Submitted:
22 October 2025
Posted:
27 October 2025
You are already at the latest version
Abstract
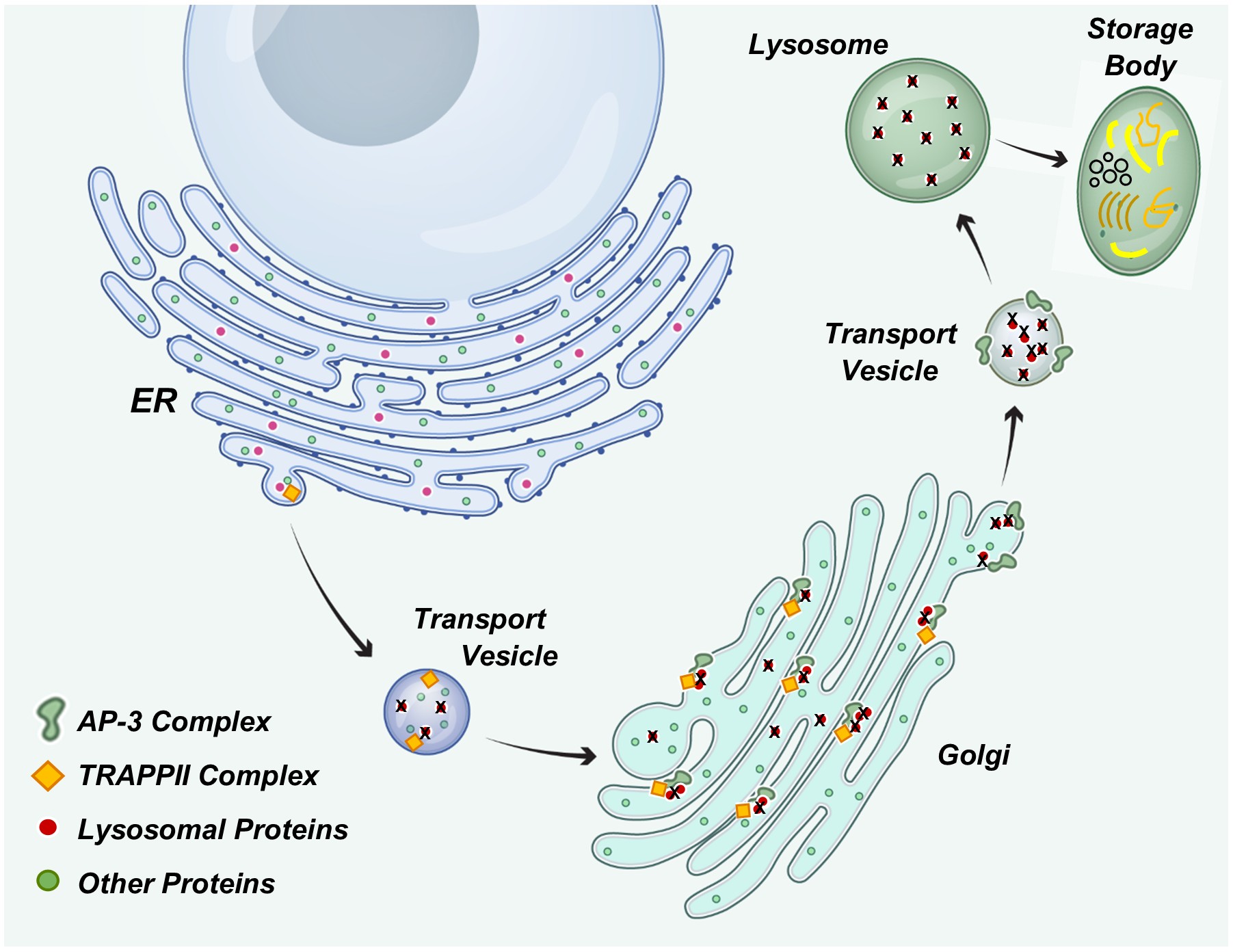
Keywords:
1. Introduction
2. Materials and Methods
2.1. Neurological Examination
2.2. Tissue Collection, Processing, and Microscopic Analyses
2.3. Molecular Genetic Analyses
3. Results
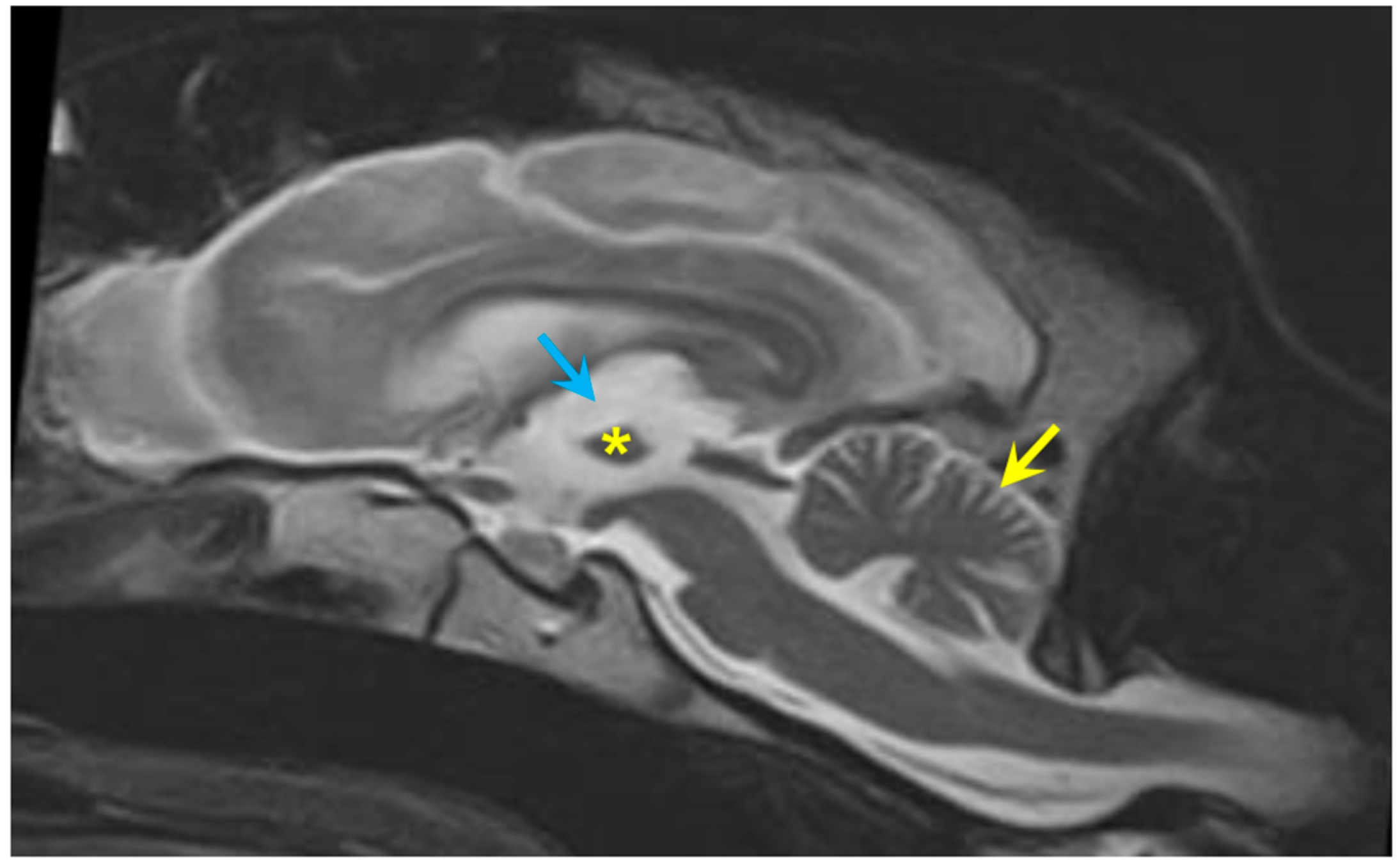
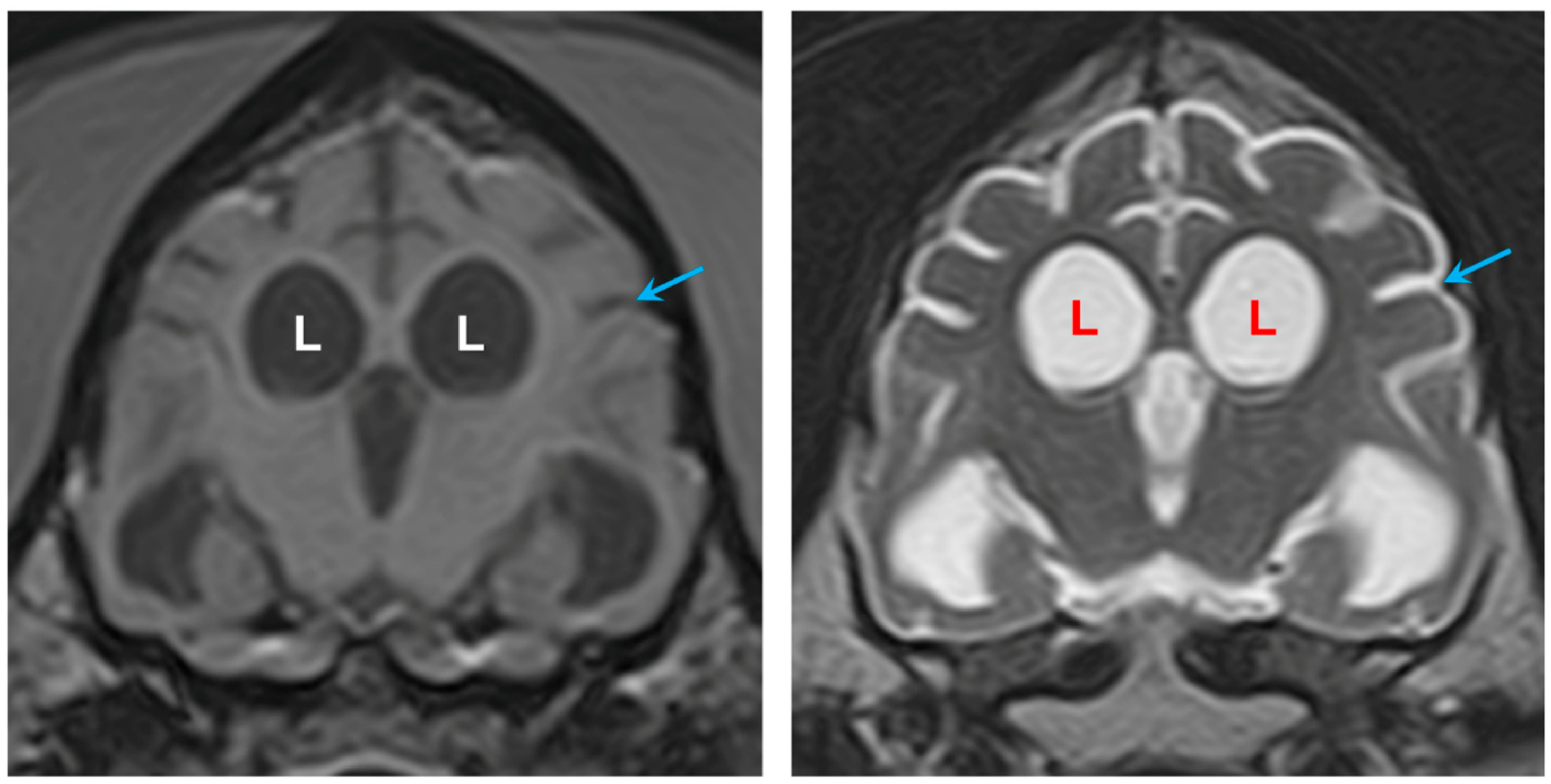
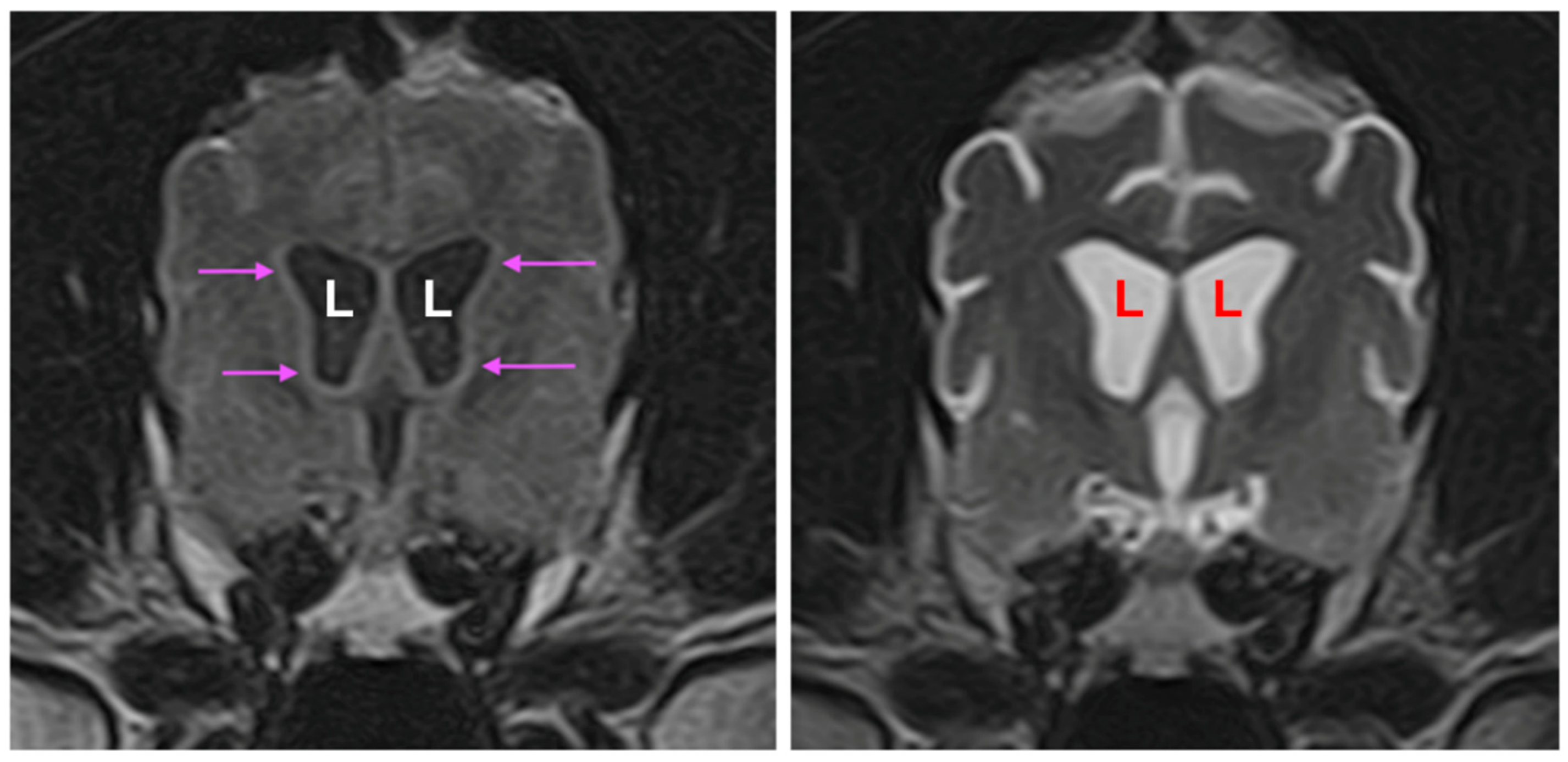
3.1. Disease Phenotype
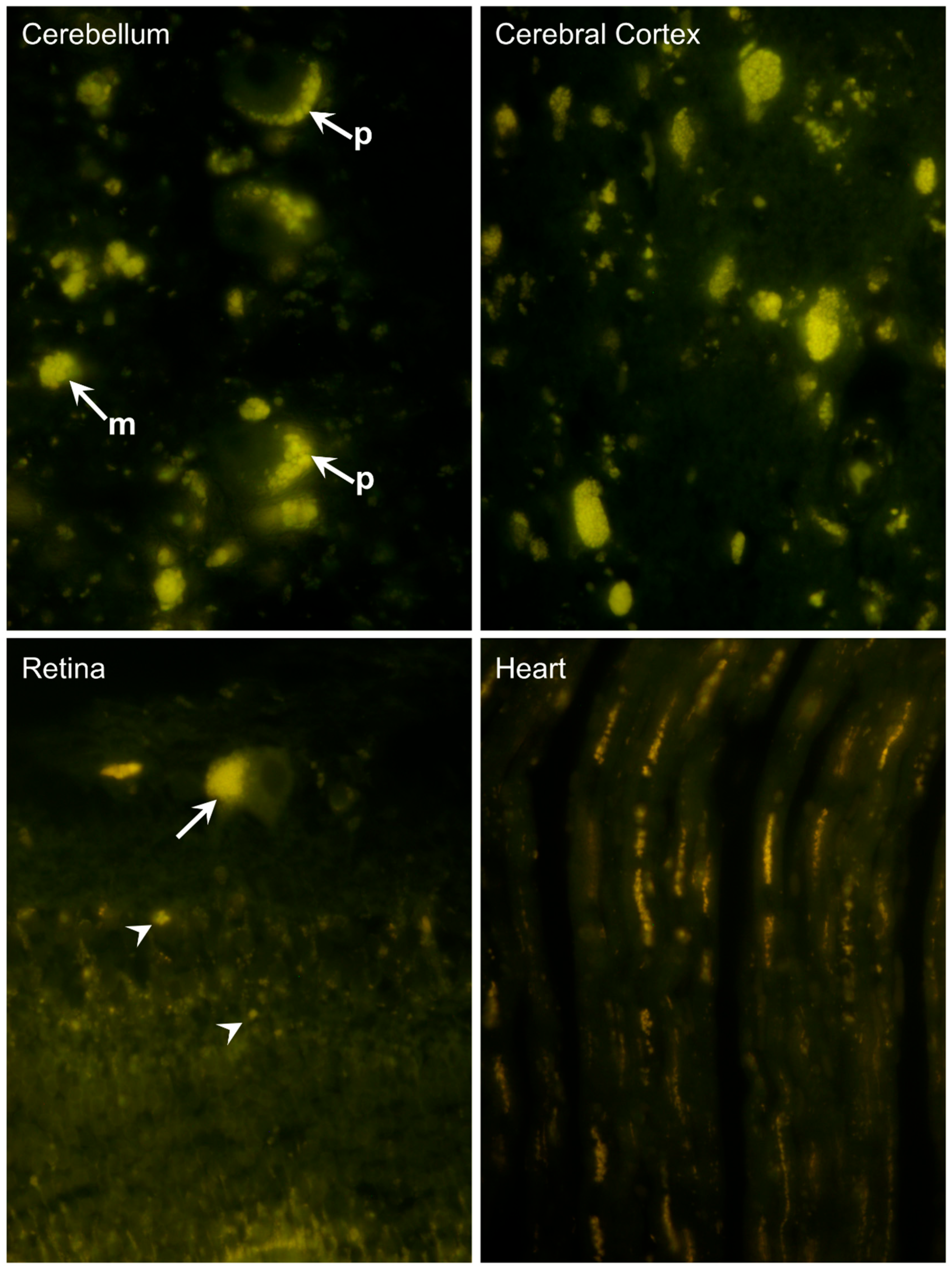
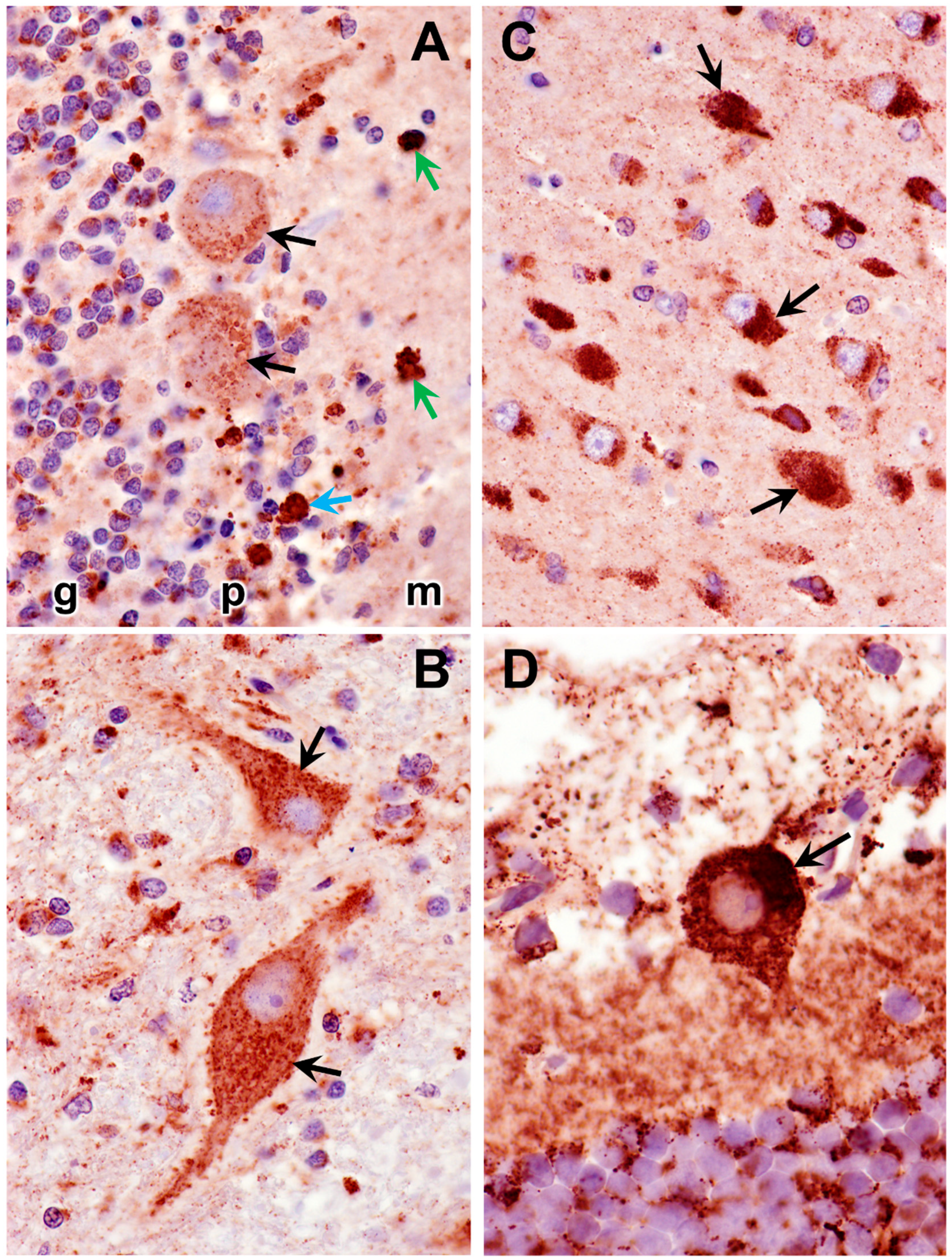
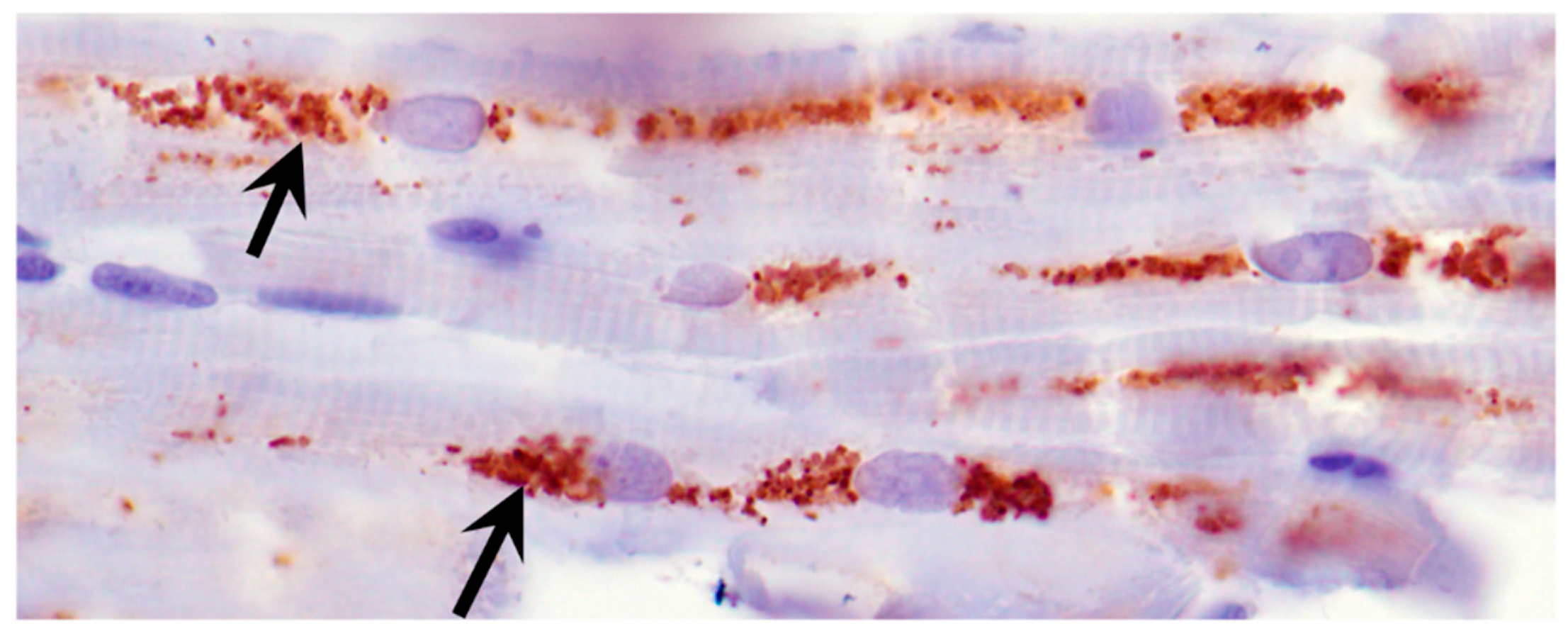
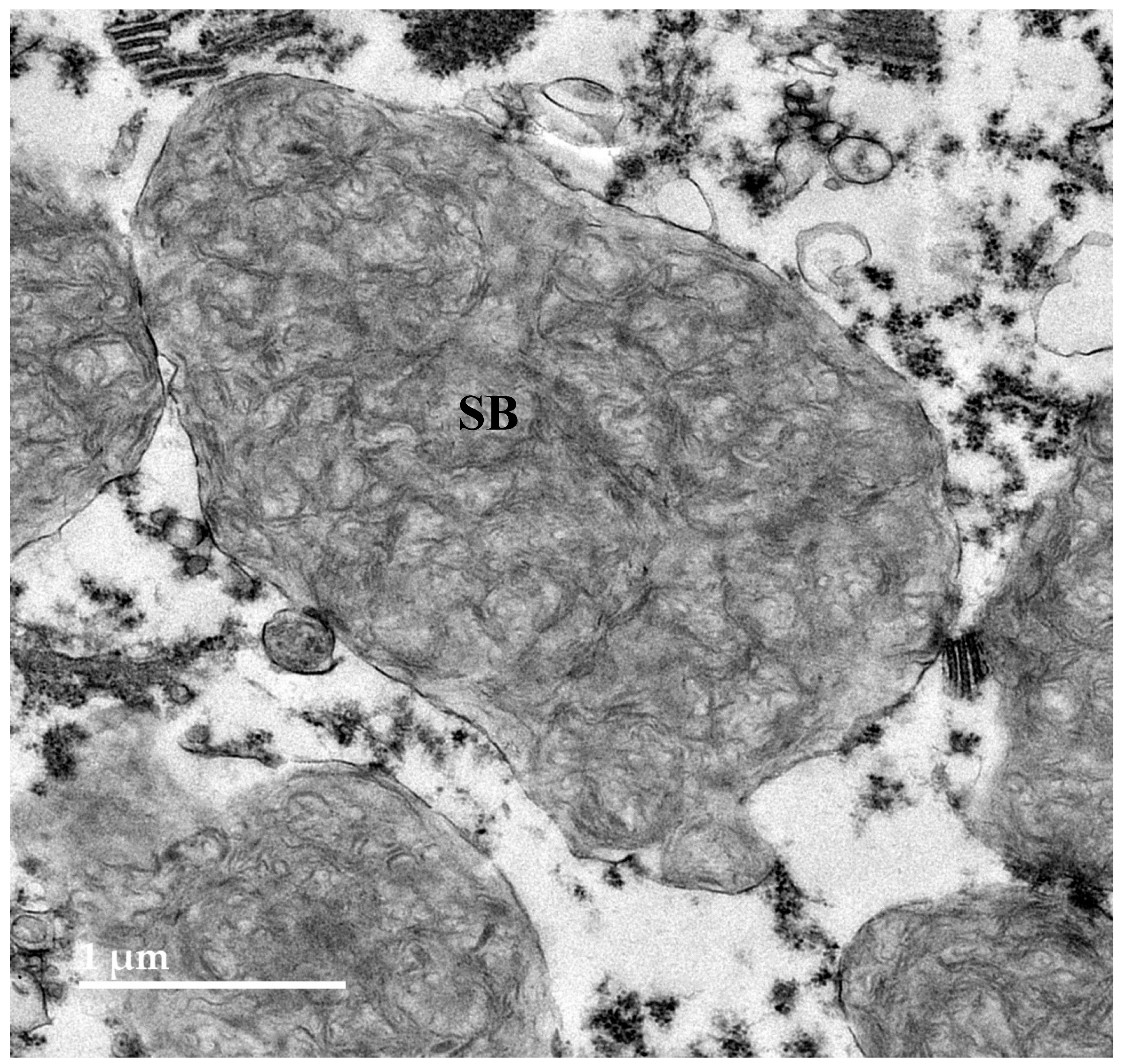
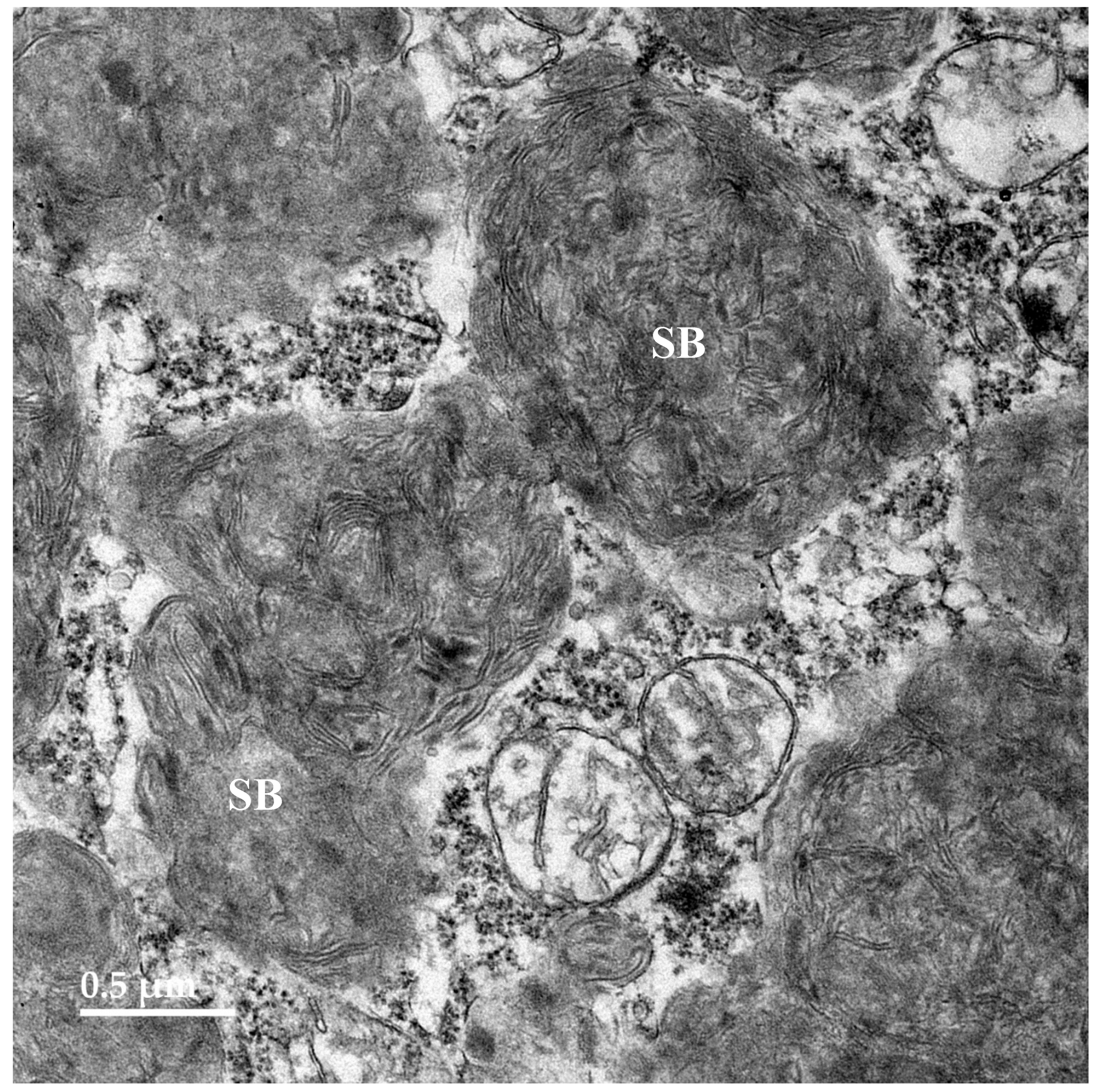
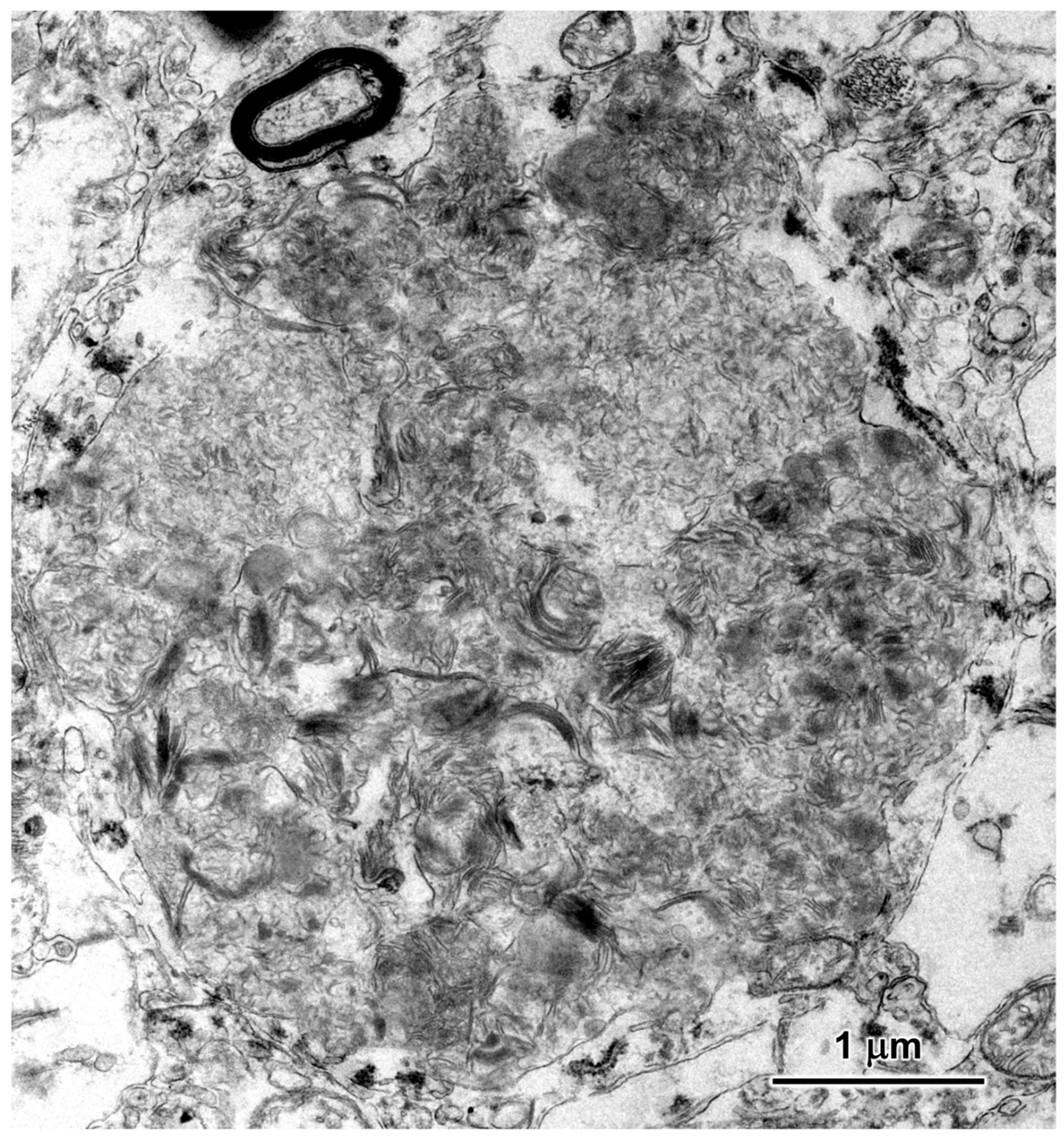
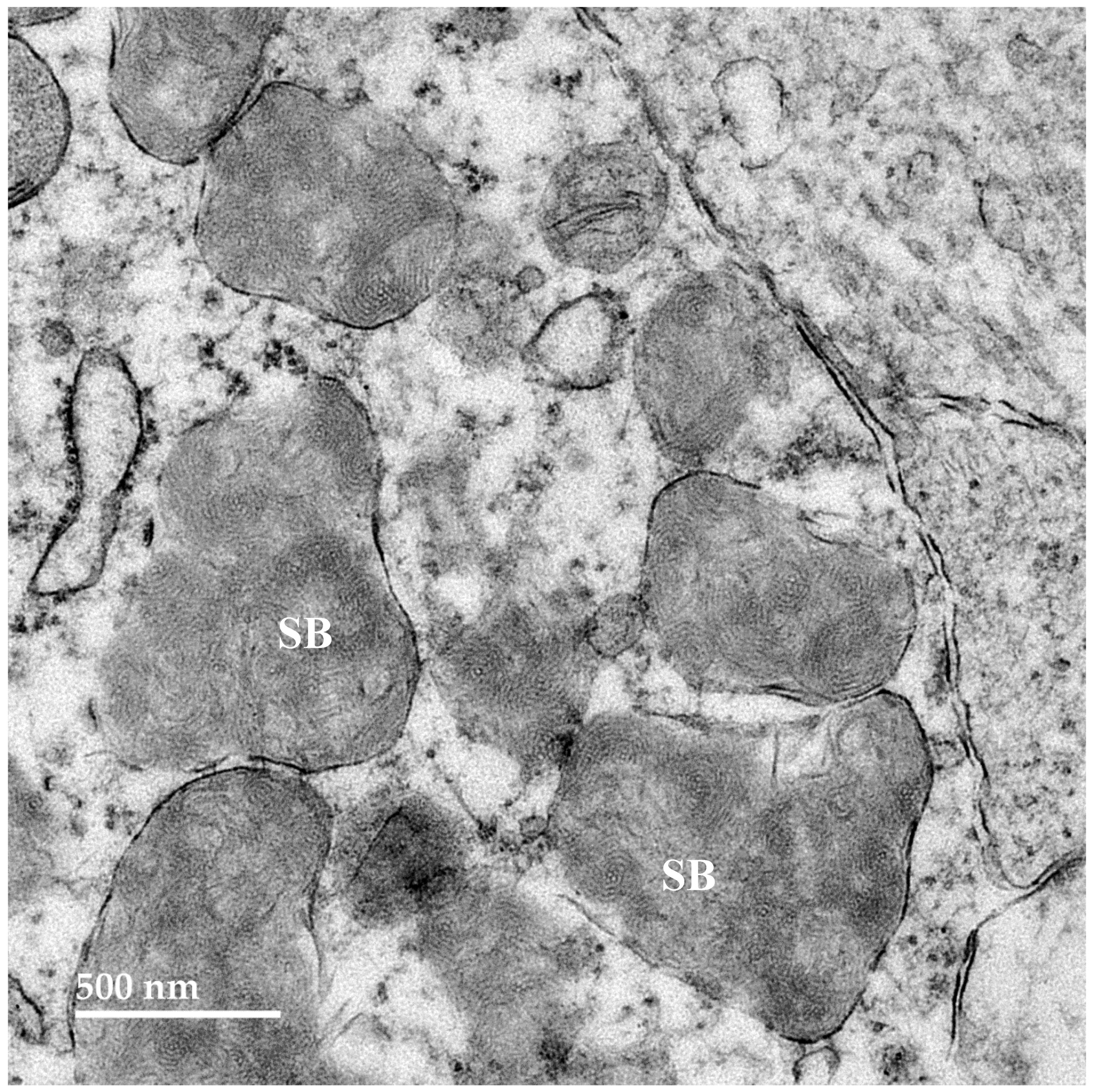
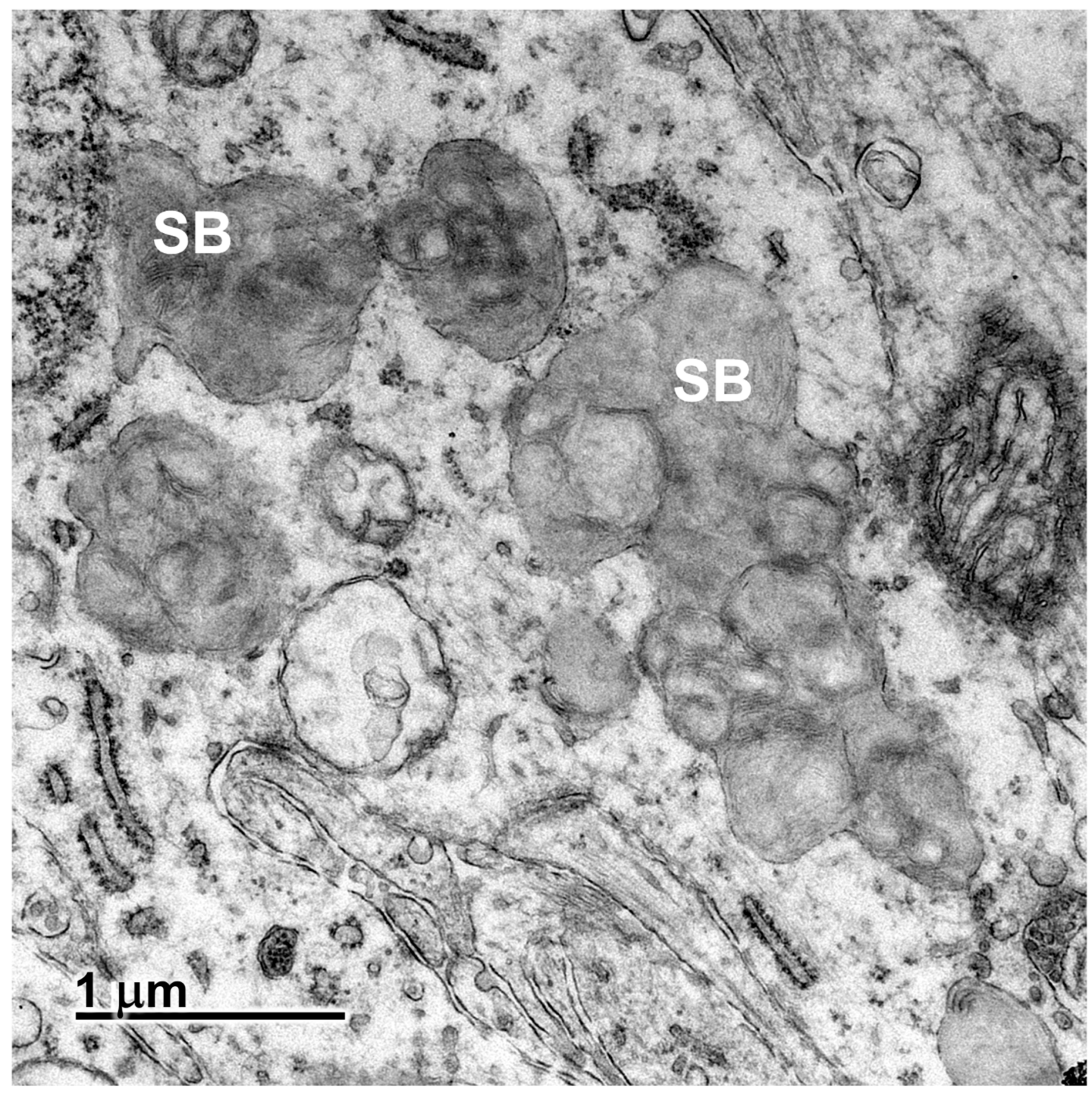
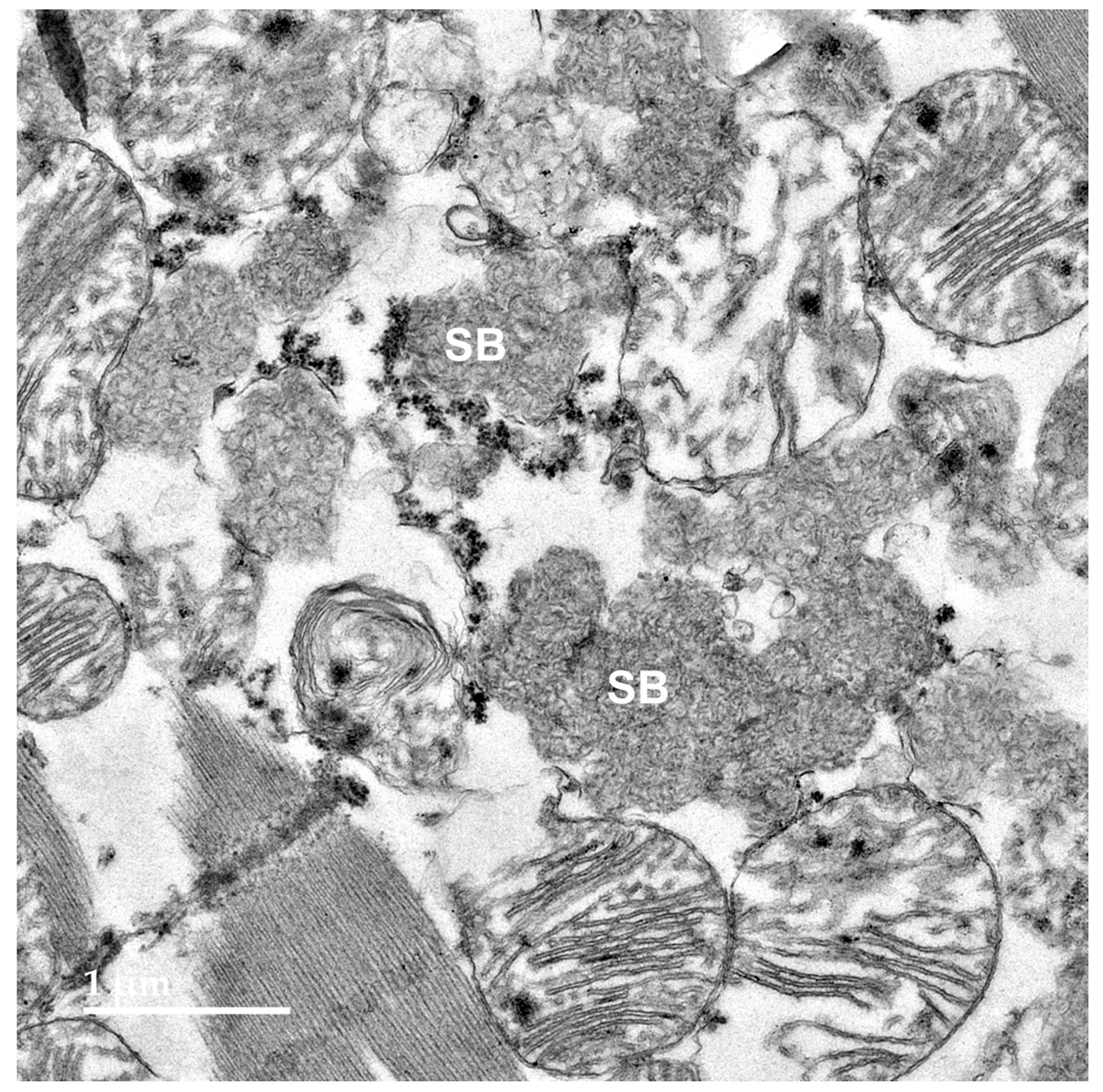
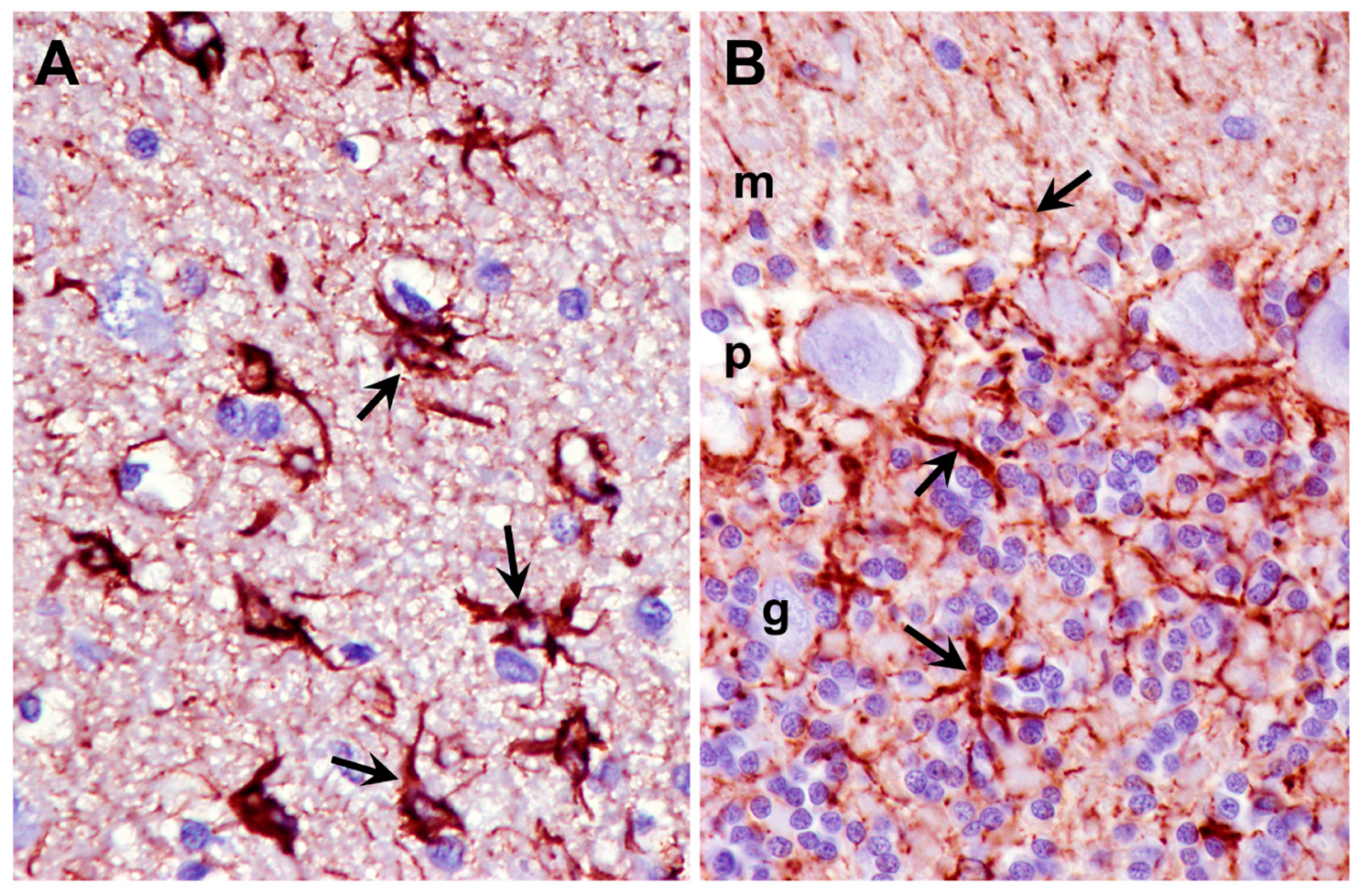
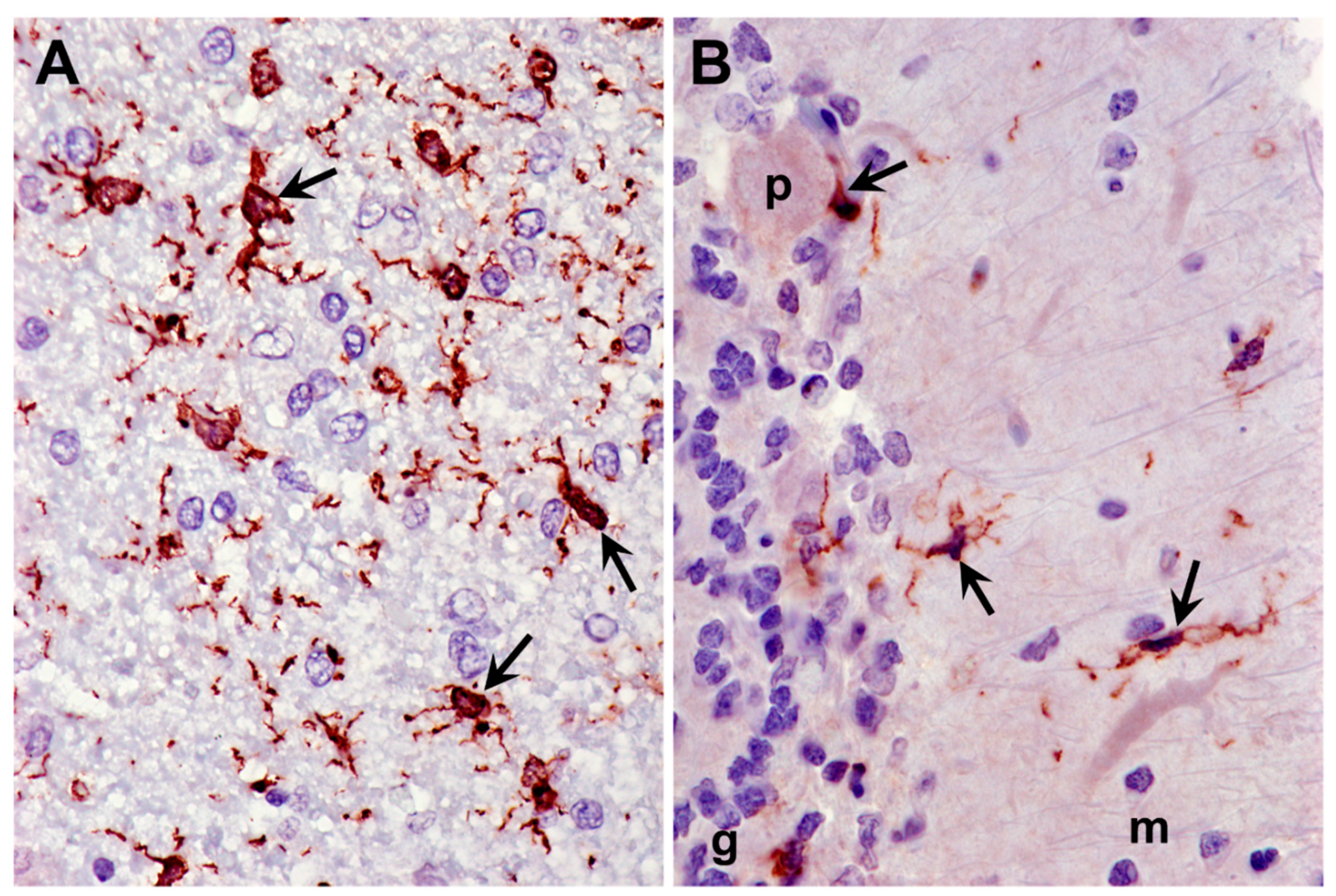
3.2. Microscopic Findings
3.3. Molecular Genetics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PBDG | Petit Bleu de Gascogne |
| NCL | Neuronal ceroid lipofuscinosis |
References
- Katz, M.L.; Rustad, E.; Robinson, G.O.; Whiting, R.E.H.; Student, J.T.; Coates, J.R.; Narfstrom, K. Canine Neuronal Ceroid Lipofuscinoses: Promising Models for Preclinical Testing of Therapeutic Interventions. Neurobiol Dis 2017, 108. [Google Scholar] [CrossRef]
