Submitted:
04 February 2025
Posted:
05 February 2025
You are already at the latest version
Abstract
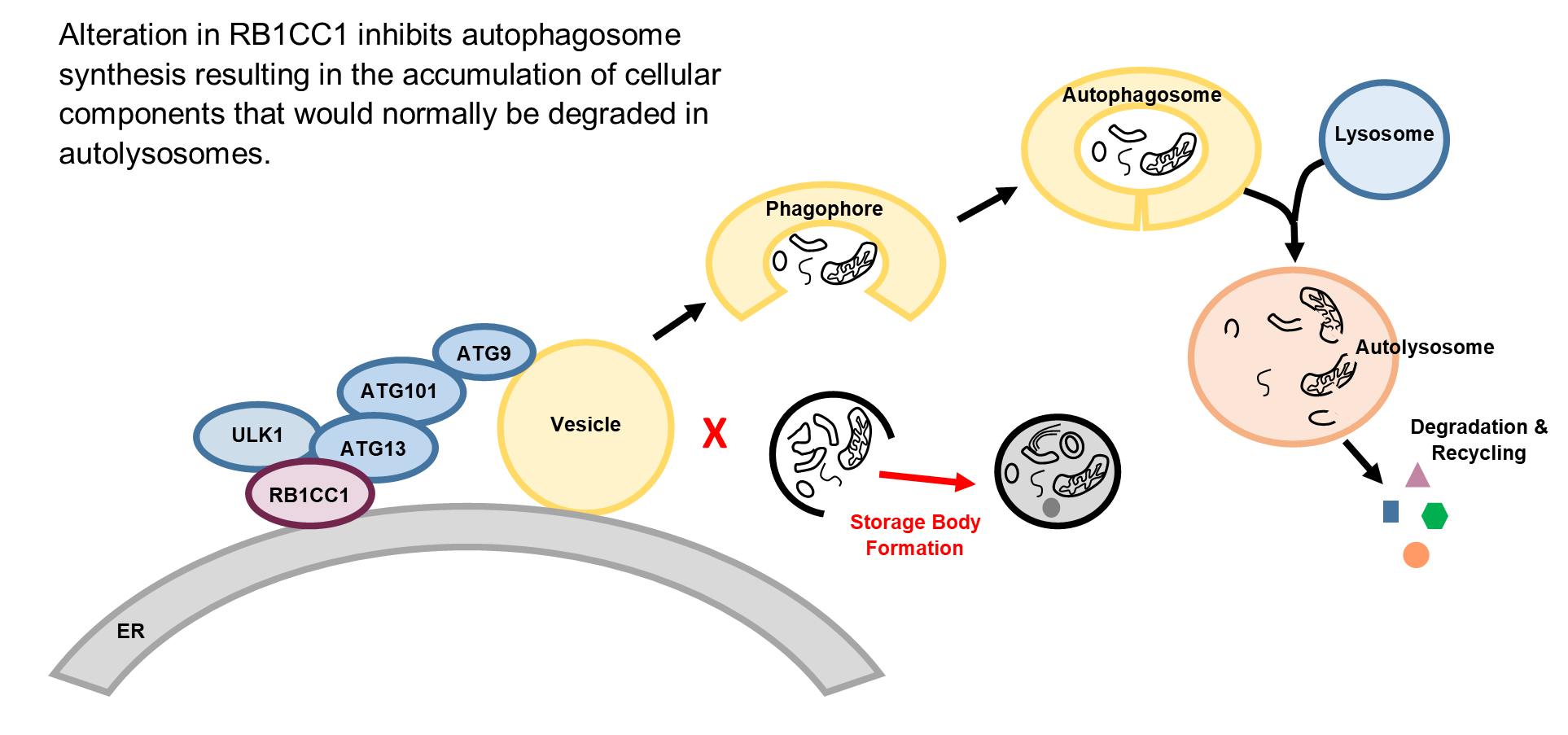
Keywords:
1. Introduction
2. Materials and Methods
2.1. Identification of Cases and Controls
2.2. Molecular Genetic Analyses
2.3. Electron Microscopy
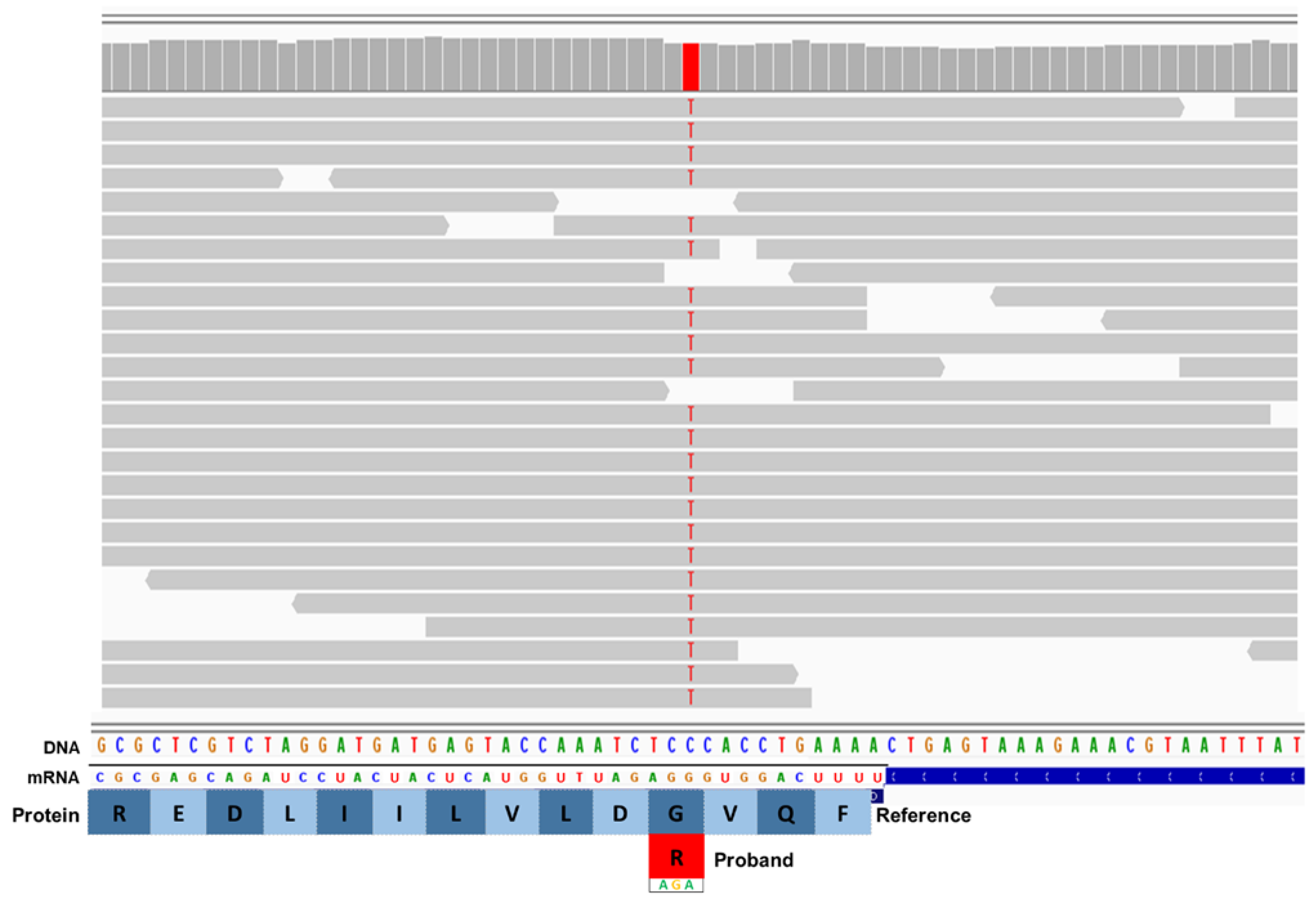
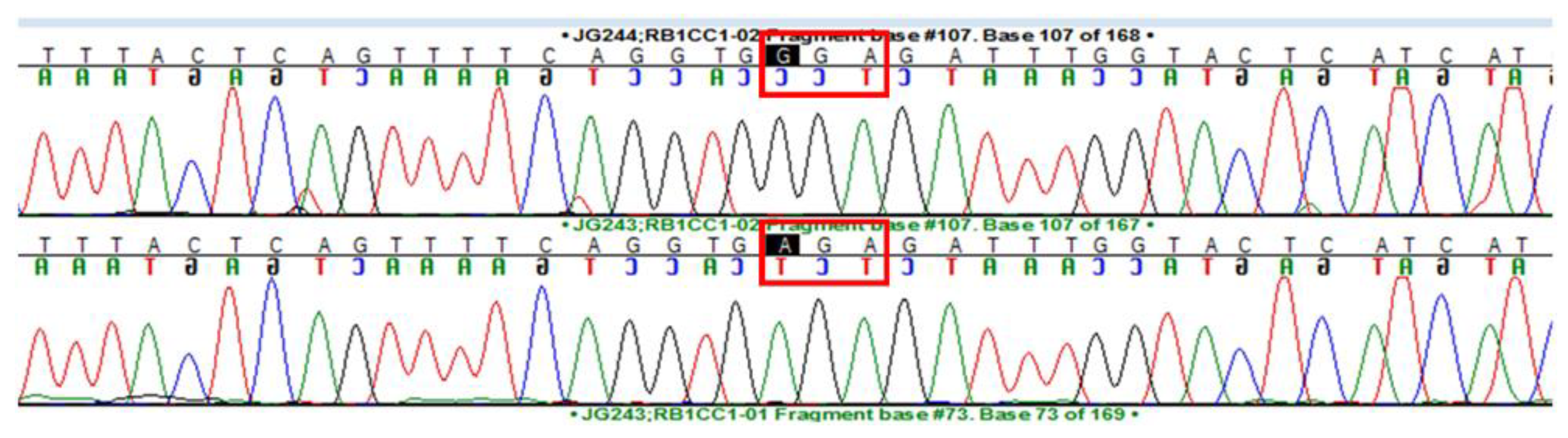
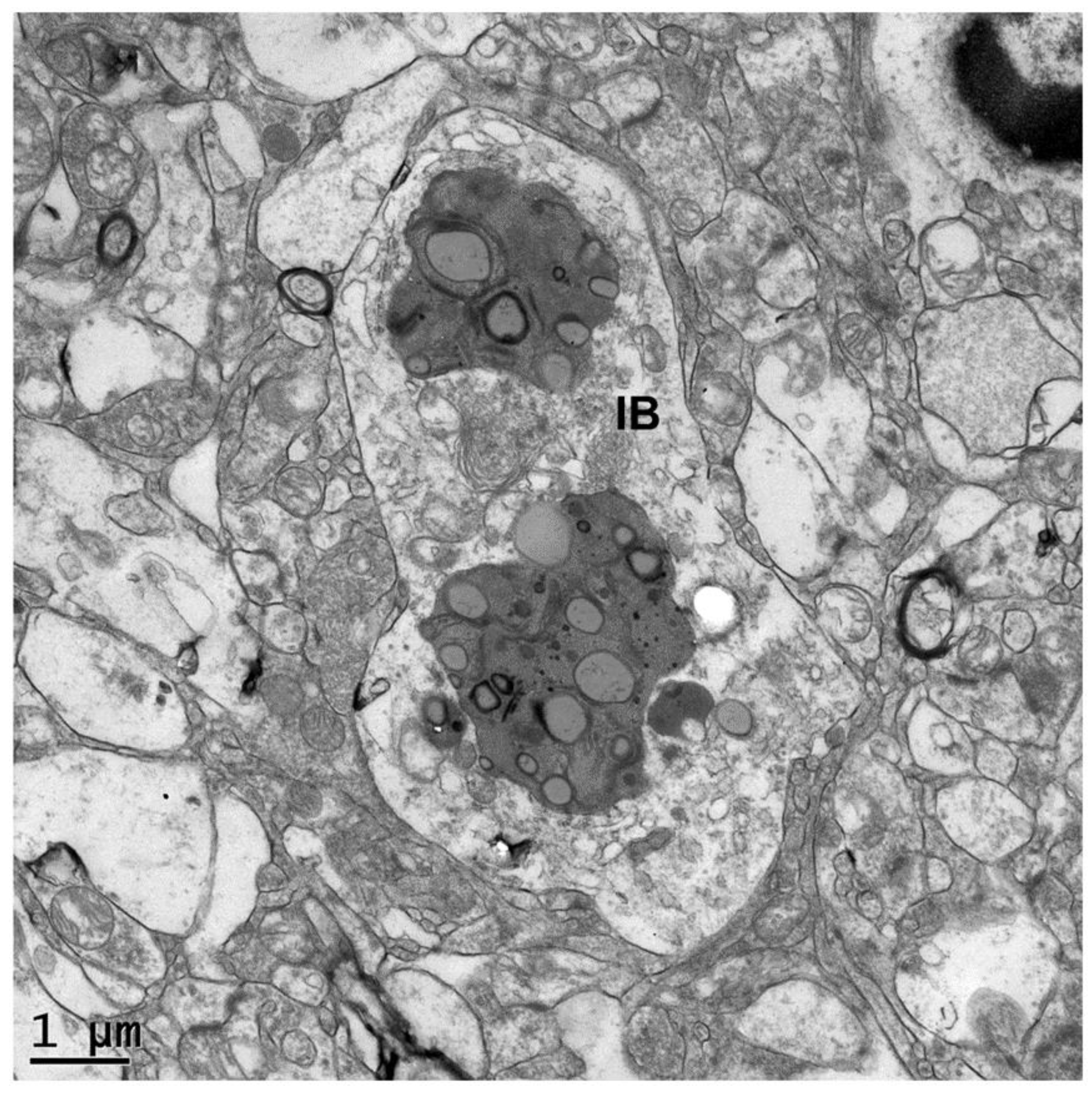
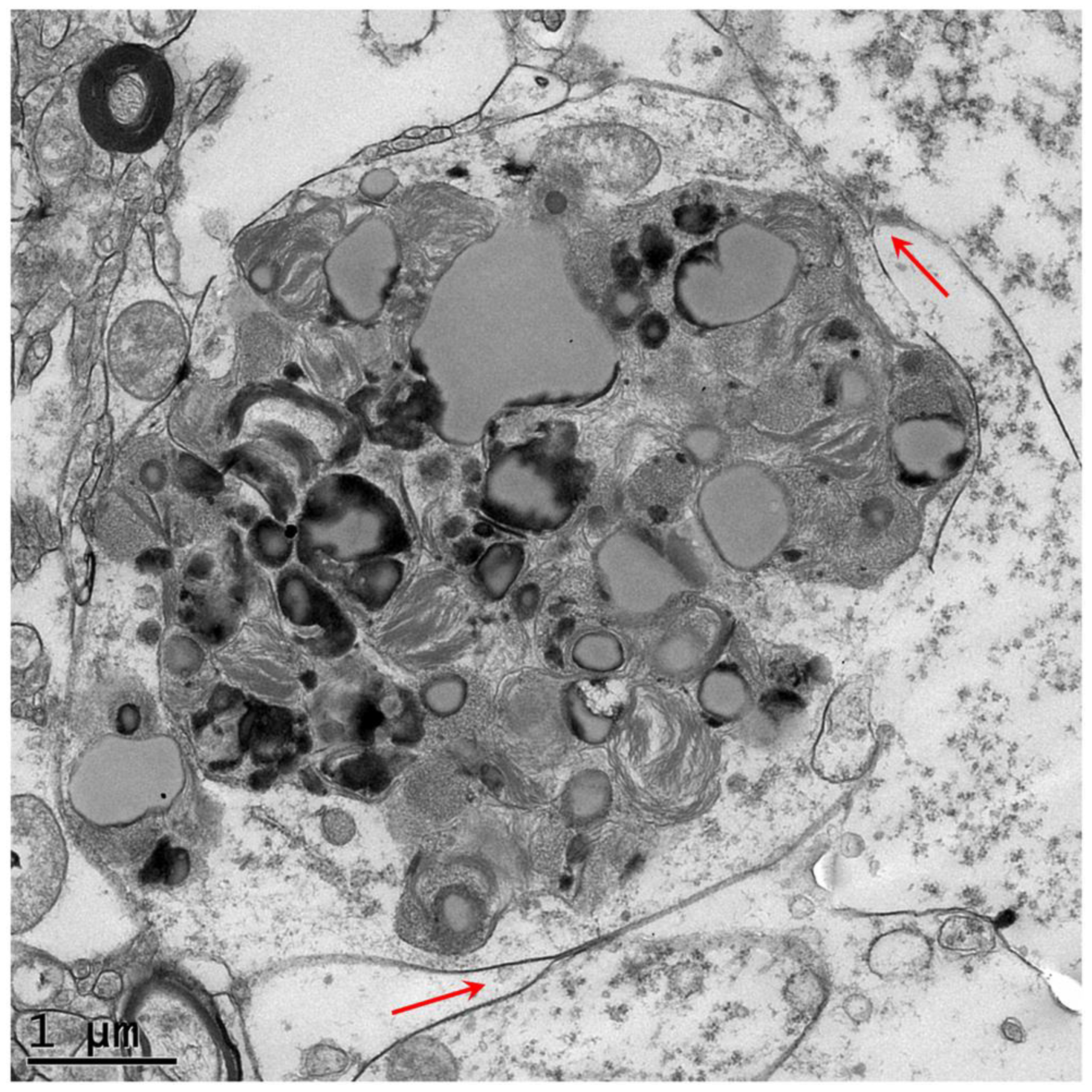
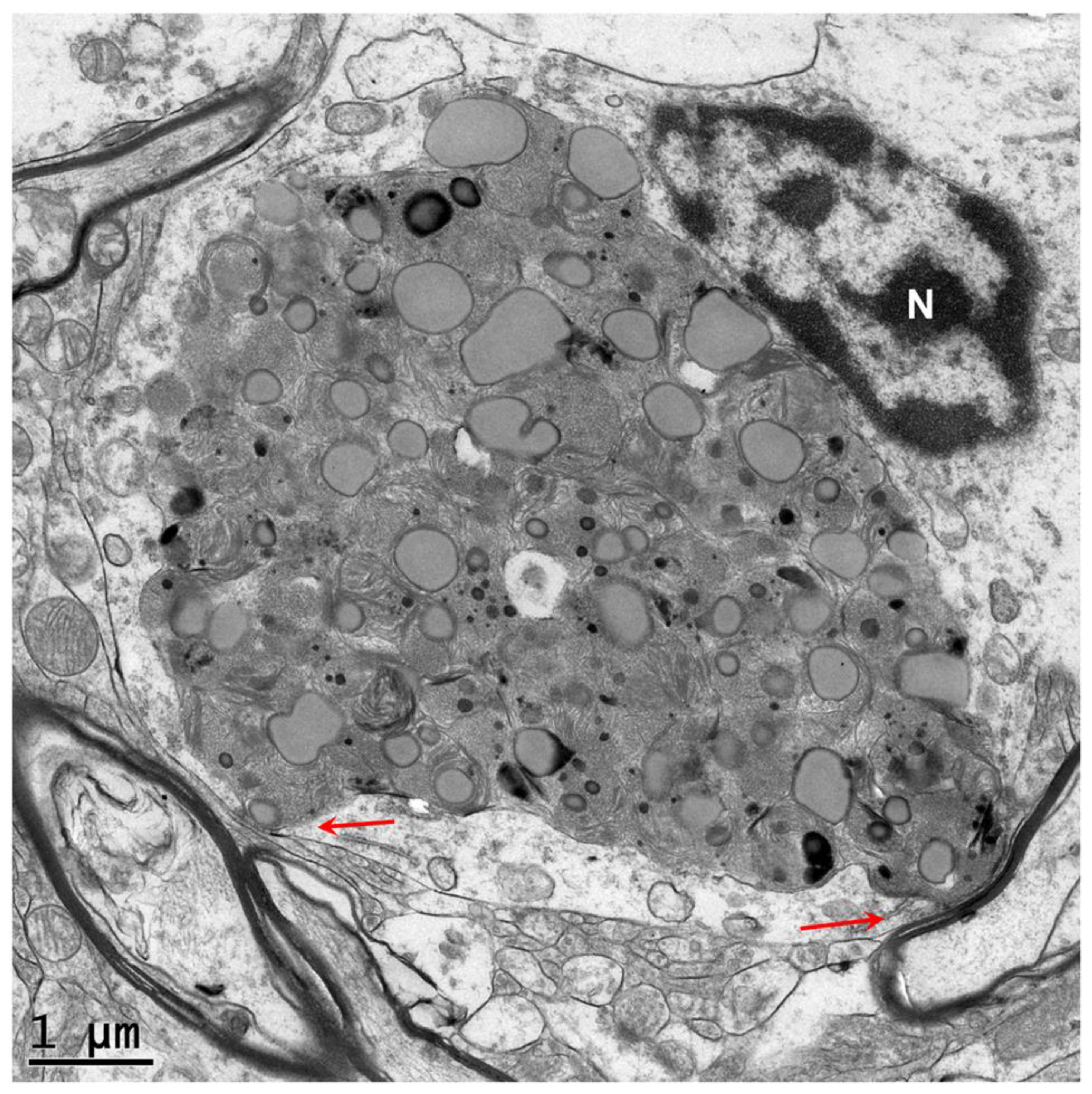
3. Results
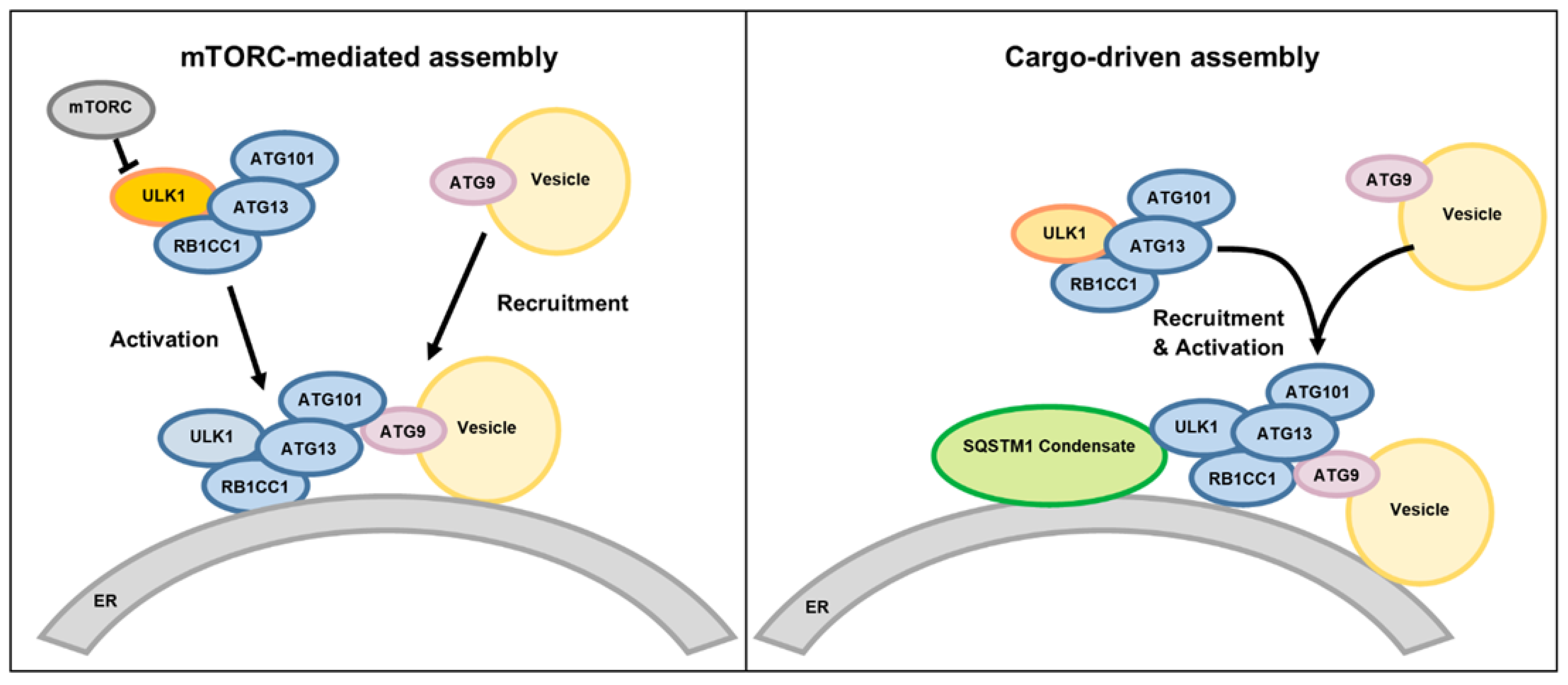
4. Discussion
Supplementary Materials
Acknowledgments
References
- Barker, E.N.; Dawson, L.J.; Rose, J.H.; Van Meervenne, S.; Frykman, O.; Rohdin, C.; Leijon, A.; Soerensen, K.E.; Jarnegren, J.; Johnson, G.C.; et al. Degenerative Encephalopathy in Nova Scotia Duck Tolling Retrievers Presenting with a Rapid Eye Movement Sleep Behavior Disorder. J Vet Intern Med 2016, 30, 1681–1689. [CrossRef]
- Katz, M.L.; Khan, S.; Awano, T.; Shahid, S.A.; Siakotos, A.N.; Johnson, G.S. A Mutation in the CLN8 Gene in English Setter Dogs with Neuronal Ceroid-Lipofuscinosis. Biochem Biophys Res Commun 2005, 327. [CrossRef]
- Zeng, R.; Coates, J.R.; Johnson, G.C.; Hansen, L.; Awano, T.; Kolicheski, A.; Ivansson, E.; Perloski, M.; Lindblad-Toh, K.; O’Brien, D.P.; et al. Breed Distribution of SOD1 Alleles Previously Associated with Canine Degenerative Myelopathy. J Vet Intern Med 2014, 28. [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly (Austin) 2012, 6, 80–92. [CrossRef]
- Ferreira, C.R.; Gahl, W.A. Lysosomal Storage Diseases. Transl Sci Rare Dis 2017, 2, 1–71. [CrossRef]
- Bullock, G.; Johnson, G.S.; Mhlanga-Mutangadura, T.; Petesch, S.C.; Thompson, S.; Goebbels, S.; Katz, M.L. Lysosomal Storage Disease Associated with a CNP Sequence Variant in Dalmatian Dogs. Gene 2022, 830, 146513. PT - Journal Article. [CrossRef]
- Furuta, A.; Kikuchi, H.; Fujita, H.; Yamada, D.; Fujiwara, Y.; Kabuta, T.; Nishino, I.; Wada, K.; Uchiyama, Y. Property of Lysosomal Storage Disease Associated with Midbrain Pathology in the Central Nervous System of Lamp-2-Deficient Mice. Am J Pathol 2015, 185, 1713–1723. [CrossRef]
- Alers, S.; Wesselborg, S.; Stork, B. ATG13: Just a Companion, or an Executor of the Autophagic Program?. Autophagy 2014, 10, 944–956. [CrossRef]
- Noda, N.N.; Mizushima, N. Atg101: Not Just an Accessory Subunit in the Autophagy-Initiation Complex. Cell Struct Funct 2016, 41, 13–20. [CrossRef]
