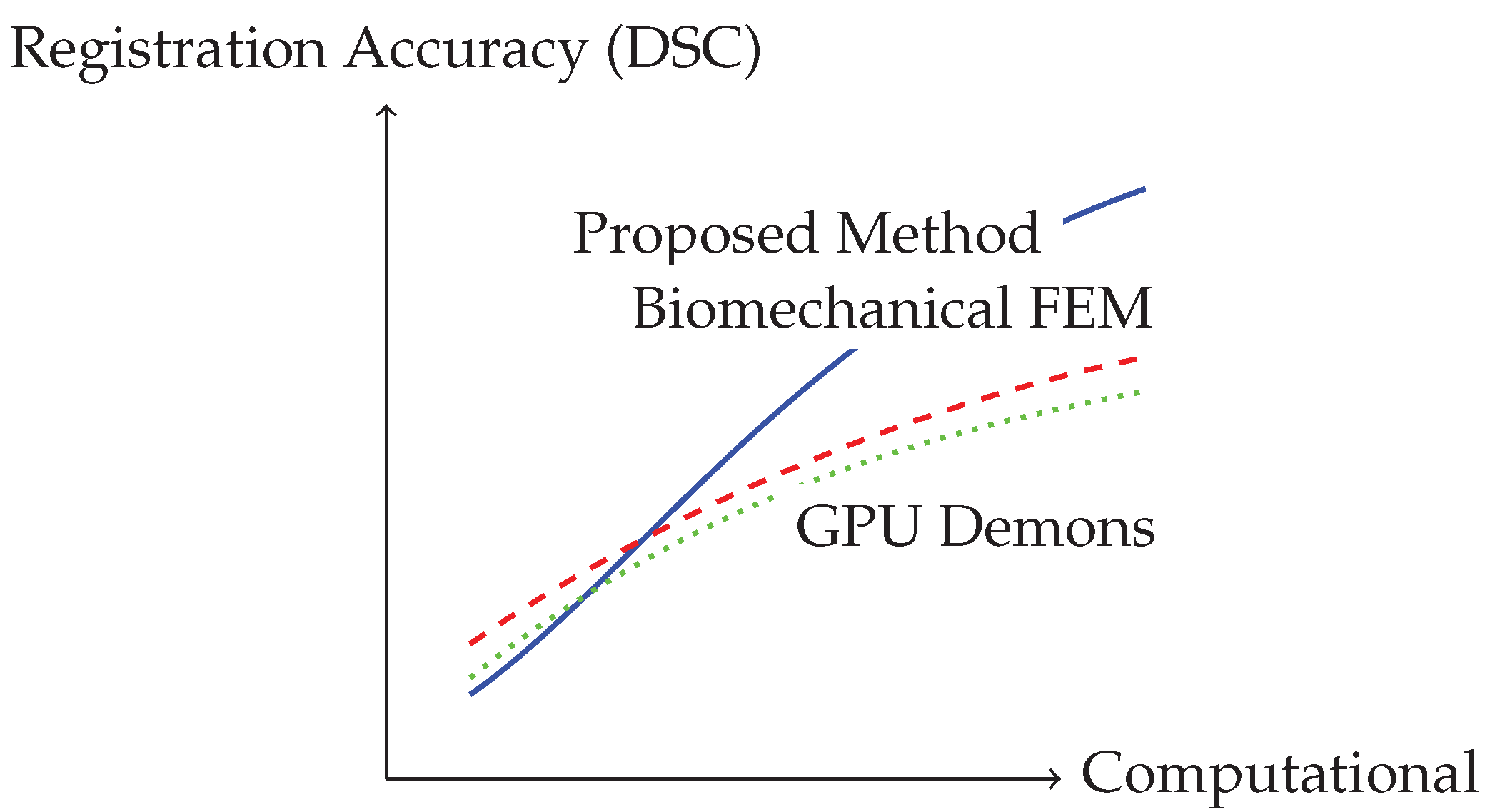
Figure 2.
