Submitted:
21 October 2025
Posted:
21 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Samples Description
2.2. Wet Chemistry
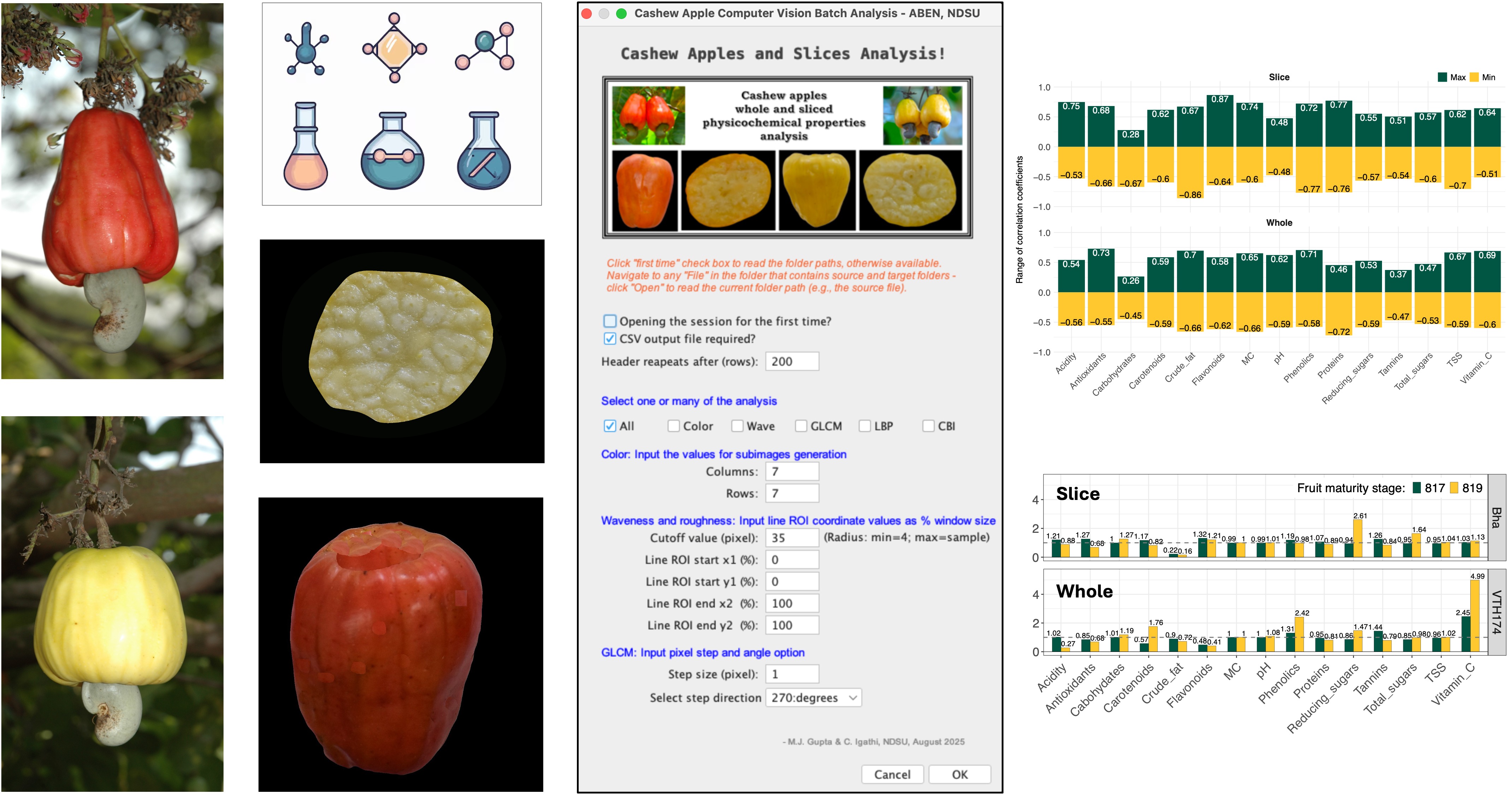
2.3. Computer Vision Methodology
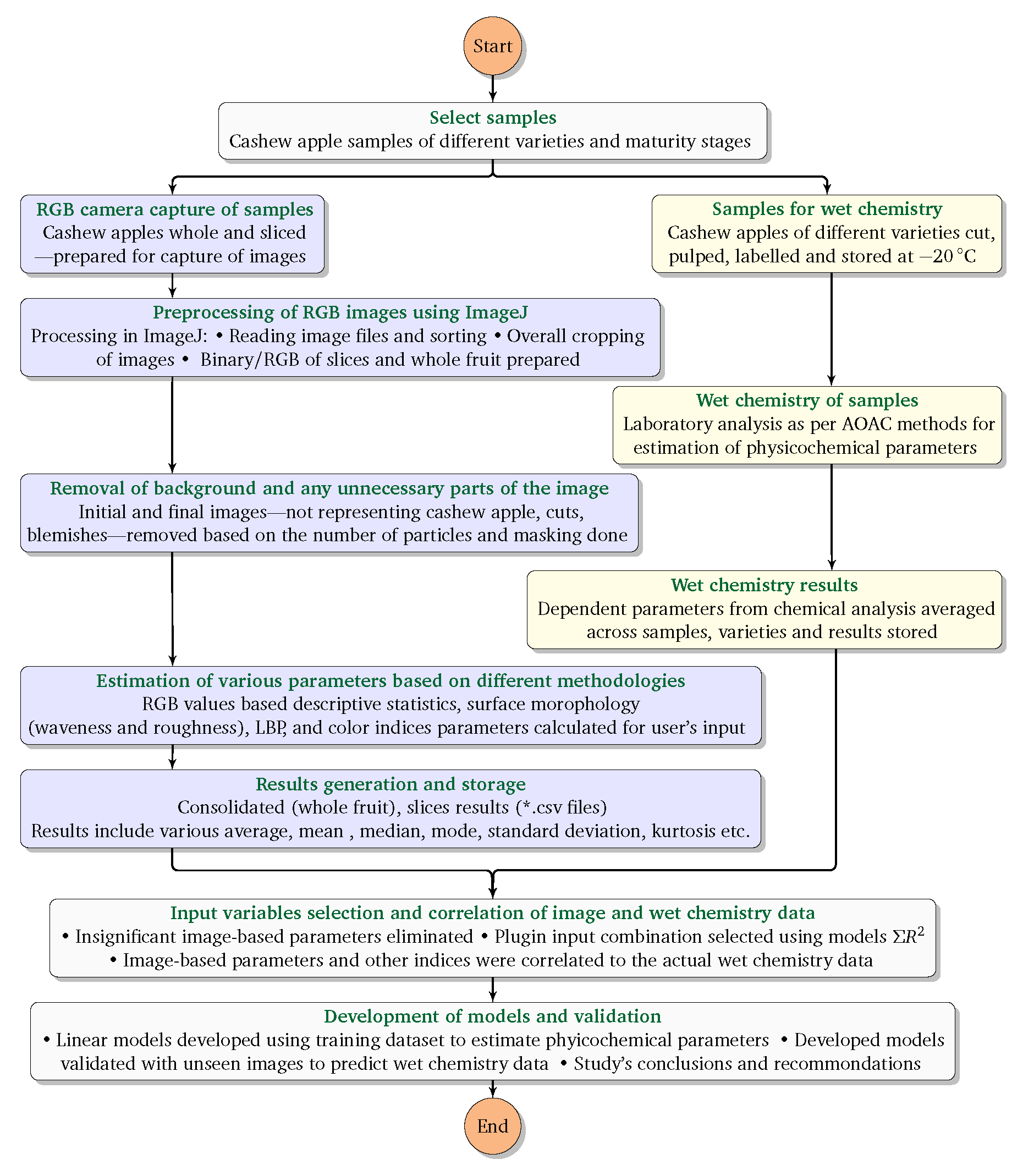
2.3.1. Image Acquisition
2.3.2. Image Preprocessing
2.3.3. Image-based parameters
(i) Color-based analysis
(ii) Surface morphology—waveness and roughness
(iii) Gray level co-occurrence matrix (GLCM) method
(iii) Local binary pattern (LBP) method
(iv) Color indices
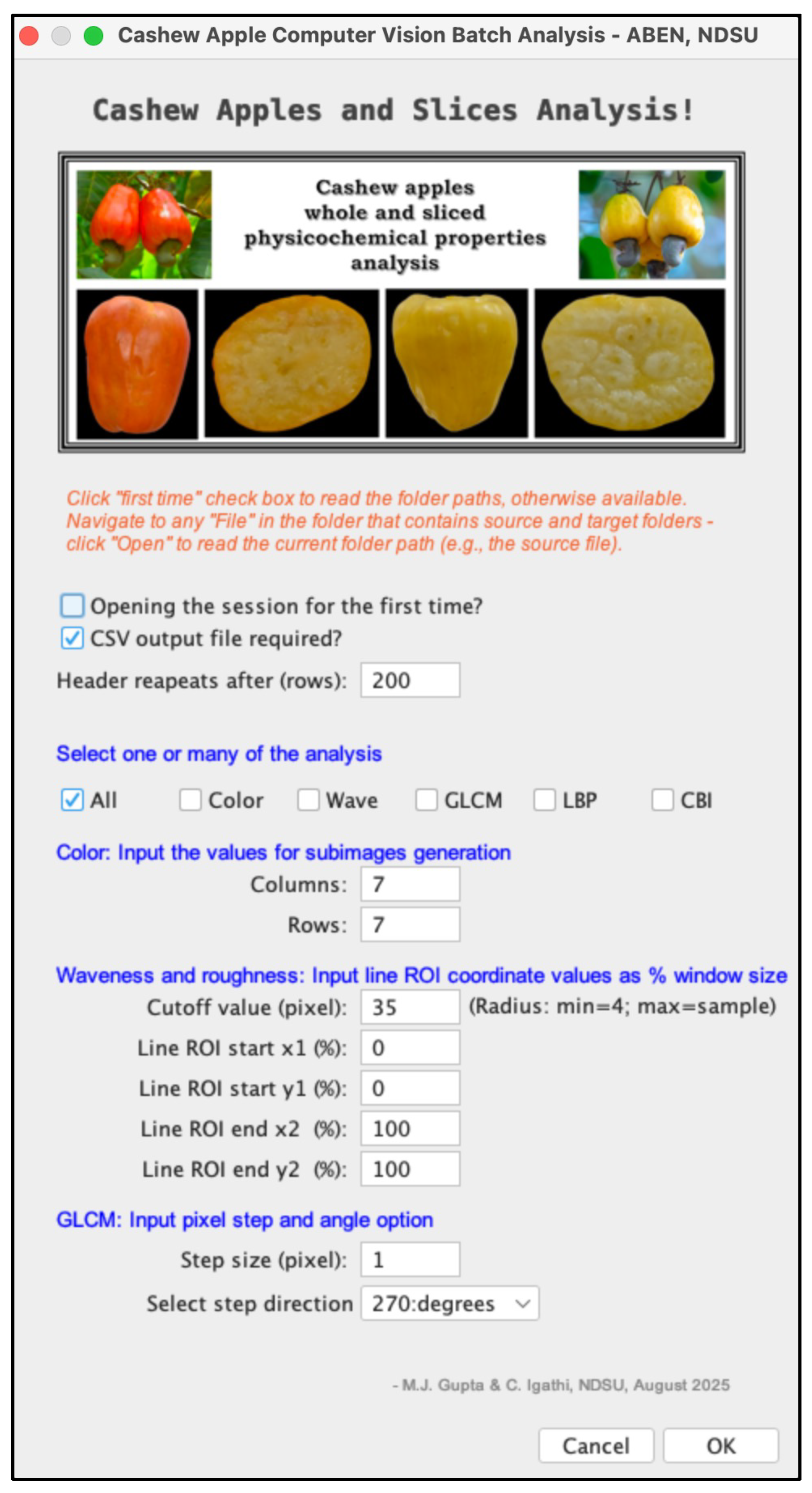
2.3.4. The plugins input panel or front panel
2.3.5. The Cashew Apples and Slices Analysis plugin
2.4. Data analysis for predicting the wet chemistry from image-based parameters
2.4.1. Correlation analysis and visualization
2.4.2. Range and ranking of correlation coefficients
2.4.3. Wet chemistry prediction models development
2.4.4. Wet chemistry prediction and validation
3. Results and discussion
3.1. Results: Variation in wet chemistry results within samples and varieties
3.2. Independent image-based variables considered for analysis
3.3. Plugin input variables combination selection
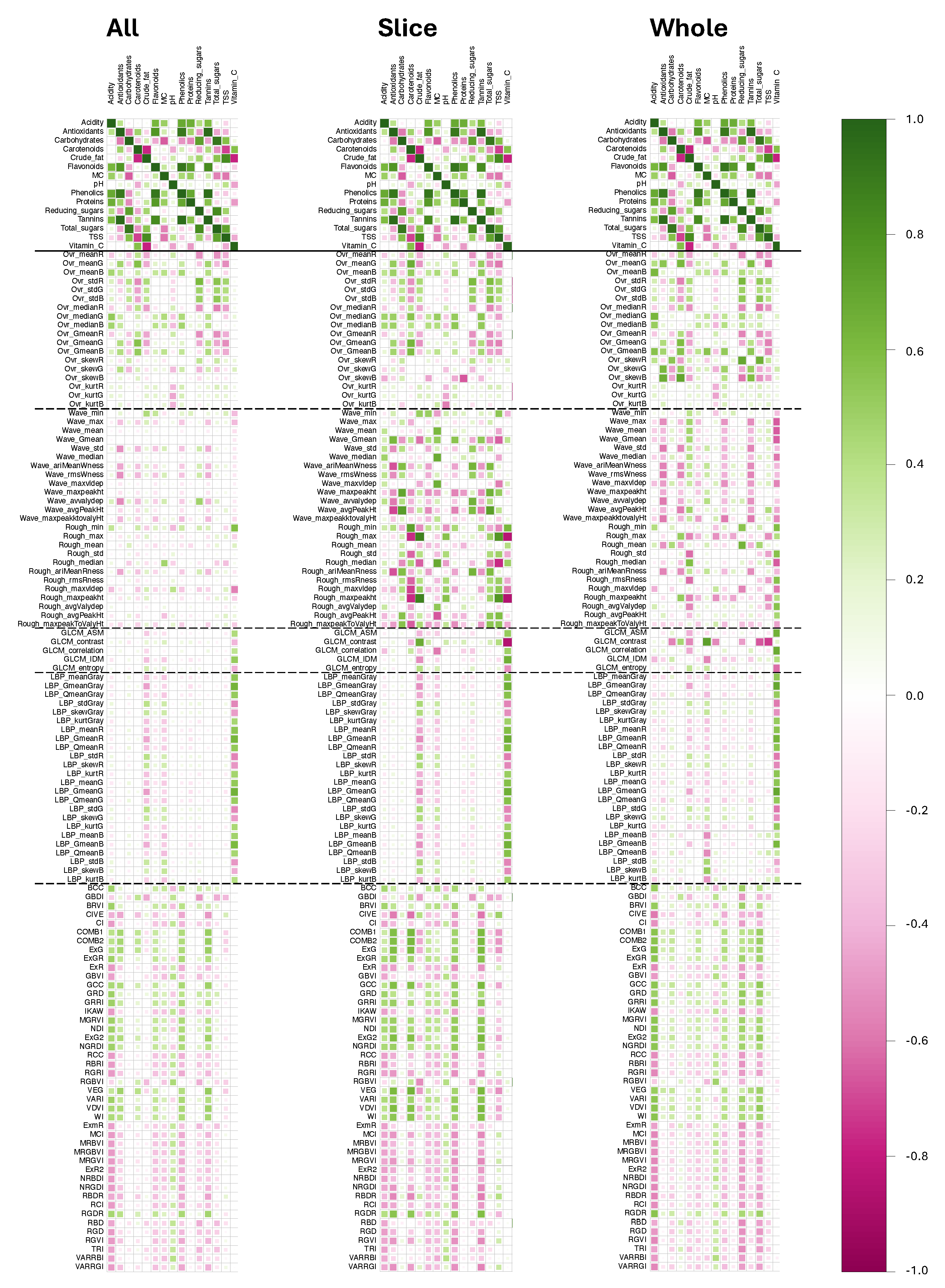
3.4. Visualizing Correlation—Correlation Diagram And Absolute Correlation Coefficients
3.4.1. Interrelationships among wet chemistry parameters
3.4.2. Correlation between wet chemistry and image-based parameters
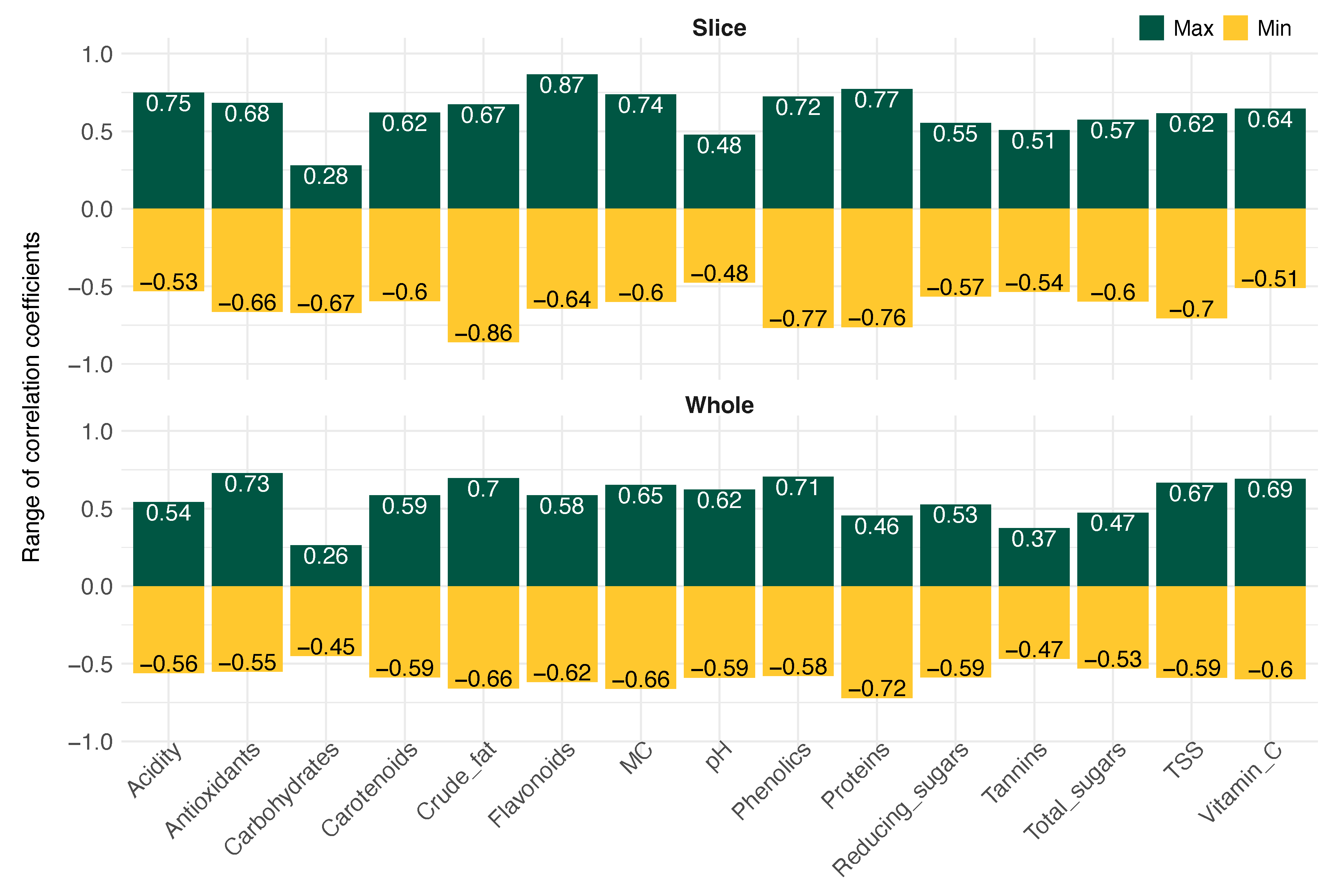
3.5. Correlation analysis of wet chemistry with image-based parameters—value ranges
3.6. Top ranking image-based parameters correlation coefficients
3.7. Wet Chemistry physicochemical properties Prediction Model Development
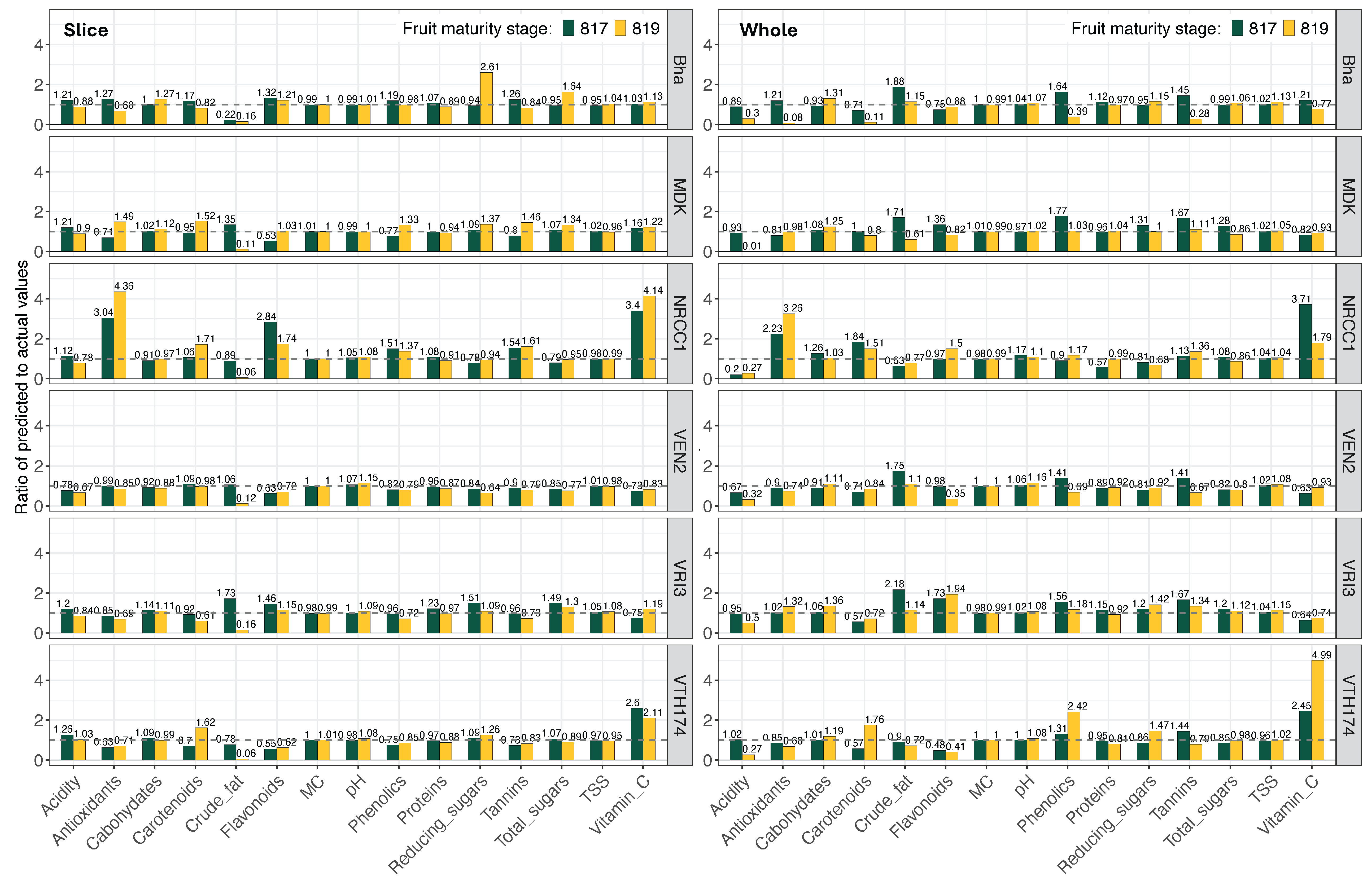
3.8. Wet Chemistry physicochemical properties Prediction Models Validation
3.8.1. Study’s Limitations and Suggestions for Future Work
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Appendix A
Appendix A.1. Physicochemical properties
(i) Moisture, protein, and fat
(ii) Total soluble solids, pH, and titratable acidity determination
(iii) Total carbohydrate
(iv) Total sugar
(v) Reducing sugar
(vi) Vitamin C (ascorbic acid) estimation
Appendix A.2. Biochemical Properties
(i) Total phenolic content (TPC)
(ii) Total flavonoids content (TFC)
(iii) Antioxidant activity using DPPH assay
(iv) Tannin content
(v) Carotene estimation
References
- Maciel, M.L.; Hansen, T.J.; Aldinger, S.B.; Labows, J.N. Flavor chemistry of cashew apple juice. Journal of Agricultural and Food Chemistry 1986, 34, 923–927. [Google Scholar] [CrossRef]