- Yamashita, S.-I.; Kanki, T. How Autophagy Eats Large Mitochondria: Autophagosome Formation Coupled with Mitochondrial Fragmentation. Autophagy 2017, 13, 980–981. [CrossRef]
- Corona Velazquez, A.F.; Jackson, W.T. So Many Roads: The Multifaceted Regulation of Autophagy Induction. Mol Cell Biol 2018, 38. [CrossRef]
- Yamano, K.; Youle, R.J. Two Different Axes CALCOCO2-RB1CC1 and OPTN-ATG9A Initiate PRKN-Mediated Mitophagy. Autophagy 2020, 16, 2105–2107. [CrossRef]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy Genes in Biology and Disease. Nat Rev Genet 2023, 24, 382–400. [CrossRef]
- Kim, M.; Sandford, E.; Gatica, D.; Qiu, Y.; Liu, X.; Zheng, Y.; Schulman, B.A.; Xu, J.; Semple, I.; Ro, S.-H.; et al. Mutation in ATG5 Reduces Autophagy and Leads to Ataxia with Developmental Delay. Elife 2016, 5. [CrossRef]
- Collier, J.J.; Guissart, C.; Olahova, M.; Sasorith, S.; Piron-Prunier, F.; Suomi, F.; Zhang, D.; Martinez-Lopez, N.; Leboucq, N.; Bahr, A.; et al. Developmental Consequences of Defective ATG7-Mediated Autophagy in Humans. N Engl J Med 2021, 384, 2406–2417. [CrossRef]
- Almannai, M.; Marafi, D.; Abdel-Salam, G.M.H.; Zaki, M.S.; Duan, R.; Calame, D.; Herman, I.; Levesque, F.; Elbendary, H.M.; Hegazy, I.; et al. El-Hattab-Alkuraya Syndrome Caused by Biallelic WDR45B Pathogenic Variants: Further Delineation of the Phenotype and Genotype. Clin Genet 2022, 101, 530–540. [CrossRef]
- Seidahmed, M.Z.; Hamad, M.H.; AlBakheet, A.; Elmalik, S.A.; AlDrees, A.; Al-Sufayan, J.; Alorainy, I.; Ghozzi, I.M.; Colak, D.; Salih, M.A.; et al. Ancient Founder Mutation in RUBCN: A Second Unrelated Family Confirms Salih Ataxia (SCAR15). BMC Neurol 2020, 20, 207. [CrossRef]
- Guo, A.; Lun, P.; Chen, J.; Li, Q.; Chang, K.; Li, T.; Pan, D.; Zhang, J.; Zhou, J.; Wang, K.; et al. Association Analysis of Risk Genes Identified by SCHEMA with Schizophrenia in the Chinese Han Population. Psychiatr Genet 2022, 32, 188–193. [CrossRef]
- Lappas, A.S.; Ioannou, M.; Christodoulou, N.G. Histopathological Evidence of Cellular Alterations in the Dentate Gyrus Is Associated with Aberrant RB1CC1-ATG16L1 Expression in the Hippocampus among Older Adults with Chronic Schizophrenia: A Pilot Post-Mortem Study. Schizophr Res 2025, 275, 14–24. [CrossRef]
- Yao, J.; Jia, L.; Khan, N.; Lin, C.; Mitter, S.K.; Boulton, M.E.; Dunaief, J.L.; Klionsky, D.J.; Guan, J.-L.; Thompson, D.A.; et al. Deletion of Autophagy Inducer RB1CC1 Results in Degeneration of the Retinal Pigment Epithelium. Autophagy 2015, 11, 939–953. [CrossRef]