- Mole, S.E.; Williams, R.E.; Goebel, H.H. The Neuronal Ceroid Lipofuscinoses (Batten Disease); Mole, S.E., Willimas, R.E., Goebel, H.H., Eds.; 2nd ed.; Oxford University Press: Oxford, 2011. [Google Scholar]
- Butz, E.S.; Chandrachud, U.; Mole, S.E.; Cotman, S.L. Moving towards a New Era of Genomics in the Neuronal Ceroid Lipofuscinoses. Biochim Biophys Acta Mol Basis Dis 2020, 1866, 165571. [Google Scholar] [CrossRef]
- Mole, S.E.; Anderson, G.; Band, H.A.; Berkovic, S.F.; Cooper, J.D.; Kleine Holthaus, S.-M.; McKay, T.R.; Medina, D.L.; Rahim, A.A.; Schulz, A.; et al. Clinical Challenges and Future Therapeutic Approaches for Neuronal Ceroid Lipofuscinosis. Lancet Neurol 2019, 18, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Kaminiow, K.; Kozak, S.; Paprocka, J. Recent Insight into the Genetic Basis, Clinical Features, and Diagnostic Methods for Neuronal Ceroid Lipofuscinosis. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, B.; Zou, M.; Peng, B.; Rao, Y. Neuronal Ceroid Lipofuscinosis-Concepts, Classification, and Avenues for Therapy. CNS Neurosci Ther 2025, 31, e70261. [Google Scholar] [CrossRef]
- Dowson, J.H. Autofluorescence Emission Spectra of Neuronal Lipopigment in a Case of Adult-Onset Ceroidosis (Kufs’ Disease). Acta Neuropathol 1983, 59, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Dowson, J.H. The Evaluation of Autofluorescence Emission Spectra Derived from Neuronal Lipopigment. J Microsc 1982, 128, 261–270. [Google Scholar] [CrossRef]