- Das, I.; Arora, A. Post-harvest processing technology for cashew apple—A review. Journal of Food Engineering 2017, 194, 87–98. [Google Scholar] [CrossRef]
- Pushpalatha, P.; Sobhana, A.; Mini, C. Processing and product diversification in cashew apple. Advances in Cashew Production Technology 2015, pp. 109–116.
- Sri Lakshmi, C.; Prema, A. Resource-use efficiency in raw cashew nut production in Kerala, India. Asian Journal of Agricultural Extension, Economics and Sociology 2025, 43, 92–98. [Google Scholar]
- FAOSTAT crops and livestock database, 2025.
- Ganesh, S.; Kannan, M.; Jawaharlal, M. Cashew industry-an outlook. In Proceedings of the I International Symposium on Cashew Nut 1080, 2011, pp. 89–95.
- Anoopkumar, A.; Gopinath, C.; Annadurai, S.; Abdullah, S.; Tarafdar, A.; Hazeena, S.H.; Rajasekharan, R.; Kuriakose, L.L.; Aneesh, E.M.; de Souza Vandenberghe, L.P.; et al. Biotechnological valorisation of cashew apple: Prospects and challenges in synthesising wide spectrum of products with market value. Bioresource Technology Reports 2024, 25, 101742. [Google Scholar] [CrossRef]
- Gnagne, A.A.G.B.; Soro, D.; Ouattara, Y.; Koui, E.; Koffi, E. A literature review of cashew apple processing. African Journal of Food, Agriculture, Nutrition and Development 2023, 23, 22452–22469. [Google Scholar] [CrossRef]
- Trevisan, M.; Pfundstein, B.; Haubner, R.; Würtele, G.; Spiegelhalder, B.; Bartsch, H.; Owen, R. Characterization of alkyl phenols in cashew (Anacardium occidentale) products and assay of their antioxidant capacity. Food and Chemical toxicology 2006, 44, 188–197. [Google Scholar] [CrossRef]
- Honorato, T.L.; Rabelo, M.C.; Gonçalves, L.R.B.; Pinto, G.A.S.; Rodrigues, S. Fermentation of cashew apple juice to produce high added value products. World Journal of Microbiology and Biotechnology 2007, 23, 1409–1415. [Google Scholar] [CrossRef]
- dos Santos Lima, F.C.; da Silva, F.L.H.; Gomes, J.P.; da Silva Neto, J.M. Chemical composition of the cashew apple bagasse and potential use for ethanol production. Advances in Chemical Engineering and Science 2012, 2, 519–523. [Google Scholar] [CrossRef]
- Dedehou, E.; Dossou, J.; Anihouvi, V.; Soumanou, M.M. A review of cashew (Anacardium occidentale L.) apple: Effects of processing techniques, properties and quality of juice. African Journal of Biotechnology 2016, 15, 2637–2648. [Google Scholar]
- Pascal, A.D.C.; Virginie, G.; Diane, B.F.T.; Estelle, K.R.; Félicien, A.; Valentin, W.D.; Dominique, S.K.C. Nutritional profile and chemical composition of juices of two cashew apple’s varieties of Benin. Chemistry Journal 2018, 4, 91–96. [Google Scholar]