- Wang, C.; Chen, S.; Yeo, S.; Karsli-Uzunbas, G.; White, E.; Mizushima, N.; Virgin, H.W.; Guan, J.-L. Elevated P62/SQSTM1 Determines the Fate of Autophagy-Deficient Neural Stem Cells by Increasing Superoxide. J Cell Biol 2016, 212, 545–560. [CrossRef]
- Chen, S.; Wang, C.; Yeo, S.; Liang, C.-C.; Okamoto, T.; Sun, S.; Wen, J.; Guan, J.-L. Distinct Roles of Autophagy-Dependent and -Independent Functions of FIP200 Revealed by Generation and Analysis of a Mutant Knock-in Mouse Model. Genes Dev 2016, 30, 856–869. [CrossRef]
- Zhu, Y.-F.; Yu, R.-H.; Zhou, S.; Tang, P.-P.; Zhang, R.; Wu, Y.-X.; Xu, R.; Wei, J.-M.; Wang, Y.-Y.; Zhang, J.-L.; et al. TAX1BP1 and FIP200 Orchestrate Non-Canonical Autophagy of P62 Aggregates for Mouse Neural Stem Cell Maintenance. Zool Res 2024, 45, 937–950. [CrossRef]
- Wang, C.; Yeo, S.; Haas, M.A.; Guan, J.-L. Autophagy Gene FIP200 in Neural Progenitors Non-Cell Autonomously Controls Differentiation by Regulating Microglia. J Cell Biol 2017, 216, 2581–2596. [CrossRef]
- Liang, C.-C.; Wang, C.; Peng, X.; Gan, B.; Guan, J.-L. Neural-Specific Deletion of FIP200 Leads to Cerebellar Degeneration Caused by Increased Neuronal Death and Axon Degeneration. J Biol Chem 2010, 285, 3499–3509. [CrossRef]
- Deneubourg, C.; Ramm, M.; Smith, L.J.; Baron, O.; Singh, K.; Byrne, S.C.; Duchen, M.R.; Gautel, M.; Eskelinen, E.-L.; Fanto, M.; et al. The Spectrum of Neurodevelopmental, Neuromuscular and Neurodegenerative Disorders Due to Defective Autophagy. Autophagy 2022, 18, 496–517. [CrossRef]
- Hahn, K.; Rohdin, C.; Jagannathan, V.; Wohlsein, P.; Baumgartner, W.; Seehusen, F.; Spitzbarth, I.; Grandon, R.; Drogemuller, C.; Jaderlund, K.H. TECPR2 Associated Neuroaxonal Dystrophy in Spanish Water Dogs. PLoS One 2015, 10, e0141824. [CrossRef]
- Tamim-Yecheskel, B.-C.; Fraiberg, M.; Kokabi, K.; Freud, S.; Shatz, O.; Marvaldi, L.; Subic, N.; Brenner, O.; Tsoory, M.; Eilam-Altstadter, R.; et al. A Tecpr2 Knockout Mouse Exhibits Age-Dependent Neuroaxonal Dystrophy Associated with Autophagosome Accumulation. Autophagy 2021, 17, 3082–3095. [CrossRef]
- Fraiberg, M.; Tamim-Yecheskel, B.-C.; Kokabi, K.; Subic, N.; Heimer, G.; Eck, F.; Nalbach, K.; Behrends, C.; Ben-Zeev, B.; Shatz, O.; et al. Lysosomal Targeting of Autophagosomes by the TECPR Domain of TECPR2. Autophagy 2021, 17, 3096–3108. [CrossRef]
- Lei, L.; Tzekov, R.; Li, H.; McDowell, J.H.; Gao, G.; Smith, W.C.; Tang, S.; Kaushal, S. Inhibition or Stimulation of Autophagy Affects Early Formation of Lipofuscin-Like Autofluorescence in the Retinal Pigment Epithelium Cell. Int J Mol Sci 2017, 18. [CrossRef]