- Katz, M.L.; Robison Jr., W. G. What Is Lipofuscin? Defining Characteristics and Differentiation from Other Autofluorescent Lysosomal Storage Bodies. Arch Gerontol Geriatr 2002, 34. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.L.; Robinson Jr., W. G.; Herrmann, R.K.; Groome, A.B.; Bieri, J.G. Lipofuscin Accumulation Resulting from Senescence and Vitamin E Deficiency: Spectral Properties and Tissue Distribution. Mech Ageing Dev 1984, 25. [Google Scholar] [CrossRef]
- Bullock, G.; Johnson, G.S.; Mhlanga-Mutangadura, T.; Petesch, S.C.; Thompson, S.; Goebbels, S.; Katz, M.L. Lysosomal Storage Disease Associated with a CNP Sequence Variant in Dalmatian Dogs. Gene 2022, 830, 146513. [Google Scholar] [CrossRef]
- Katz, M.L.; Cook, J.; Vite, C.H.; Campbell, R.S.; Coghill, L.M.; Lyons, L.A. Beta-Mannosidosis in a Domestic Cat Associated with a Missense Variant in MANBA. Gene 2024, 893, 147941. [Google Scholar] [CrossRef]
- Bullock, G.; Johnson, G.S.; Pattridge, S.G.; Mhlanga-Matangadura, T.; Guo, T.; Cook, J.; Campbell, R.S.; Vite, C.H.; Katz, M.L. A Homozygous MAN2B1 Missense Mutation in a Doberman Pinscher Dog with Neurodegeneration, Cytoplasmic Vacuoles, Autofluorescent Storage Granules and an α-Mannosidase Deficiency. Genes (Basel), In Press. 2023. [Google Scholar]