- Reina, L.J.C.; Durán-Aranguren, D.D.; Forero-Rojas, L.F.; Tarapuez-Viveros, L.F.; Durán-Sequeda, D.; Carazzone, C.; Sierra, R. Chemical composition and bioactive compounds of cashew (Anacardium occidentale) apple juice and bagasse from Colombian varieties. Heliyon 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Akyereko, Y.G.; Yeboah, G.B.; Wireko-Manu, F.D.; Alemawor, F.; Mills-Robertson, F.; Odoom, W. Nutritional value and health benefits of cashew apple. JSFA reports 2023, 3, 110–118. [Google Scholar] [CrossRef]
- Prasertsri, P.; Roengrit, T.; Kanpetta, Y.; Tong-Un, T.; Muchimapura, S.; Wattanathorn, J.; Leelayuwat, N. Cashew apple juice supplementation enhanced fat utilization during high-intensity exercise in trained and untrained men. Journal of the International Society of Sports Nutrition 2013, 10, 13. [Google Scholar] [CrossRef]
- Aidoo, R.; Kwofie, E.M.; Ngadi, M.O. Circularity of cashew apples: examining the product-process pathways, techno-functional, nutritional/phytomolecular qualities for food applications. ACS Food Science & Technology 2022, 2, 1051–1066. [Google Scholar]
- da Silveira Vasconcelos, M.; Gomes-Rochette, N.F.; de Oliveira, M.L.M.; Nunes-Pinheiro, D.C.S.; Tome, A.R.; Maia de Sousa, F.Y.; Pinheiro, F.G.M.; Moura, C.F.H.; Miranda, M.R.A.; Mota, E.F.; et al. Anti-inflammatory and wound healing potential of cashew apple juice (Anacardium occidentale L.) in mice. Experimental Biology and Medicine 2015, 240, 1648–1655. [Google Scholar] [CrossRef]
- Salehi, B.; Gültekin-Özgüven, M.; Kirkin, C.; Özçelik, B.; Morais-Braga, M.F.B.; Carneiro, J.N.P.; Bezerra, C.F.; Silva, T.G.d.; Coutinho, H.D.M.; Amina, B.; et al. Antioxidant, antimicrobial, and anticancer effects of anacardium plants: an ethnopharmacological perspective. Frontiers in Endocrinology 2020, 11, 295. [Google Scholar] [CrossRef]
- Attri, B. Effect of initial sugar concentration on the physico-chemical characteristics and sensory qualities of cashew apple wine. Natural product radiance 2009, 8, 374–379. [Google Scholar]
- Kouassi, E.K.A.; Soro, Y.; Vaca-Garcia, C.; Yao, K.B. Chemical composition and specific lipids profile of the cashew apple bagasse. Rasayan Journal of Chemistry 2018, 11, 386–391. [Google Scholar]
- Okpanachi, U.; Attah, S.; Shaahu, D. A comparative study between vitamins and amino acid profile of sun-dried red and yellow cashew pulp. Int. J. An. Biol 2015, 1, 23. [Google Scholar]
- Runjala, S.; Kella, L. Cashew apple (Anacardium occidentale L.) therapeutic benefits, processing and product development: An overview. The Pharma Innovation 2017, 6, 260. [Google Scholar]