- Liu, A.; Guo, E.; Yang, J.; Yang, Y.; Liu, S.; Jiang, X.; Hu, Q.; Dirsch, O.; Dahmen, U.; Zhang, C.; et al. Young Plasma Reverses Age-Dependent Alterations in Hepatic Function through the Restoration of Autophagy. Aging Cell 2018, 17. [CrossRef]
- Nian, F.-S.; Li, L.-L.; Cheng, C.-Y.; Wu, P.-C.; Lin, Y.-T.; Tang, C.-Y.; Ren, B.-S.; Tai, C.-Y.; Fann, M.-J.; Kao, L.-S.; et al. Rab18 Collaborates with Rab7 to Modulate Lysosomal and Autophagy Activities in the Nervous System: An Overlapping Mechanism for Warburg Micro Syndrome and Charcot-Marie-Tooth Neuropathy Type 2B. Mol Neurobiol 2019, 56, 6095–6105. [CrossRef]
- Sarraf, S.A.; Shah, H. V; Kanfer, G.; Pickrell, A.M.; Holtzclaw, L.A.; Ward, M.E.; Youle, R.J. Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. Mol Cell 2020, 80, 779-795.e10. [CrossRef]
- Li, W.-W.; Wang, H.-J.; Tan, Y.-Z.; Wang, Y.-L.; Yu, S.-N.; Li, Z.-H. Reducing Lipofuscin Accumulation and Cardiomyocytic Senescence of Aging Heart by Enhancing Autophagy. Exp Cell Res 2021, 403, 112585. [CrossRef]
- Borner, J.H.; Rawashdeh, O.; Rami, A. Exacerbated Age-Related Hippocampal Alterations of Microglia Morphology, Beta-Amyloid and Lipofuscin Deposition and Presenilin Overexpression in <ovid:I>Per1</Ovid:I><ovid:Sup>-/-</Ovid:Sup>-Mice. Antioxidants (Basel) 2021, 10. [CrossRef]
- Hyttinen, J.M.T.; Koskela, A.; Blasiak, J.; Kaarniranta, K. Autophagy in Drusen Biogenesis Secondary to Age-Related Macular Degeneration. Acta Ophthalmol 2024, 102, 759–772. [CrossRef]
- Wang, X.-L.; Gao, Y.-X.; Yuan, Q.-Z.; Zhang, M. NLRP3 and Autophagy in Retinal Ganglion Cell Inflammation in Age-Related Macular Degeneration: Potential Therapeutic Implications. Int J Ophthalmol 2024, 17, 1531–1544. [CrossRef]
- Sosulski, M.L.; Gongora, R.; Danchuk, S.; Dong, C.; Luo, F.; Sanchez, C.G. Deregulation of Selective Autophagy during Aging and Pulmonary Fibrosis: The Role of TGFbeta1. Aging Cell 2015, 14, 774–783. [CrossRef]
- Li, W.-W.; Wang, H.-J.; Tan, Y.-Z.; Wang, Y.-L.; Yu, S.-N.; Li, Z.-H. Reducing Lipofuscin Accumulation and Cardiomyocytic Senescence of Aging Heart by Enhancing Autophagy. Exp Cell Res 2021, 403, 112585. [CrossRef]
- Winicki, N.M.; Nanavati, A.P.; Morrell, C.H.; Moen, J.M.; Axsom, J.E.; Krawczyk, M.; Petrashevskaya, N.N.; Beyman, M.G.; Ramirez, C.; Alfaras, I.; et al. A Small Erythropoietin Derived Non-Hematopoietic Peptide Reduces Cardiac Inflammation, Attenuates Age Associated Declines in Heart Function and Prolongs Healthspan. Front Cardiovasc Med 2022, 9, 1096887. [CrossRef]