- Ellezam, B.; Kaseka, M.L.; Nguyen, D.K.; Michaud, J. SCA34 Caused by ELOVL4 L168F Mutation Is a Lysosomal Lipid Storage Disease Sharing Pathology Features with Neuronal Ceroid Lipofuscinosis and Peroxisomal Disorders. Acta Neuropathol 2023, 146, 337–352. [Google Scholar] [CrossRef]
- Poet, M.; Kornak, U.; Schweizer, M.; Zdebik, A.A.; Scheel, O.; Hoelter, S.; Wurst, W.; Schmitt, A.; Fuhrmann, J.C.; Planells-Cases, R.; et al. Lysosomal Storage Disease upon Disruption of the Neuronal Chloride Transport Protein ClC-6. Proc Natl Acad Sci U S A 2006, 103, 13854–13859. [Google Scholar] [CrossRef]
- Beck-Wödl, S.; Harzer, K.; Sturm, M.; Buchert, R.; Rieß, O.; Mennel, H.D.; Latta, E.; Pagenstecher, A.; Keber, U. Homozygous TBC1 Domain-Containing Kinase (TBCK) Mutation Causes a Novel Lysosomal Storage Disease - a New Type of Neuronal Ceroid Lipofuscinosis (CLN15)? Acta Neuropathol Commun 2018, 6, 145. [Google Scholar] [CrossRef]
- Abitbol, M.; Jagannathan, V.; Laurent, N.; Noblet, E.; Dutil, G.F.; Troupel, T.; de Dufaure de Citres, C.; Gache, V.; Blot, S.; Escriou, C.; et al. A PNPLA8 Frameshift Variant in Australian Shepherd Dogs with Hereditary Ataxia. Anim Genet 2022, 53, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, I.; Jagannathan, V.; Bartenschlager, F.; Stein, V.M.; Gruber, A.D.; Leeb, T.; Katz, M.L. ATP13A2 