- Sivaprakasam, S.; Chinnaiyan, U.; Varatharajan, S.; Uthayasuryan, E.; Chandrasekaran, M.; Paramasivam, S.; Ramasamy, P. Unveiling the physicochemical attributes and antioxidant potential of cashew apple juice varieties. Applied Fruit Science 2025, 67, 74. [Google Scholar] [CrossRef]
- Kiatti, D.d.; Serrapica, F.; Musco, N.; Di Palo, R.; Calabrò, S. Potential use of tropical and subtropical fruits by-products in pig diet: In vitro two-step evaluation. Animals 2025, 15, 1454. [Google Scholar] [CrossRef]
- Preethi, P.; Rajkumar, A.; Shamsudheen, M.; Nayak, M. Prospects of cashew apple—A compilation report. Technical Bulletin 2019, 2, 1–28. [Google Scholar]
- Codjia, F.S.; Dabadé, D.S.; Agbobatinkpo, P.; Collombel, I.; Achir, N.; Azokpota, P.; Dossou, J. Fermented cashew apple beverages: Current state of knowledge and prospects. Beverages 2025, 11, 49. [Google Scholar] [CrossRef]
- Mtashobya, L.A.; Mgeni, S.T.; Emmanuel, J.K. Potential contributions of cashew apple juice to nutrition, medicine and bioethanol generation: An outlook. Natural Product Communications 2025, 20, 1934578X251357414. [Google Scholar] [CrossRef]
- Levy, H.M.; Rodrigues, T.H.S.; de Albuquerque, T.L.; Sant’Ana, H.B.; Feitosa, F.X.; Rocha, M.V.P. Biosynthesis of biolubricant catalyzed by lipase B from Candida antarctica immobilized on nanocomposite of cashew apple bagasse lignin. Renewable Energy 2025, 243, 122452. [Google Scholar] [CrossRef]
- Osei, E.D.; Amotoe-Bondzie, A.; Ataa Pokuah, A.; Laar, W.S.; Afoakwah, N.A.; Ivanišová, E. Cashew apple pomace: Chemical composition and applications in functional food product development—A review. Food Science & Nutrition 2025, 13, e70185. [Google Scholar] [CrossRef]
- Cormier, R.; Michodjehourn, S.; Fulcrand, B. Clarification of cashew apple juice and commercial applications. Oxfarm Quebec, Benin, West Africa 2008, pp. 1–9.
- Ogunjobi, M.; Ogunwolu, S. Physicochemical and sensory properties of cassava flour biscuits supplemented with cashew apple powder. Journal of Food Technology 2010, 8, 24–29. [Google Scholar]
- Mohammadzadeh, Z.; Shojaeiyan, A.; Mahfeli, M.; Ayyari, M.; Tohidfar, M.; Mokhtassi-Bidgoli, A.; Atighi, M.R. Predictive modeling of CIELAB color parameters in okra accessions based on phytochemical composition and antioxidant activity: A non-destructive Imagej and RSM approach. LWT 2025, 228, 118080. [Google Scholar] [CrossRef]
- Jinorose, M.; Prachayawarakorn, S.; Soponronnarit, S. A novel image-analysis based approach to evaluate some physicochemical and cooking properties of rice kernels. Journal of Food Engineering 2014, 124, 184–190. [Google Scholar] [CrossRef]