- Terman, A.; Kurz, T.; Gustafsson, B.; Brunk, U.T. The Involvement of Lysosomes in Myocardial Aging and Disease. Curr Cardiol Rev 2008, 4, 107–115. [CrossRef]
- Turco, E.; Witt, M.; Abert, C.; Bock-Bierbaum, T.; Su, M.-Y.; Trapannone, R.; Sztacho, M.; Danieli, A.; Shi, X.; Zaffagnini, G.; et al. FIP200 Claw Domain Binding to P62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 2019, 74, 330-346.e11. [CrossRef]
- Turco, E.; Witt, M.; Abert, C.; Bock-Bierbaum, T.; Su, M.-Y.; Trapannone, R.; Sztacho, M.; Danieli, A.; Shi, X.; Zaffagnini, G.; et al. How RB1CC1/FIP200 Claws Its Way to Autophagic Engulfment of SQSTM1/P62-Ubiquitin Condensates. Autophagy 2019, 15, 1475–1477. [CrossRef]
- Zhang, M.; Wang, Y.; Gong, X.; Wang, Y.; Zhang, Y.; Tang, Y.; Zhou, X.; Liu, H.; Huang, Y.; Zhang, J.; et al. Mechanistic Insights into the Interactions of TAX1BP1 with RB1CC1 and Mammalian ATG8 Family Proteins. Proc Natl Acad Sci U S A 2024, 121, e2315550121. [CrossRef]
- Fu, T.; Zhang, M.; Zhou, Z.; Wu, P.; Peng, C.; Wang, Y.; Gong, X.; Li, Y.; Wang, Y.; Xu, X.; et al. Structural and Biochemical Advances on the Recruitment of the Autophagy-Initiating ULK and TBK1 Complexes by Autophagy Receptor NDP52. Sci Adv 2021, 7. [CrossRef]
- Assar, E.A.; Tumbarello, D.A. Loss of the Essential Autophagy Regulators FIP200 or Atg5 Leads to Distinct Effects on Focal Adhesion Composition and Organization. Front Cell Dev Biol 2020, 8, 733. [CrossRef]
- Yeo, S.K.; Guan, J.-L. Regulation of Immune Checkpoint Blockade Efficacy in Breast Cancer by FIP200: A Canonical-Autophagy-Independent Function. Cell Stress 2020, 4, 216–217. [CrossRef]
- Liu, H.; Wang, C.; Yi, F.; Yeo, S.; Haas, M.; Tang, X.; Guan, J.-L. Non-Canonical Function of FIP200 Is Required for Neural Stem Cell Maintenance and Differentiation by Limiting TBK1 Activation and P62 Aggregate Formation. Sci Rep 2021, 11, 23907. [CrossRef]
- Xue, X.; Ma, L.; Zhang, X.; Xu, X.; Guo, S.; Wang, Y.; Qiu, S.; Cui, J.; Guo, W.; Yu, Y.; et al. Tumour Cells Are Sensitised to Ferroptosis via RB1CC1-Mediated Transcriptional Reprogramming. Clin Transl Med 2022, 12, e747. [CrossRef]
- Yang, Y.; Klionsky, D.J. A Novel Role of ATG9A and RB1CC1/FIP200 in Mediating Cell-Death Checkpoints to Repress TNF Cytotoxicity. Autophagy 2023, 19, 1617–1618. [CrossRef]