Missense Variant in Australian Cattle Dogs with Late Onset Neuronal Ceroid Lipofuscinosis. Mol Genet Metab 2019, 127, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Rietmann, S.J.; Loderstedt, S.; Matiasek, K.; Kiefer, I.; Jagannathan, V.; Leeb, T. Intragenic Duplication Disrupting the Reading Frame of MFSD8 in Small Swiss Hounds with Neuronal Ceroid Lipofuscinosis. Anim Genet 2024, 55, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, K.K.L.; Skedsmo, F.S.; Hultman, J.; Jansen, J.H.; Lingaas, F. Neuronal Ceroid Lipofuscinosis in a Schapendoes Dog Is Caused by a Missense Variant in CLN6. Anim Genet 2024, 55, 612–620. [Google Scholar] [CrossRef]
- Kolicheski, A.; Johnson, G.S.; Villani, N.A.; O’Brien, D.P.; Mhlanga-Mutangadura, T.; Wenger, D.A.; Mikoloski, K.; Eagleson, J.S.; Taylor, J.F.; Schnabel, R.D.; et al. GM2 Gangliosidosis in Shiba Inu Dogs with an In-Frame Deletion in HEXB. J Vet Intern Med 2017, 31. [Google Scholar] [CrossRef]
- Mansour, T.A.; Woolard, K.D.; Vernau, K.L.; Ancona, D.M.; Thomasy, S.M.; Sebbag, L.; Moore, B.A.; Knipe, M.F.; Seada, H.A.; Cowan, T.M.; et al. Whole Genome Sequencing for Mutation Discovery in a Single Case of Lysosomal Storage Disease (MPS Type 1) in the Dog. Sci Rep 2020, 10, 6558, doi:https://dx.doi.org/10.1038/s41598-020-63451-4 PT - Case Reports, Journal Article, Research Support, Non-U.S. Gov’t.