- Sabzi, S.; Nadimi, M.; Abbaspour-Gilandeh, Y.; Paliwal, J. Non-destructive estimation of physicochemical properties and detection of ripeness level of apples using machine vision. International Journal of Fruit Science 2022, 22, 628–645. [Google Scholar] [CrossRef]
- Cárdenas-Pérez, S.; Chanona-Pérez, J.; Méndez-Méndez, J.V.; Calderón-Domínguez, G.; López-Santiago, R.; Perea-Flores, M.J.; Arzate-Vázquez, I. Evaluation of the ripening stages of apple (Golden Delicious) by means of computer vision system. Biosystems Engineering 2017, 159, 46–58. [Google Scholar] [CrossRef]
- Abalone, R.; Cassinera, A.; Gaston, A.; Lara, M. Some physical properties of amaranth seeds. Biosystems Engineering 2004, 89, 109–117. [Google Scholar] [CrossRef]
- Adiga, J.D.; Muralidhara, B.M.; Preethi, P.; Savadi, S. Phenological growth stages of the cashew tree (Anacardium occidentale L.) according to the extended BBCH scale. Annals of Applied Biology 2019, 175, 246–252. [Google Scholar] [CrossRef]
- Gupta, M.J.; Nishad, J.; Igathinathane, C. Assessment of Physicochemical Properties of Cashew Apple through Computer Vision — Original Images, Data, and R Script. Mendeley Data, V1, 2025. [CrossRef]
- Pawlus, P.; Reizer, R.; Wieczorowski, M. Functional importance of surface texture parameters. Materials 2021, 14, 5326. [Google Scholar] [CrossRef]
- Chinga, G. Waveness Roughness. ImageJ Plugins https://imagej.net/ij/plugins/waveness-roughness.html, 2018. Accessed: 2025-09-09.
- Haralick, R.M.; Shanmugam, K. Computer classification of reservoir sandstones. IEEE Transactions on Geoscience Electronics 1973, 11, 171–177. [Google Scholar] [CrossRef]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image texture feature extraction using GLCM approach. International journal of scientific and research publications 2013, 3, 1–5. [Google Scholar]
- Song, Y.; Sa, J.; Luo, Y.; Zhang, Z. A comprehensively improved local binary pattern framework for texture classification. Multimedia Tools and Applications 2025, 84, 22043–22068. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Zhang, H.; Xu, W.; Peng, J.; Wang, X.; Chen, P.; Li, P.; Lu, F.; Yan, J.; et al. Monitoring Autumn Phenology in Understory Plants with a Fine-Resolution Camera. Remote Sensing 2025, 17, 1025. [Google Scholar] [CrossRef]
- Quille-Mamani, J.; Porras-Jorge, R.; Saravia-Navarro, D.; Herrera, J.; Chavez-Galarza, J.; Arbizu, C.I. Prediction of biometric variables through multispectral images obtained from UAV in beans (Phaseolus vulgaris L.) during ripening stage 2021.