- Chen, P.; Duan, Y.; Lu, X.; Chen, L.; Zhang, W.; Wang, H.; Hu, R.; Liu, S. RB1CC1 Functions as a Tumor-Suppressing Gene in Renal Cell Carcinoma via Suppression of PYK2 Activity and Disruption of TAZ-Mediated PDL1 Transcription Activation. Cancer Immunol Immunother 2021, 70, 3261–3275. [CrossRef]
- Wang, L.; Song, K.; Hao, W.; Wu, Y.; Patil, G.; Hua, F.; Sun, Y.; Huang, C.; Ritchey, J.; Jones, C.; et al. FIP200 Restricts RNA Virus Infection by Facilitating RIG-I Activation. Commun Biol 2021, 4, 921. [CrossRef]
- Yang, Y.; Klionsky, D.J. A Novel Role of ATG9A and RB1CC1/FIP200 in Mediating Cell-Death Checkpoints to Repress TNF Cytotoxicity. Autophagy 2023, 19, 1617–1618. [CrossRef]
- Okamoto, T.; Yeo, S.K.; Hao, M.; Copley, M.R.; Haas, M.A.; Chen, S.; Guan, J.-L. FIP200 Suppresses Immune Checkpoint Therapy Responses in Breast Cancers by Limiting AZI2/TBK1/IRF Signaling Independent of Its Canonical Autophagy Function. Cancer Res 2020, 80, 3580–3592. [CrossRef]
- Yeo, S.K.; Wang, C.; Guan, J.-L. Role of FIP200 in Inflammatory Processes beyond Its Canonical Autophagy Function. Biochem Soc Trans 2020, 48, 1599–1607. [CrossRef]
- Goodwin, J.M.; Dowdle, W.E.; DeJesus, R.; Wang, Z.; Bergman, P.; Kobylarz, M.; Lindeman, A.; Xavier, R.J.; McAllister, G.; Nyfeler, B.; et al. Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9. Cell Rep 2017, 20, 2341–2356. [CrossRef]
- Yang, Y.; Klionsky, D.J. A Novel Role of ATG9A and RB1CC1/FIP200 in Mediating Cell-Death Checkpoints to Repress TNF Cytotoxicity. Autophagy 2023, 19, 1617–1618. [CrossRef]
- Abbi, S.; Ueda, H.; Zheng, C.; Cooper, L.A.; Zhao, J.; Christopher, R.; Guan, J.-L. Regulation of Focal Adhesion Kinase by a Novel Protein Inhibitor FIP200. Mol Biol Cell 2002, 13, 3178–3191. [CrossRef]
- Melkoumian, Z.K.; Peng, X.; Gan, B.; Wu, X.; Guan, J.-L. Mechanism of Cell Cycle Regulation by FIP200 in Human Breast Cancer Cells. Cancer Res 2005, 65, 6676–6684. [CrossRef]
- Choi, J.D.; Ryu, M.; Ae Park, M.; Jeong, G.; Lee, J.-S. FIP200 Inhibits Beta-Catenin-Mediated Transcription by Promoting APC-Independent Beta-Catenin Ubiquitination. Oncogene 2013, 32, 2421–2432. [CrossRef]
- Wang, C.; Liang, C.-C.; Bian, Z.C.; Zhu, Y.; Guan, J.-L. FIP200 Is Required for Maintenance and Differentiation of Postnatal Neural Stem Cells. Nat Neurosci 2013, 16, 532–542. [CrossRef]
- Watanabe, R.; Chano, T.; Inoue, H.; Isono, T.; Koiwai, O.; Okabe, H. Rb1cc1 Is Critical for Myoblast Differentiation through Rb1 Regulation. Virchows Arch 2005, 447, 643–648. [CrossRef]