- Faller, K.M.E.; Ridyard, A.E.; Gutierrez-Quintana, R.; Rupp, A.; Kun-Rodrigues, C.; Orme, T.; Tylee, K.L.; Church, H.J.; Guerreiro, R.; Bras, J. A Deletion of IDUA Exon 10 in a Family of Golden Retriever Dogs with an Attenuated Form of Mucopolysaccharidosis Type I. J Vet Intern Med 2020, 34, 1813–1824. [Google Scholar] [CrossRef]
- Keller, S.H.; Johnson, G.S.; Bullock, G.; Mhlanga-Mutangadura, T.; Schwartz, M.; Pattridge, S.G.; Guo, J.; Kortz, G.D.; Katz, M.L. Homozygous CNP Mutation and Neurodegeneration in Weimaraners: Myelin Abnormalities and Accumulation of Lipofuscin-like Inclusions. Genes (Basel) 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.L.; Khan, S.; Awano, T.; Shahid, S.A.; Siakotos, A.N.; Johnson, G.S. A Mutation in the CLN8 Gene in English Setter Dogs with Neuronal Ceroid-Lipofuscinosis. Biochem Biophys Res Commun 2005, 327. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bullock, G.; O’Brien, D.P.; Johnson, G.S.; Katz, M.L. An RB1CC1 Missense Variant in Nova Scotia Duck Tolling Retrievers with Degenerative Encephalopathy. Genes (Basel) 2025, 16. [Google Scholar] [CrossRef]
- Jaffey, J.A.; Bullock, G.; Guo, J.; Mhlanga-Mutangadura, T.; O’Brien, D.P.; Coates, J.R.; Morrissey, R.; Hutchison, R.; Donnelly, K.S.; Cohn, L.A.; et al. Novel Homozygous ADAMTS2 Variants and Associated Disease Phenotypes in Dogs with Dermatosparactic Ehlers-Danlos Syndrome. Genes (Basel) 2022, 13. [Google Scholar] [CrossRef]
- Kronfeld, D.S.; Donoghue, S.; Glickman, L.T. Body Condition and Energy Intakes of Dogs in a Referral Teaching Hospital. J Nutr 1991, 121, S157–8. [Google Scholar] [CrossRef]
- Laflamme, D.P. Development and Validation of a Body Condition Score System for Dogs. Canine Pract 1997, 22, 10–15. [Google Scholar]
- Sanders, D.N.; Farias, F.H.; Johnson, G.S.; Chiang, V.; Cook, J.R.; O’Brien, D.P.; Hofmann, S.L.; Lu, J.-Y.; Katz, M.L. A Mutation in Canine PPT1 Causes Early Onset Neuronal Ceroid Lipofuscinosis in a Dachshund. Mol Genet Metab 2010, 100. [Google Scholar] [CrossRef]
- Katz, M.L.; Eldred, G.E.; Siakotos, A.N.; Koppang, N. Characterization of Disease-Specific Brain Fluorophores in Ceroid-Lipofuscinosis. Am J Med Genet Suppl 1988, 5. [Google Scholar] [CrossRef] [PubMed]
- Bensaoula, T.; Shibuya, H.; Katz, M.L.; Smith, J.E.; Johnson, G.S.; John, S.K.; Milam, A.H. Histopathologic and Immunocytochemical Analysis of the Retina and Ocular Tissues in Batten Disease. Ophthalmology 2000, 107. [Google Scholar] [CrossRef]
- Gilliam, D.; Kolicheski, A.; Johnson, G.S.; Mhlanga-Mutangadura, T.; Taylor, J.F.; Schnabel, R.D.; Katz, M.L. Golden Retriever Dogs with Neuronal Ceroid Lipofuscinosis Have a Two-Base-Pair Deletion and Frameshift in CLN5. Mol Genet Metab 2015, 115. [Google Scholar] [CrossRef]
- Awano, T.; Katz, M.L.; O’Brien, D.P.; Sohar, I.; Lobel, P.; Coates, J.R.; Khan, S.; Johnson, G.C.; Giger, U.; Johnson, G.S. A Frame Shift Mutation in Canine TPP1 (the Ortholog of Human CLN2) in a Juvenile Dachshund with Neuronal Ceroid Lipofuscinosis. Mol Genet Metab 2006, 89. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.L.; Farias, F.H.; Sanders, D.N.; Zeng, R.; Khan, S.; Johnson, G.S.; O’Brien, D.P. A Missense Mutation in Canine CLN6 in an Australian Shepherd with Neuronal Ceroid Lipofuscinosis. J Biomed Biotechnol 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Johnson, G.S.; Cook, J.; Harris, O.K.; Mhlanga-Mutangadura, T.; Schnabel, R.D.; Jensen, C.A.; Katz, M.L. Neuronal Ceroid Lipofuscinosis in a German Shorthaired Pointer Associated with a Previously Reported CLN8 Nonsense Variant. Mol Genet Metab Rep 2019, 21, 100521. [Google Scholar] [CrossRef]
- Villani, N.A.; Bullock, G.; Michaels, J.R.; Yamato, O.; O’Brien, D.P.; Mhlanga-Mutangadura, T.; Johnson, G.S.; Katz, M.L. A Mixed Breed Dog with Neuronal Ceroid Lipofuscinosis Is Homozygous for a CLN5 Nonsense Mutation Previously Identified in Border Collies and Australian Cattle Dogs. Mol Genet Metab 2019, 127, 107–115. [Google Scholar] [CrossRef]
- Guo, J.; Johnson, G.S.; Brown, H.A.; Provencher, M.L.; da Costa, R.C.; Mhlanga-Mutangadura, T.; Taylor, J.F.; Schnabel, R.D.; O’Brien, D.P.; Katz, M.L. A CLN8 Nonsense Mutation in the Whole Genome Sequence of a Mixed Breed Dog with Neuronal eroid Lipofuscinosis and Australian Shepherd Ancestry. Mol Genet Metab 2014, 112. [Google Scholar] [CrossRef]
- Guo, J.; O’Brien, D.P.; Mhlanga-Mutangadura, T.; Olby, N.J.; Taylor, J.F.; Schnabel, R.D.; Katz, M.L.; Johnson, G.S. A Rare Homozygous MFSD8 Single-Base-Pair Deletion and Frameshift in the Whole Genome Sequence of a Chinese Crested Dog with Neuronal Ceroid Lipofuscinosis. BMC Vet Res 2015, 10, 960. [Google Scholar] [CrossRef]