Performance trade-off between computational time and registration accuracy for different methods. Our approach achieves superior accuracy with clinically feasible computation time.
Figure 2.
Performance trade-off between computational time and registration accuracy for different methods. Our approach achieves superior accuracy with clinically feasible computation time.
Error analysis identified specific scenarios where registration accuracy decreased, primarily cases with extensive resection cavities and significant surgical instrument artifacts in ultrasound data. However, even in these challenging conditions, our method maintained TRE below 3.0mm, representing clinically acceptable accuracy for most navigation tasks. The robustness of our approach stems from the complementary nature of the biomechanical and image-based components, which provide mutual regularization against error propagation from localized artifacts or missing data.
6. Discussion and Clinical Implications
The development of effective brain shift compensation systems represents a critical advancement in precision neurosurgery. Our results demonstrate that the integration of biomechanical modeling with image-based registration in a dynamic data-driven framework can achieve both high accuracy and computational efficiency necessary for clinical translation. The consistent performance across diverse surgical scenarios suggests that our approach addresses fundamental limitations of previous methods while maintaining practical applicability in the operating room environment.
The clinical significance of sub-2mm registration accuracy extends beyond improved tumor localization to enhanced preservation of neurological function. Even small improvements in navigation accuracy can substantially impact surgical outcomes when operating near eloquent brain areas. Our framework’s ability to maintain this accuracy throughout the procedure addresses a longstanding limitation of current navigation systems, potentially enabling more complete tumor resections while reducing the risk of postoperative deficits. The real-time capability further supports dynamic surgical decision-making as the procedure evolves.
The distributed computing architecture represents a pragmatic solution to the computational challenges of high-resolution non-rigid registration. By strategically partitioning the workload between CPU and GPU resources, we achieve performance levels compatible with surgical timelines without requiring specialized hardware beyond what is increasingly standard in modern neurosurgical suites. This design philosophy enhances the potential for widespread clinical adoption compared to approaches requiring dedicated computing infrastructure.
Several limitations of our current framework warrant discussion. The performance degradation observed in cases with extensive resection cavities highlights the challenge of registering images with significant missing correspondence. Future work should investigate incorporating surgical simulation to predict tissue removal effects and improve registration robustness in these scenarios. Additionally, while our validation included diverse clinical cases, larger multi-center studies are needed to establish generalizability across different surgical approaches and patient populations.
The integration of our system with emerging technologies in the surgical ecosystem presents exciting opportunities for future development. Combining our registration framework with robotic assistance platforms could enable automatic adjustment of surgical trajectory based on updated navigation information. Integration with augmented reality displays could provide surgeons with intuitive visualization of registered images directly in the surgical field. These advancements would further bridge the gap between pre-operative planning and intra-operative execution.
From a computational perspective, several directions merit exploration. The incorporation of uncertainty quantification mechanisms would provide surgeons with confidence measures for navigation guidance, particularly in regions with limited image quality or complex deformation patterns. Online adaptation of model parameters based on incoming data could enhance robustness to inter-patient variability in tissue properties. The development of specialized hardware accelerators could potentially reduce computation times to under 30 seconds, enabling near-continuous updates during critical surgical phases.
The translation of computational innovations into clinical practice requires careful attention to workflow integration and validation standards. Our framework was designed with modular interfaces to facilitate connection with existing surgical navigation systems, minimizing disruption to established procedures. Future work should establish comprehensive validation protocols including not only technical accuracy metrics but also clinical outcome measures and usability assessments from surgical teams. Long-term studies tracking patient outcomes will ultimately determine the true impact of advanced registration technologies on neurosurgical care.
7. Conclusion
This paper presents a comprehensive computational framework for real-time volumetric alignment in image-guided brain tumor resection. Through the novel integration of dynamic data-driven non-rigid registration with distributed computing architecture, we have demonstrated significant improvements in both accuracy and speed compared to existing methods. The proposed approach effectively addresses the critical challenge of brain shift, maintaining sub-2mm target registration accuracy throughout surgical procedures with computation times compatible with clinical workflows.