- Chano, T.; Saeki, Y.; Serra, M.; Matsumoto, K.; Okabe, H. Preferential Expression of RB1-Inducible Coiled-Coil 1 in Terminal Differentiated Musculoskeletal Cells. Am J Pathol 2002, 161, 359–364. [CrossRef]
- Wei, H.; Gan, B.; Wu, X.; Guan, J.-L. Inactivation of FIP200 Leads to Inflammatory Skin Disorder, but Not Tumorigenesis, in Conditional Knock-out Mouse Models. J Biol Chem 2009, 284, 6004–6013. [CrossRef]
- Liu, F.; Guan, J.-L. FIP200, an Essential Component of Mammalian Autophagy Is Indispensible for Fetal Hematopoiesis. Autophagy 2011, 7, 229–230. [CrossRef]
- Wei, H.; Wei, S.; Gan, B.; Peng, X.; Zou, W.; Guan, J.-L. Suppression of Autophagy by FIP200 Deletion Inhibits Mammary Tumorigenesis. Genes Dev 2011, 25, 1510–1527. [CrossRef]
- Bae, H.; Guan, J.-L. Suppression of Autophagy by FIP200 Deletion Impairs DNA Damage Repair and Increases Cell Death upon Treatments with Anticancer Agents. Mol Cancer Res 2011, 9, 1232–1241. [CrossRef]
- Ma, D.; Molusky, M.M.; Song, J.; Hu, C.-R.; Fang, F.; Rui, C.; Mathew, A. V; Pennathur, S.; Liu, F.; Cheng, J.-X.; et al. Autophagy Deficiency by Hepatic FIP200 Deletion Uncouples Steatosis from Liver Injury in NAFLD. Mol Endocrinol 2013, 27, 1643–1654. [CrossRef]
- Liu, F.; Fang, F.; Yuan, H.; Yang, D.; Chen, Y.; Williams, L.; Goldstein, S.A.; Krebsbach, P.H.; Guan, J.-L. Suppression of Autophagy by FIP200 Deletion Leads to Osteopenia in Mice through the Inhibition of Osteoblast Terminal Differentiation. J Bone Miner Res 2013, 28, 2414–2430. [CrossRef]
- Li, Y.; Gan, C.; Zhang, S.; Zhou, X.; Li, X.; Wei, Y.; Yang, J.; Wu, M. FIP200 Is Involved in Murine Pseudomonas Infection by Regulating HMGB1 Intracellular Translocation. Cell Physiol Biochem 2014, 33, 1733–1744. [CrossRef]
- Yao, J.; Jia, L.; Khan, N.; Lin, C.; Mitter, S.K.; Boulton, M.E.; Dunaief, J.L.; Klionsky, D.J.; Guan, J.-L.; Thompson, D.A.; et al. Deletion of Autophagy Inducer RB1CC1 Results in Degeneration of the Retinal Pigment Epithelium. Autophagy 2015, 11, 939–953. [CrossRef]
- Chen, S.; Wang, C.; Yeo, S.; Liang, C.-C.; Okamoto, T.; Sun, S.; Wen, J.; Guan, J.-L. Distinct Roles of Autophagy-Dependent and -Independent Functions of FIP200 Revealed by Generation and Analysis of a Mutant Knock-in Mouse Model. Genes Dev 2016, 30, 856–869. [CrossRef]
- Wang, C.; Yeo, S.; Haas, M.A.; Guan, J.-L. Autophagy Gene FIP200 in Neural Progenitors Non-Cell Autonomously Controls Differentiation by Regulating Microglia. J Cell Biol 2017, 216, 2581–2596. [CrossRef]