- Beniaich, A.; Silva, M.L.N.; Avalos, F.A.P.; de Menezes, M.D.; Cândido, B.M. Determination of vegetation cover index under different soil management systems of cover plants by using an unmanned aerial vehicle with an onboard digital photographic camera. Semina: Ciências Agrárias 2019, 40, 49–66. [Google Scholar] [CrossRef]
- Ju, Z.; Liu, C.; Yuan, Y.; Wang, Y.; Liu, G. Coloration potential, anthocyanin accumulation, and enzyme activity in fruit of commercial apple cultivars and their F1 progeny. Scientia Horticulturae 1999, 79, 39–50. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.; Burgos-Artizzu, X.; Ribeiro, A. Automatic segmentation of relevant textures in agricultural images. Computers and Electronics in Agriculture 2011, 75, 75–83. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Computers and electronics in agriculture 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Strati, V.; Albéri, M.; Barbagli, A.; Boncompagni, S.; Casoli, L.; Chiarelli, E.; Colla, R.; Colonna, T.; Elek, N.I.; Galli, G.; et al. Advancing Grapevine Disease Detection Through Airborne Imaging: A Pilot Study in Emilia-Romagna (Italy). Remote Sensing 2025, 17, 2465. [Google Scholar] [CrossRef]
- Jiménez-Muñoz, J.C.; Sobrino, J.A.; Plaza, A.; Guanter, L.; Moreno, J.; Martínez, P. Comparison between fractional vegetation cover retrievals from vegetation indices and spectral mixture analysis: Case study of PROBA/CHRIS data over an agricultural area. Sensors 2009, 9, 768–793. [Google Scholar] [CrossRef]
- Banerjee, K.; Dutta, S.; Das, B.; Roy, D.; Sen, S.; Mandal, B.P.; Chatterjee, A. Crop type discrimination through low cost proximal RGB imaging and multivariate analysis. Arabian Journal of Geosciences 2025, 18, 31. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Plant species identification, size, and enumeration using machine vision techniques on near-binary images. In Proceedings of the Optics in Agriculture and Forestry. SPIE, 1993, Vol. 1836, pp. 208–219.
- Hussain, S.; Teshome, F.T.; Tulu, B.B.; Awoke, G.W.; Hailegnaw, N.S.; Bayabil, H.K. Leaf area index (LAI) prediction using machine learning and UAV based vegetation indices. European Journal of Agronomy 2025, 168, 127557. [Google Scholar] [CrossRef]
- Ritchie, G.; Sullivan, D.; Vencill, W.; Bednarz, C.; Hook, J. Sensitivities of normalized difference vegetation index and a green/red ratio index to cotton ground cover fraction. Crop science 2010, 50, 1000–1010. [Google Scholar] [CrossRef]
- Aynalem, H.M.; Righetti, T.L.; Reed, B.M. Non-destructive evaluation of in vitro-stored plants: a comparison of visual and image analysis. In Vitro Cellular & Developmental Biology-Plant 2006, 42, 562–567. [Google Scholar]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. The New Phytologist 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Xiaoqin, W.; Miaomiao, W.; Shaoqiang, W.; Yundong, W. Extraction of vegetation information from visible unmanned aerial vehicle images. Trans. Chin. Soc. Agric. Eng. 2015, 31. [Google Scholar]
- Song, H.; Asghari, M.; Zahedipour-Sheshglani, P.; Aljanabi, S.M.A.K.; Diao, E.; Xiang, X.; Qian, S.; Liang, X. Impact of exogenous cinnamic acid on some quality indices, phenolic compounds, main phenolic biosynthesis enzymes and senescence rate of strawberry fruit. Food Chemistry 2025, 487, 144726. [Google Scholar] [CrossRef]
- Hasanzadeh, B.; Abbaspour-Gilandeh, Y.; Soltani-Nazarloo, A.; Hernández-Hernández, M.; Gallardo-Bernal, I.; Hernández-Hernández, J.L. Non-destructive detection of fruit quality parameters using hyperspectral imaging, multiple regression analysis and artificial intelligence. Horticulturae 2022, 8, 598. [Google Scholar] [CrossRef]
- Paulo, D.J.; Neves, C.M.; Wessel, D.F.; Neves, J.C. WildFruiP: Estimating Fruit Physicochemical Parameters from Images Captured in the Wild. In Proceedings of the Iberoamerican Congress on Pattern Recognition, 2023, pp. 314–326.
- Andrés-Bello, A.; Barreto-Palacios, V.; García-Segovia, P.; Mir-Bel, J.; Martínez-Monzó, J. Effect of pH on color and texture of food products. Food Engineering Reviews 2013, 5, 158–170. [Google Scholar] [CrossRef]
- Akkaya, E. Characterization of Physicochemical, Colour and Textural Properties of Turkish Type Cheeses. Dicle Üniversitesi Veteriner Fakültesi Dergisi 2024, 17, 137–142. [Google Scholar] [CrossRef]
- of Official Analytical Chemists, T.A. Official methods of analysis of the Association of Official Analytical Chemists; Vol. 11, The Association, 2000.
- Sadasivam, S.; Manikam, A. Biochemical Methods. Third Print. New Age International, New Delhi 2010.
- Hedge, J.; Hofreiter, B. Methods in Carbohydrate Chemistry, Vol. 17. Whistler, RL and BeMiller, JN Ed 1962, 17, 420.
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical chemistry 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Ramful, D.; Tarnus, E.; Aruoma, O.I.; Bourdon, E.; Bahorun, T. Polyphenol composition, vitamin C content and antioxidant capacity of Mauritian citrus fruit pulps. Food research international 2011, 44, 2088–2099. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. [14] Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. In Methods in enzymology; Elsevier, 1999; Vol. 299, pp. 152–178.