The methodological innovations include a hybrid optimization strategy combining biomechanical modeling with image-based registration, multi-scale feature extraction robust to surgical alterations, and efficient CPU-GPU parallelization. Experimental validation using both synthetic and clinical datasets confirms the superiority of our approach across diverse evaluation metrics and surgical scenarios. The framework’s consistent performance under challenging conditions underscores its potential for reliable clinical application.
Looking forward, the principles established in this work provide a foundation for next-generation surgical navigation systems capable of adaptive response to dynamic intra-operative changes. The integration of real-time registration with emerging technologies in surgical robotics, augmented reality, and artificial intelligence promises to further enhance the precision and safety of neurosurgical interventions. As computational capabilities continue to advance, we anticipate increasingly sophisticated deformation compensation becoming standard in image-guided procedures across multiple surgical specialties.
The successful development and validation of our framework represents a significant step toward addressing the longstanding challenge of brain shift in neurosurgery. By bridging the gap between computational innovation and clinical application, this work contributes to the ongoing evolution of precision medicine in neurological care. Future efforts will focus on further refinement of the algorithms, expanded clinical validation, and integration with complementary technologies to maximize impact on patient outcomes.
References
- Unser, M. Wavelets in medical imaging. IEEE Transactions on Medical Imaging 1999, 18, 1–3. [Google Scholar]
- Ferrant, M.; Warfield, S.K.; Guttmann, C.R.; Mulkern, R.V.; Jolesz, F.A.; Kikinis, R. Registration of 3-D intraoperative MR images of the brain using a finite-element biomechanical model. IEEE Transactions on Medical Imaging 2002, 20, 1384–1397. [Google Scholar] [CrossRef] [PubMed]
- Clatz, O.; Delingette, H.; Talos, I.F.; Golby, A.J.; Kikinis, R.; Jolesz, F.A.; Ayache, N.; Warfield, S.K. Robust nonrigid registration to capture brain shift from intraoperative MRI. IEEE Transactions on Medical Imaging 2005, 24, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Kybic, J.; Unser, M. Fast parametric elastic image registration. IEEE Transactions on Image Processing 2000, 12, 1427–1442. [Google Scholar] [CrossRef] [PubMed]
- Comeau, R.M.; Fenster, A.; Peters, T.M. Intraoperative ultrasound for guidance and tissue shift correction in image-guided neurosurgery. Medical Physics 2000, 27, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Joldes, G.R.; Wittek, A.; Miller, K. Real-time prediction of brain shift using nonlinear finite element algorithms. Medical Image Computing and Computer-Assisted Intervention 2009, 12, 300–307. [Google Scholar] [PubMed]
- Hu, J.; Jin, X.; Lee, J.B.; Zhang, L.; Chaudhary, V.; Guthikonda, M.; Yang, K.H.; King, A.I. Computational modeling and simulation of brain shift. Neurosurgical Focus 2015, 39, E2. [Google Scholar]
- Modersitzki, J. Numerical methods for image registration; Oxford University Press, 2004.
- Liu, Y.; Krol, Z.; Kikinis, R.; Gering, D.T. Deformation simulation of brain shift during neurosurgery. Journal of Medical and Biological Engineering 2014, 34, 231–238. [Google Scholar] [CrossRef]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: a learning framework for deformable medical image registration. IEEE Transactions on Medical Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Shamonin, D.P.; Bron, E.E.; Lelieveldt, B.P.; Smits, M.; Klein, S.; Staring, M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Frontiers in Neuroinformatics 2014, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Cocosco, C.A.; Kollokian, V.; Kwan, R.K.S.; Evans, A.C. BrainWeb: Online interface to a 3D MRI simulated brain database. NeuroImage 1997, 5, S425. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).