- Wang, B.; Iyengar, R.; Li-Harms, X.; Joo, J.H.; Wright, C.; Lavado, A.; Horner, L.; Yang, M.; Guan, J.-L.; Frase, S.; et al. The Autophagy-Inducing Kinases, ULK1 and ULK2, Regulate Axon Guidance in the Developing Mouse Forebrain via a Noncanonical Pathway. Autophagy 2018, 14, 796–811. [CrossRef]
- Oestreich, A.K.; Chadchan, S.B.; Medvedeva, A.; Lydon, J.P.; Jungheim, E.S.; Moley, K.H.; Kommagani, R. The Autophagy Protein, FIP200 (RB1CC1) Mediates Progesterone Responses Governing Uterine Receptivity and Decidualization . Biol Reprod 2020, 102, 843–851. [CrossRef]
- Li, D.; Vogel, P.; Li-Harms, X.; Wang, B.; Kundu, M. ATG14 and RB1CC1 Play Essential Roles in Maintaining Muscle Homeostasis. Autophagy 2021, 17, 2576–2585. [CrossRef]
- Wang, L.; Song, K.; Hao, W.; Wu, Y.; Patil, G.; Hua, F.; Sun, Y.; Huang, C.; Ritchey, J.; Jones, C.; et al. FIP200 Restricts RNA Virus Infection by Facilitating RIG-I Activation. Commun Biol 2021, 4, 921. [CrossRef]
- Xue, X.; Ma, L.; Zhang, X.; Xu, X.; Guo, S.; Wang, Y.; Qiu, S.; Cui, J.; Guo, W.; Yu, Y.; et al. Tumour Cells Are Sensitised to Ferroptosis via RB1CC1-Mediated Transcriptional Reprogramming. Clin Transl Med 2022, 12, e747. [CrossRef]
- Yang, Y.; White, E. Autophagy in PDGFRA<ovid:Sup>+</Ovid:Sup> Mesenchymal Cells Is Required for Intestinal Homeostasis and Mammalian Survival. Autophagy 2023, 19, 726–728. [CrossRef]
- Yang, F.; Kalantari, S.; Ruan, B.; Sun, S.; Bian, Z.; Guan, J.-L. Autophagy Inhibition Prevents Lymphatic Malformation Progression to Lymphangiosarcoma by Decreasing Osteopontin and Stat3 Signaling. Nat Commun 2023, 14, 978. [CrossRef]
- Zhu, Y.-F.; Yu, R.-H.; Zhou, S.; Tang, P.-P.; Zhang, R.; Wu, Y.-X.; Xu, R.; Wei, J.-M.; Wang, Y.-Y.; Zhang, J.-L.; et al. TAX1BP1 and FIP200 Orchestrate Non-Canonical Autophagy of P62 Aggregates for Mouse Neural Stem Cell Maintenance. Zool Res 2024, 45, 937–950. [CrossRef]
- Li, L.; Wang, G.; Hu, J.-S.; Zhang, G.-Q.; Chen, H.-Z.; Yuan, Y.; Li, Y.-L.; Lv, X.-J.; Tian, F.-Y.; Pan, S.-H.; et al. RB1CC1-Enhanced Autophagy Facilitates PSCs Activation and Pancreatic Fibrogenesis in Chronic Pancreatitis. Cell Death Dis 2018, 9, 952. [CrossRef]
- Huang, Y.; Bodnar, D.; Chen, C.-Y.; Sanchez-Andrade, G.; Sanderson, M.; Shi, J.; Meilleur, K.G.; Hurles, M.E.; Gerety, S.S.; Tsai, E.A.; et al. Rare Genetic Variants Impact Muscle Strength. Nat Commun 2023, 14, 3449. [CrossRef]
- Li, D.; Vogel, P.; Li-Harms, X.; Wang, B.; Kundu, M. ATG14 and RB1CC1 Play Essential Roles in Maintaining Muscle Homeostasis. Autophagy 2021, 17, 2576–2585. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