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food chemistry 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT-Food science and Technology 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Smit, C.; Joslyn, M.; Lukton, A. Determination of tannins and related polyphenols in foods. Analytical Chemistry 1955, 27, 1159–1162. [Google Scholar] [CrossRef]
- Rodriguez-Amaya, D.B.; Raymundo, L.; Lee, T.; Simpson, K.; Chichester, C. Carotenoid pigment changes in ripening Momordica charantia fruits. Annals of Botany 1976, 40, 615–624. [Google Scholar] [CrossRef]



| Methodology | Applicable parameter variables | Total: 159 |
|---|---|---|
| Wet chemistry * | Acidity (titrable), Antioxidant activity, Carbohydrates (total), Carotene (content), Fat crude, Flavonoids (total), Moisture (content), pH, Phenolic (total content), Protein, | |
| Reducing sugars, Tannin (content), Total soluble solids, Total sugars, and Vitamin C (Section 2.2). | Total: 15 | |
| Color grid **,† | Mean R, Mean G, Mean B, Standard deviation of R, Standard deviation of G, Standard deviation of B, Median R, Median G, Median B, Geometric mean R, Geometric | |
| mean G, Geometric mean B, Maximum R, Maximum G, Maximum B, Minimum R, Minimum G, Minimum B, Number of R values, Number of G values, Number of B | ||
| values, Skewness R, Skewness G, Skewness B, Kutosis R, Kutosis G, and Kurtosis B (Section 2.3.3.i). | Total: 27 | |
| Waveness and roughness ** | Descriptive statistics parameters of waveness plot (Number of waveness plot data points, Minimum waveness, Maximum waveness, Mean waveness, Geometric mean | |
| waveness, Standard deviation waveness, Median waveness, Geometric mean waveness, Standard deviation of waveness), Waveness parameters calculated based on | ||
| formulae from references (Arithmetic mean waveness, Root mean square waveness, Maximum valley depth of waveness plot, Maximum peak height of waveness plot, | ||
| Average valley depth of waveness plot, Average peak height of waveness plot, Maximum peak to valley height of waveness plot, Descriptive statistics parameters of | ||
| roughness plot (Number of roughness plot data points, Minimum roughness, Maximum roughness, Mean roughness, Geometric mean roughness, Standard deviation | ||
| roughness, Median roughness), Waveness parameters calculated based on formulas from references (Arithmetic mean roughness, Root mean square roughness, Maximum | ||
| valley depth of roughness plot, Maximum peak height of roughness plot, Average valley depth of roughness plot, Average peak height of roughness curve, and Maximum | ||
| peak to valley height of roughness plot) (Section 2.3.3.ii). | Total: 28 | |
| GLCM ** | Angular second moment, Contrast, Correlation, Inverse difference moment, and Entropy (Section 2.3.3.iii). | Total: 5 |
| LBP **,‡ | Number of gray pixels analyzed, Minimum gray, Maximum gray, Mean gray, Geometric mean gray, Quartile mean gray, Standard deviation gray, Median gray, Skewness | |
| gray, Kurtosis gray, Number of R pixels analyzed, Minimum R, Maximum R, Mean R, Geometric mean R, Quartile mean R, Standard deviation R, Median R, Skewness R, | ||
| Kurtosis R, Number of G pixels analyzed, Minimum G, Maximum G, Mean G, Geometric mean G, Quartile mean G, Standard deviation G, Median G, Skewness G, | ||
| Kurtosis G, Number of B pixels analyzed, Minimum B, Maximum B, Mean B, Geometric mean B, Quartile mean B, Standard deviation B, Median B, Skewness B, and | ||
| Kurtosis B (Section 2.3.3.iv). | Total: 40 | |
| Color indices ** | Color indices derived from overall R, G, and B values (27 existing + 17 new) are given in Table 2. | Total: 44 |
| Vegetation Index | Equation | Reference |
|---|---|---|
| Blue chromatic coordinate (BCC) | [45] | |
| Green-blue difference index (GBDI) | [46] | |
| Blue red vegetation index (BRVI) | [47] | |
| Color index of vegetation (CIVE) | [48,49] | |
| Coloration index (CI) | [50] | |
| Combination1 (COMB1) | [51] | |
| Combination2 (COMB2) | [51] | |
| Excess green (ExG ) | [45,48] | |
| Excess green minus excess red (ExGR) | [48,52] | |
| Excess red (ExR) | [48] | |
| Green-blue vegetation index (GBVI) | [53,54] | |
| Green chromatic coordinate (GCC) | [45] | |
| Green-red difference (GRD) | [46,48] | |
| Green-red ratio index (GRRI) | [48,55] | |
| Kawashima index (IKAW) | [47,48] | |
| Modified green-red vegetation index (MGRVI) | [46,48] | |
| Normalized difference index (NDI) | [56] | |
| Normalized excess green (ExG2) | [45] | |
| Normalized green-red difference index (NGRDI) | [57,58] | |
| Red chromatic coordinate (RCC) | [45] | |
| Red blue ratio index (RBRI) | [59] | |
| Red green ratio index (RGRI) | [60] | |
| Red green blue vegetation index (RGBVI) | [46] | |
| Vegetativen (VEG) | [46] | |
| Visible atmospherically resistant index (VARI) | [61] | |
| Visible-band difference vegetation index (VDVI) | [62] | |
| Woebbecke index (WI) | [45] | |
| Excess modified red (ExmR ) | † | |
| Modified coloration index (MCI) | † | |
| Modified red-blue vegetation index (MRBVI) | † | |
| Modified red-green-blue vegetation index (MRGBVI) | † | |
| Modified red-green vegetation index (MRGVI) | † | |
| Normalized excess red (ExR2) | † | |
| Normalized red-blue difference index (NRBDI) | † | |
| Normalized red-green difference index (NRGDI) | † | |
| Red-blue difference ratio (RBDR) | † | |
| Red color index (RCI) | † | |
| Red-green difference ratio (RGDR) | † | |
| Red-blue difference (RBD) | † | |
| Red-green difference (RGD) | † | |
| Red-green vegetation index (RGVI) | † | |
| Triangular redness index (TRI) | † | |
| Visible atmospherically resistant red-blue index (VARRBI) | † | |
| Visible atmospherically resistant red-green index (VARRGI) | † |
| Wet chemistry | Cashew apple varieties and maturity stages | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bhaskara | MDK | NRCC-1 | Vengurla-2 | VRI-3 | VTH-174 | |||||||
| 817 | 819 | 817 | 819 | 817 | 819 | 817 | 819 | 817 | 819 | 817 | 819 | |