- Tieze, S.M.; Esqueda, A.; McAllister, R.; Lagator, M.; Yucel, B.; Sun, E.; Lam, T.T.; Lockyer, N.; Gupta, K.; Chandra, S.S. Molecular Elucidation of Brain Lipofuscin in Aging and Neuronal Ceroid Lipofuscinosis. Res Sq 2025. [CrossRef]
- Corcuera-Delgado, C.T.; Ramirez-Ristori, A.G.; Perez-Munoz, E.; Mendizabal-Rodriguez, M.E.; Villarroel, C.E. Clinical, Pathological, and Molecular Findings in a Mexican Patient With Neuronal Ceroid Lipofuscinosis Type 2: Support for Pathogenicity of the c.1226 G>T Variant and for Presence of Cherry-Red Spot in This Disease. Pediatr Dev Pathol 2025, 28, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Tomiyasu, H.; Takahashi, W.; Ohta, T.; Yoshii, F.; Shibuya, M.; Shinohara, Y. [An Autopsy Case of Juvenile Neuronal Ceroid-Lipofuscinosis with Dilated Cardiomyopathy]. Rinsho Shinkeigaku 2000, 40, 350–357. [Google Scholar]
- Katz, M.L.; Christianson, J.S.; Norbury, N.E.; Gao, C.-L.; Siakotos, A.N.; Koppang, N. Lysine Methylation of Mitochondrial ATP Synthase Subunit c Stored in Tissues of Dogs with Hereditary Ceroid Lipofuscinosis. Journal of Biological Chemistry 1994, 269. [Google Scholar] [CrossRef]
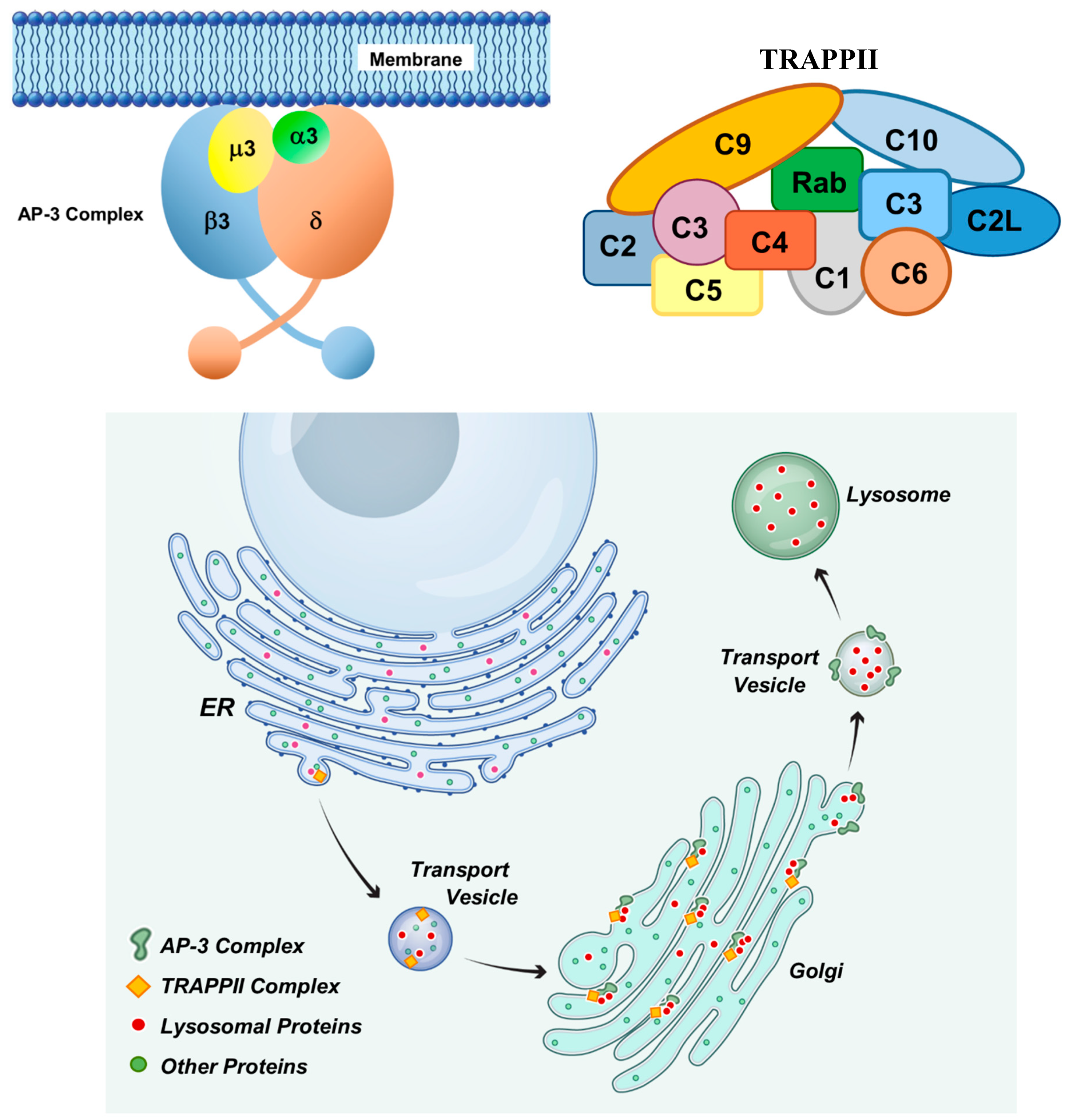
- Simpson, F.; Peden, A.A.; Christopoulou, L.; Robinson, M.S. Characterization of the Adaptor-Related Protein Complex, AP-3. J Cell Biol 1997, 137, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Islam, M.N.; Xu, T.; Song, E. AP-3 Adaptor Complex-Mediated Vesicle Trafficking. Biophys Rep 2021, 7, 91–100. [Google Scholar]
- Odorizzi, G.; Cowles, C.R.; Emr, S.D. The AP-3 Complex: A Coat of Many Colours. Trends Cell Biol 1998, 8, 282–288. [Google Scholar] [CrossRef]
- Yip, C.K.; Berscheminski, J.; Walz, T. Molecular Architecture of the TRAPPII Complex and Implications for Vesicle Tethering. Nat Struct Mol Biol 2010, 17, 1298–1304. [Google Scholar] [CrossRef]
- Hall, R.; Sawant, V.; Gu, J.; Sikora, T.; Rollo, B.; Velasco, S.; Kim, J.; Segev, N.; Christodoulou, J.; Van Bergen, N.J. TRAPPopathies: Severe Multisystem Disorders Caused by Variants in Genes of the Transport Protein Particle (TRAPP) Complexes. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef]
- Mbimba, T.; Hussein, N.J.; Najeed, A.; Safadi, F.F. TRAPPC9: Novel Insights into Its Trafficking and Signaling Pathways in Health and Disease (Review). Int J Mol Med 2018, 42, 2991–2997. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Traub, L.M. Signals for Sorting of Transmembrane Proteins to Endosomes and Lysosomes. Annu Rev Biochem 2003, 72, 395–447. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, F.; Ohno, H. Adaptor Protein Complexes as the Key Regulators of Protein Sorting in the Post-Golgi Network. Cell Struct Funct 2003, 28, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Z.A.; Gleeson, P.A. Cargo Sorting at the Trans-Golgi Network for Shunting into Specific Transport Routes: Role of Arf Small G Proteins and Adaptor Complexes. Cells 2019, 8. [Google Scholar] [CrossRef]
- Barrowman, J.; Bhandari, D.; Reinisch, K.; Ferro-Novick, S. TRAPP Complexes in Membrane Traffic: Convergence through a Common Rab. Nat Rev Mol Cell Biol 2010, 11, 759–763. [Google Scholar] [CrossRef]
- Palmer, D.N.; Fearnley, I.M.; Walker, J.E.; Hall, N.A.; Lake, B.D.; Wolfe, L.S.; Haltia, M.; Martinus, R.D.; Jolly, R.D. Mitochondrial ATP Synthase Subunit c Storage in the Ceroid-Lipofuscinoses (Batten Disease). Am J Med Genet 1992, 42, 561–567. [Google Scholar] [CrossRef]
- Palmer, D.N. The Relevance of the Storage of Subunit c of ATP Synthase in Different Forms and Models of Batten Disease (NCLs). Biochim Biophys Acta Mol Basis Dis 2015, 1852, 2287–2291. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, I.M.; Walker, J.E.; Martinus, R.D.; Jolly, R.D.; Kirkland, K.B.; Shaw, G.J.; Palmer, D.N. The Sequence of the Major Protein Stored in Ovine Ceroid Lipofuscinosis Is Identical with That of the Dicyclohexylcarbodiimide-Reactive Proteolipid of Mitochondrial ATP Synthase. Biochem J 1990, 268, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Aslanger, A.D.; Goncu, B.; Duzenli, O.F.; Yucesan, E.; Sengenc, E.; Yesil, G. Biallelic Loss of TRAPPC9 Function Links Vesicle Trafficking Pathway to Autosomal Recessive Intellectual Disability. J Hum Genet 2022, 67, 279–284. [Google Scholar] [CrossRef]
- Kramer, J.; Beer, M.; Bode, H.; Winter, B. Two Novel Compound Heterozygous Mutations in the TRAPPC9 Gene Reveal a Connection of Non-Syndromic Intellectual Disability and Autism Spectrum Disorder. Front Genet 2020, 11, 972. [Google Scholar] [CrossRef] [PubMed]
- Bolat, H.; Unsel-Bolat, G.; Derin, H.; Sen, A.; Ceylaner, S. Distinct Autism Spectrum Disorder Phenotype and Hand-Flapping Stereotypes: Two Siblings with Novel Homozygous Mutation in TRAPPC9 Gene and Literature Review. Mol Syndromol 2022, 13, 263–269. [Google Scholar] [CrossRef]
- Uctepe, E.; Yesilyurt, A.; Esen, F.N.; Tumer, S.; Mancilar, H.; Sonmez, F.M. TRAPPC9-Related Intellectual Disability: Report of Two New Cases and Review of the Literature. Mol Syndromol 2023, 14, 485–492. [Google Scholar] [CrossRef]
- Kharrat, M.; Triki, C.; Ben Isaa, A.; Bouchaala, W.; Alila, O.; Chouchen, J.; Ghouliya, Y.; Kamoun, F.; Tlili, A.; Fakhfakh, F. Expanding the Genetic and Phenotypic Spectrum of TRAPPC9 and MID2-Related Neurodevelopmental Disabilities: Report of Two Novel Mutations, 3D-Modelling, and Molecular Docking Studies. J Hum Genet 2024, 69, 291–299. [Google Scholar] [CrossRef]
- Yu, B.; Chen, J.; Yang, S.; Wang, H.; Xiao, Y.; Liu, S. Case Report: Whole Exome Sequencing Identifies Compound Heterozygous Variants in the TRAPPC9 Gene in a Child with Developmental Delay. Front Genet 2024, 15, 1415194. [Google Scholar] [CrossRef]
- Feng, L.; Novak, E.K.; Hartnell, L.M.; Bonifacino, J.S.; Collinson, L.M.; Swank, R.T. The Hermansky-Pudlak Syndrome 1 (HPS1) and HPS2 Genes Independently Contribute to the Production and Function of Platelet Dense Granules, Melanosomes, and Lysosomes. Blood 2002, 99, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Wenham, M.; Grieve, S.; Cummins, M.; Jones, M.L.; Booth, S.; Kilner, R.; Ancliff, P.J.; Griffiths, G.M.; Mumford, A.D. Two Patients with Hermansky Pudlak Syndrome Type 2 and Novel Mutations in AP3B1. Haematologica 2010, 95, 333–337. [Google Scholar] [CrossRef]
- Alcidi, R.; Campanella, T.; Curcio, R.; Chiatti, L.; Arrivi, A.; Ferranti, L.; Carreras, G.; Barabani, M.; Pucci, G. A 29-Year-Old Man With Type 2 Hermansky-Pudlak Syndrome and Wolff-Parkinson-White Syndrome: The Hypothesis of a Potential Link Between These Two Conditions. Case Rep Med 2025, 2025, 5525411. [Google Scholar] [CrossRef]
- Alasmari, B.G.; Wafa, S.; Tahir, A.M.; Aljubran, A.; Alfaifi, A.; Alsaab, K.; Elzubair, L. Hermansky-Pudlak Syndrome Type 2: A Case Report on an Ultra-Rare Disorder. Cureus 2024, 16, e65114. [Google Scholar] [CrossRef]
- Pierson, D.M.; Ionescu, D.; Qing, G.; Yonan, A.M.; Parkinson, K.; Colby, T.C.; Leslie, K. Pulmonary Fibrosis in Hermansky-Pudlak Syndrome. a Case Report and Review. Respiration 2006, 73, 382–395. [Google Scholar] [CrossRef]
- Huizing, M.; Anikster, Y.; Gahl, W.A. Hermansky-Pudlak Syndrome and Chediak-Higashi Syndrome: Disorders of Vesicle Formation and Trafficking. Thromb Haemost 2001, 86, 233–245. [Google Scholar] [CrossRef]
- Calcagni’, A.; Staiano, L.; Zampelli, N.; Minopoli, N.; Herz, N.J.; Di Tullio, G.; Huynh, T.; Monfregola, J.; Esposito, A.; Cirillo, C.; et al. Loss of the Batten Disease Protein CLN3 Leads to Mis-Trafficking of M6PR and Defective Autophagic-Lysosomal Reformation. Nat Commun 2023, 14, 3911. [Google Scholar] [CrossRef] [PubMed]
- di Ronza, A.; Bajaj, L.; Sharma, J.; Sanagasetti, D.; Lotfi, P.; Adamski, C.J.; Collette, J.; Palmieri, M.; Amawi, A.; Popp, L.; et al. CLN8 Is an Endoplasmic Reticulum Cargo Receptor That Regulates Lysosome Biogenesis. Nat Cell Biol 2018, 20, 1370–1377. [Google Scholar] [CrossRef]
- Bajaj, L.; Sharma, J.; di Ronza, A.; Zhang, P.; Eblimit, A.; Pal, R.; Roman, D.; Collette, J.R.; Booth, C.; Chang, K.T.; et al. A CLN6-CLN8 Complex Recruits Lysosomal Enzymes at the ER for Golgi Transfer. J Clin Invest 2020, 130, 4118–4132. [Google Scholar] [CrossRef] [PubMed]
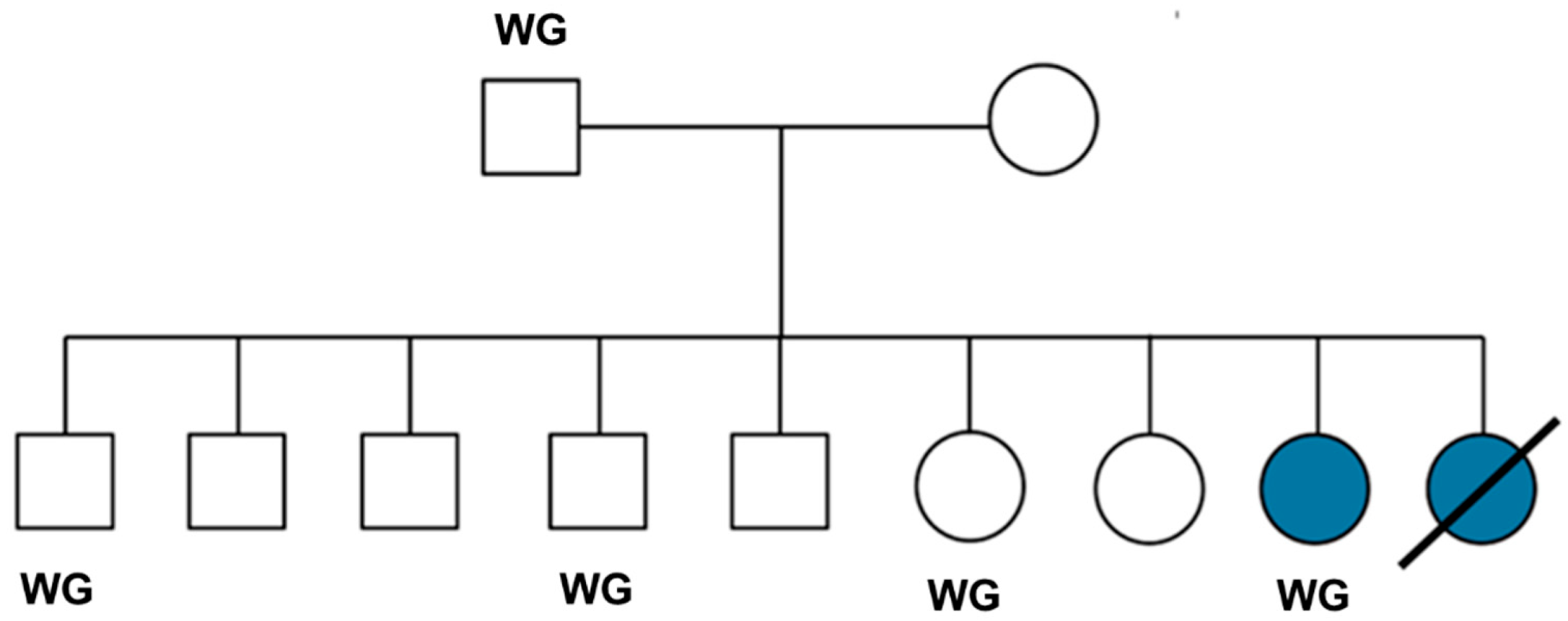

| Filtering Steps | Number of Variants |
| Proband was homozygous relative to the Dog10K_Boxer_Tasha reference | 20,399 |
| Proband was the only homozygous PBDG within its family | 2,607 |
| Sire was heterozygous | 2,403 |
| Proband was the only homozygote among 387 other dogs* | 41 |
| Proband uniquely homozygous and no heterozygotes in control cohort** | 10 |
| Chr. | Position1 | Ref2 | Alt4 | Effect | AA Change | Gene ID |
| 2 | 70241214 | T | C | Missense | C340G | ZNF683 |
| 2 | 70429672 | A | AC | 3’UTR | - | PDIK1L |
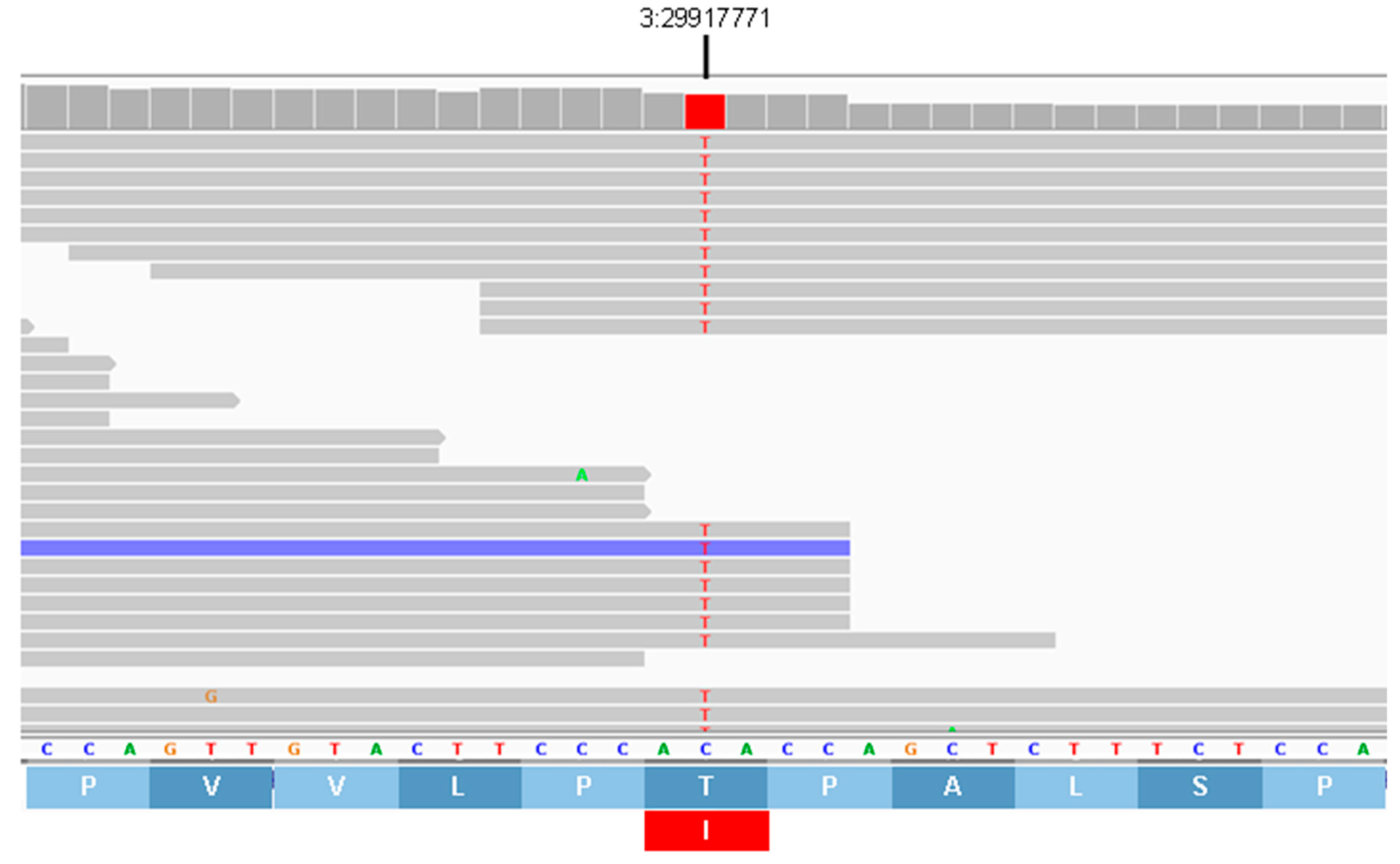
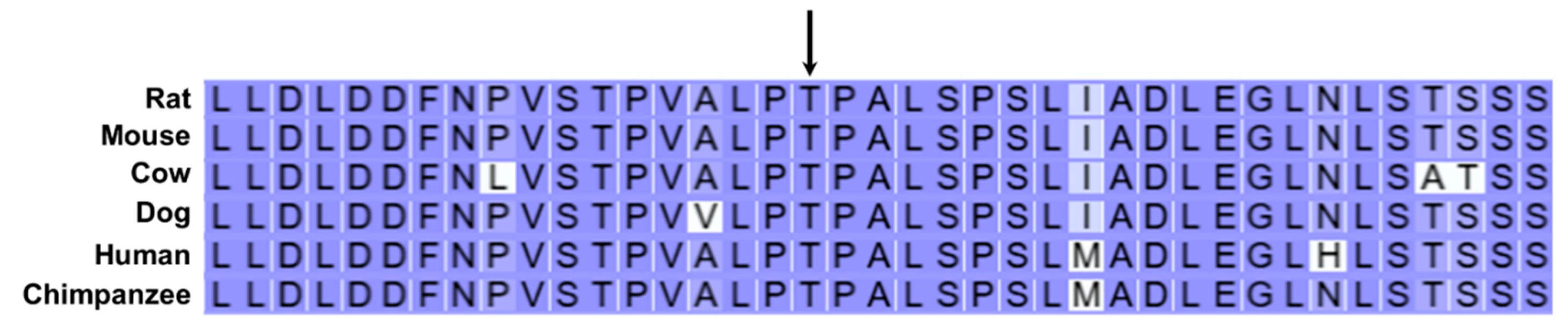
| 3 | 29917771 | C | T | Missense | T832I | AP3B1 |
| 5 | 61470177 | G | A | Stop Gained | W373* | PER3 |
| 5 | 61564260 | A | T | Missense | F190I | TNFRSF9 |
| 8 | 67681637 | C | T | 5’UTR | - | EML1 |
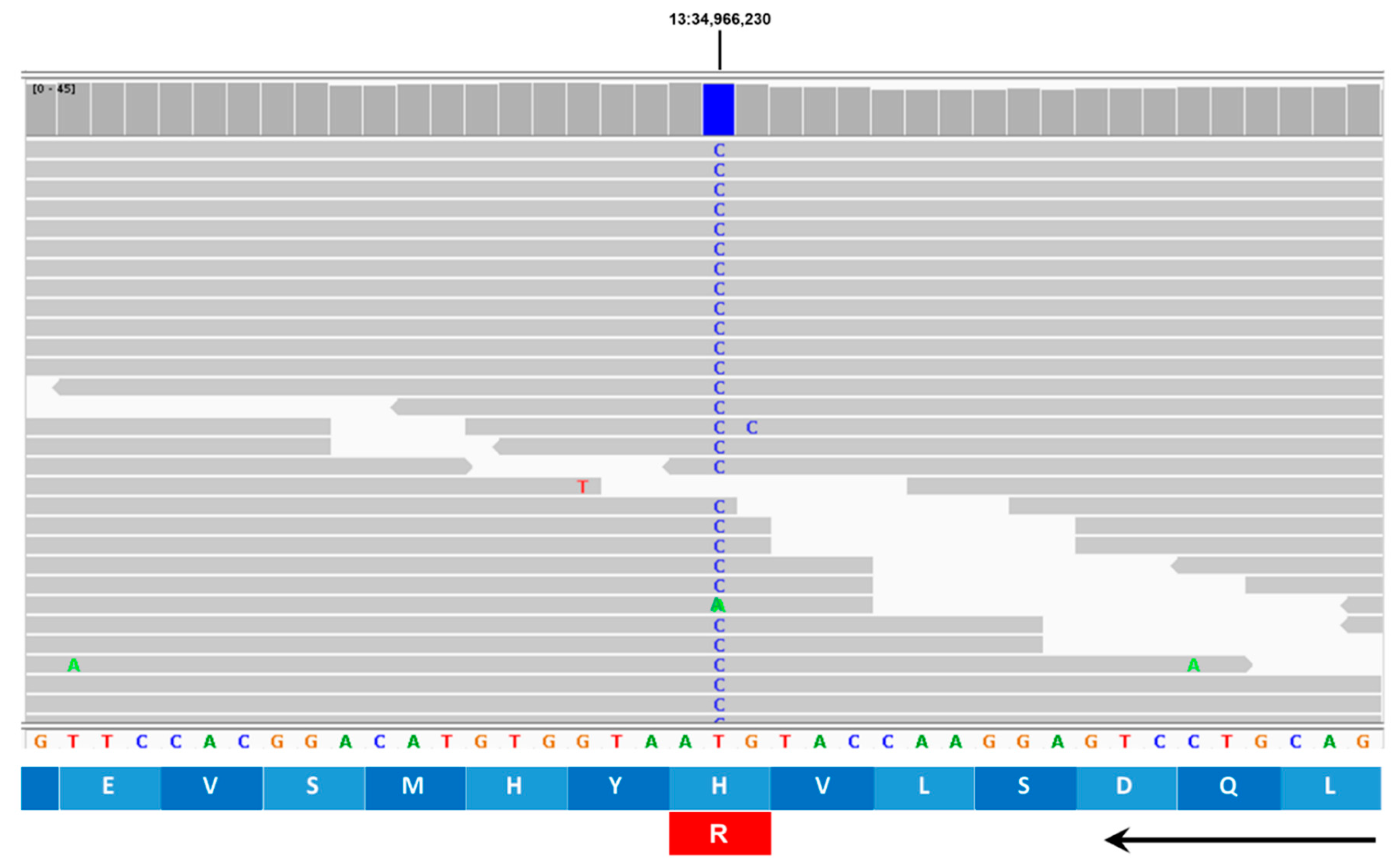
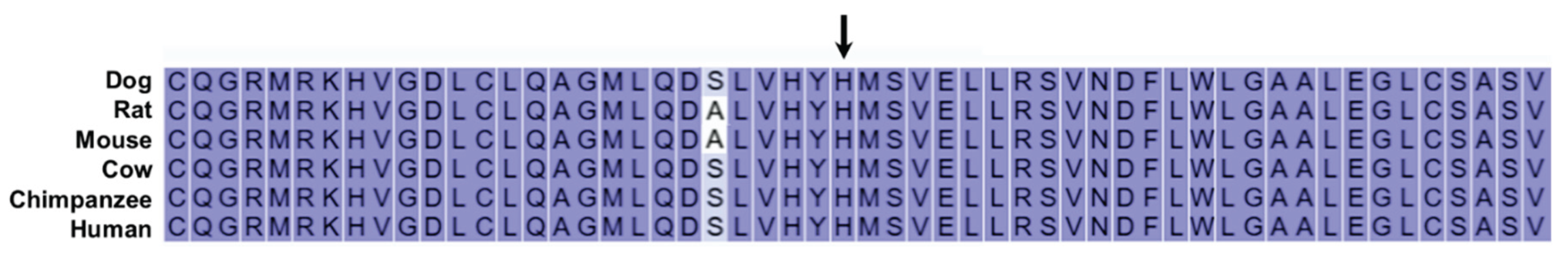
| 13 | 34966230 | T | C | Missense | H225R | TRAPPC9 |
| 13 | 37186500 | G | A | Missense | R870C | MROH6 |
| 27 | 5344292 | CTCTCCG | C | 5’UTR | - | PRMT8 |
| 27 | 6171476 | CG | C | 3’UTR | - | FGF23 |
| Number* | ||
| Genotype** | AP3B1 | TRAPPC9 |
| V/V | 3# | 1## |
| R/V | 17 | 14 |
| R/R | 21 | 26 |
| Heterozygous For: | Number* |
| AP3B1 Only | 12 |
| TRAPPC9 Only | 9 |
| Both | 5** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