| Acidity (%) | 0.14±0.0 | 0.09±0.01 | 0.12±0.0 | 0.08±0.0 | 0.16±0.0 | 0.12±0.0 | 0.25±0.0 | 0.18±0.0 | 0.16±0.0 | 0.12±0.0 | 0.160±0.0 | 0.11±0.0 |
| Antioxidants (μmol | 28.59±1.13 | 20.85±0.22 | 25.96±0.45 | 8.94±0.11 | 24.39±0.47 | 22.69±0.91 | 26.01±0.83 | 2.88±0.28 | 4.21±0.12 | 11.83±0.87 | 30.47±0.72 | 22.57±0.87 |
| Carbohydrates ( | 14.09±0.93 | 16.35±0.71 | 14.67±0.42 | 17.2±0.69 | 16.94±0.11 | 20.77±0.35 | 16.81±0.24 | 19.24±0.48 | 14.16±1.1 | 15.78±0.41 | 14.67±0.62 | 18.07±0.29 |
| Carotenes (mg | 1.5±0.01 | 1.4±0.01 | 1.41±0.02 | 0.82±0.0 | 1.29±0.0 | 0.34±0.0 | 0.74±0.0 | 1±0.0 | 0.42±0.0 | 0.86±0.0 | 0.57±0.0 | 0.78±0.0 |
| Crude fat (%) | 2.02±0.11 | 1.78±0.04 | 2.73±0.07 | 2.46±0.09 | 5.61±0.04 | 4.59±0.19 | 2.46±0.04 | 2.32±0.04 | 2.23±0.11 | 1.68±0.07 | 5.44±0.01 | 4.66±0.11 |
| Flavonoids (mg | 64.33±13.9 | 106.04±0.0 | 101.49±21.6 | 30.84±2.8 | 25.87±0.4 | 16.78±0.2 | 171.30±56.7 | 71.52±5.7 | 80.24±8.4 | 12.98±3.5 | 218.94±31.8 | 28.47±5.7 |
| MC (% w.b.) | 87.57±0.49 | 85.81±0.56 | 86.42±1 | 85.92±0.8 | 87±0.65 | 85.92±0.32 | 87.31±0.25 | 86±0.21 | 88.89±1.47 | 86.93±0.32 | 86.91±0.28 | 85.26±0.17 |
| pH | 4.56±0.01 | 4.73±0.02 | 4.87±0.02 | 5.23±0.15 | 4.49±0.02 | 4.78±0.01 | 4.29±0.09 | 4.45±0.02 | 4.53±0.01 | 4.69±0.01 | 4.49±0.0 | 4.85±0.04 |
| Phenolics (mg | 311.41±5.1 | 191.77±4.3 | 308.66±6.5 | 134.33±4.0 | 161±7.0 | 124.33±3.8 | 374.7±10.7 | 308.33±6.8 | 318.55±3.5 | 170.49±4.5 | 405.84±4.3 | 243.5±4.8 |
| Proteins (%) | 1.7±0.07 | 1.2±0.04 | 2.16±0.06 | 1.12±0.03 | 2.05±0.07 | 1.18±0.03 | 2.26±0.09 | 1.26±0.04 | 1.78±0.14 | 1.26±0.05 | 2.24±0.09 | 1.45±0.03 |
| Reducing sugars ( | 7.89±0.17 | 6.75±0.38 | 9.33±0.51 | 10.84±0.29 | 14.7±0.59 | 17.68±0.55 | 14.11±0.41 | 16.2±1.34 | 9.04±0.07 | 10.4±0.58 | 12.22±0.71 | 9.7±0.38 |
| Tannins (mg | 2.41±0.11 | 1.65±0.05 | 2.25±0.28 | 0.88±0.03 | 0.91±0.02 | 0.76±0.05 | 2.57±0.07 | 2.36±0.06 | 2.04±0.06 | 1.11±0.03 | 2.53±0.14 | 1.86±0.14 |
| Total sugars ( | 8.62±0.15 | 12.05±0.08 | 10.4±1.27 | 13.07±1.47 | 15.81±0.5 | 19.88±0.6 | 9.92±0.59 | 11.19±0.25 | 15.26±1.26 | 17.81±0.42 | 13.63±0.23 | 17.05±0.26 |
| TSS (°brix) | 2.41±0.11 | 1.65±0.05 | 2.25±0.28 | 0.88±0.03 | 0.91±0.02 | 0.76±0.05 | 2.57±0.07 | 2.36±0.06 | 2.04±0.06 | 1.11±0.03 | 2.53±0.14 | 1.86±0.14 |
| Vitamin C (mg | 98.86±5.1 | 181.71±1.5 | 154.62±3.1 | 252.2±17.4 | 27.5±2.3 | 91.12±0.29 | 197.8±12.3 | 305.5±11.5 | 150.86±2.6 | 305±2.3 | 40.62±2.3 | 69.24±7.6 |
| Selected input | Input combination | change from | |
|---|---|---|---|
| *reference (%) | |||
| Waveness cutoff value | ; 4 pix; 1 pix; 0° | 73.900* | 0.00 |
| ; 5 pix; 1 pix; 0° | 73.900 | 0.00 | |
| ; 10 pix; 1 pix; 0° | 75.003 | 1.49 | |
| ; 15 pix; 1 pix; 0° | 75.505 | 2.17 | |
| ; 20 pix; 1 pix; 0° | 75.805 | 2.58 | |
| ; 25 pix; 1 pix; 0° | 75.941 | 2.76 | |
| ; 30 pix; 1 pix; 0° | 76.014 | 2.86 | |
| ; 35 pix***; 1 pix; 0° | 76.043** | 2.90** | |
| ; 40 pix; 1 pix; 0° | 75.966 | 2.80 | |
| ; 50; pix 1 pix; 0° | 75.966 | 2.80 | |
| GCLM step size | ; 35 pix; 1 pix***; 0° | 76.043** | 2.90** |
| ; 35 pix; 2 pix; 0° | 76.023 | 2.87 | |
| ; 35 pix; 3 pix; 0° | 76.024 | 2.87 | |
| ; 35 pix; 5 pix; 0° | 76.031 | 2.88 | |
| ; 35 pix; 10 pix; 0° | 75.933 | 2.75 | |
| GCLM step direction | ; 35 pix; 1 pix; 0° | 76.043 | 2.90 |
| ; 35 pix; 1 pix; 90° | 76.434 | 3.43 | |
| ; 35 pix; 1 pix; 180° | 76.056 | 2.92 | |
| ; 35 pix; 1 pix; 270°*** | 76.437** | 3.43** | |
| Color grid size | ; 35 pix; 1 pix; 270° | 74.670 | 1.04 |
| †***; 35 pix; 1 pix; 270° | 76.437** | 3.43** | |
| ; 35 pix; 1 pix; 270° | 73.751 |
| Parameters group | N | All samples | Slice samples | Whole samples | |||
|---|---|---|---|---|---|---|---|
| Sum | Mean | Sum | Mean | Sum | Mean | ||
| Wet chemistry | 225 | 99.6 | 0.443 | 99.6 | 0.443 | 99.6 | 0.443 |
| Color grid | 270 | 61.0 | 0.226 | 67.8 | 0.251 | 72.2 | 0.267 |
| Waveness and roughness | 375 | 56.7 | 0.151 | 121.6 | 0.324 | 95.1 | 0.254 |
| GLCM | 75 | 9.6 | 0.128 | 16.0 | 0.213 | 17.5 | 0.234 |
| LBP | 360 | 59.3 | 0.165 | 68.2 | 0.189 | 74.0 | 0.206 |
| Color indices | 660 | 156.5 | 0.237 | 193.0 | 0.292 | 179.7 | 0.272 |
| Total image-based | 1740 | 343.1 | 0.197 | 466.5 | 0.268 | 438.6 | 0.252 |
| Total all methods | 1965 | 442.7 | 0.225 | 566.1 | 0.288 | 538.2 | 0.274 |
| Wet chemistry | Ranks of image-based independent variables based on correlation (r) values | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| properties | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Slice | ||||||||||
| Acidity | Wn-avPH | Wn-mxPH | Wn-amWn | Rn-amRn | Rn-xpvh | Rn-avPH | Rn-med | Wn-Gmn | Ov-stdR | Rn-mxVD |
| Antioxidants | Rn-med | Wn-med | Rn-avPH | Wn-mxVD | Wn-mn | Gl-corr | Rn-avVD | Wn-Gmn | Ov-medG | Wn-mxPH |
| Carbohydrates | Ov-skB | Wn-xPt | Rn-mn | Rn-avPH | Rn-xpvh | Wn-med | Gl-corr | Gl-cont | Rn-avVD | Wn-avPH |
| Carotenes | VDVI | GCC | ExG2 | VEG | COMB1 | COMB2 | ExG | CIVE | RGDR | Wn-avPH |
| Crude fat | Gl-cont | Rn-mxPHt | Rn-mx | L-GmnR | L-GmnG | L-GmnGr | Gl-IDM | L-GmnB | Rn-mi | Rn-mxVD |
| Flavonoids | Rn-mxPH | Rn-mx | Gl-cont | Rn-med | Wn-Gmn | L-GmnR | Gl-IDM | RGBVI | Rn-mi | L-GmnG |
| MC | Wn-avPH | Wn-mxPH | Rn-med | Wn-amWn | Rn-amRn | Ov-stdR | Ov-stdB | Rn-xpvh | Ov-medR | Ov-GmnG |
| pH | Wn-med | GRD | RGD | Ov-medG | RBRI | ExmR | TRI | RCC | ExR2 | BRVI |
| Phenolics | Rn-mx | Rn-mxPH | Rn-mi | Rn-mxVD | Ov-mnG | Rn-xpvh | Rn-amRn | Rn-std | VEG | Ov-GmnG |
| Proteins | Rn-mx | Rn-mxPH | Rn-med | Wn-Gmn | Ov-mnG | Rn-avPH | Wn-mxVD | Ov-GmnG | Rn-xpvh | Wn-mi |
| Reducing sugars | Wn-avPH | Wn-mxPH | Wn-Gmn | Ov-medB | Rn-xpvh | ExGR | Ov-medG | VARI | VARRGI | RGRI |
| Tannins | Wn-avPH | Wn-mxPH | Wn-mi | Rn-xpvh | Wn-amWn | Rn-amRn | Ov-skB | Wn-rsWn | Wn-std | Wn-Gmn |
| Total Sugars | Ov-medG | RBRI | BRVI | IKAW | NRBDI | BCC | CI | MRBVI | MRGBVI | RCC |
| TSS | Wn-avPH | Wn-amWn | Rn-amRn | Wn-Gmn | VEG | VDVI | GCC | ExG2 | COMB1 | COMB2 |
| Vitamin C | Wn-avVD | Wn-amWn | Rn-amRn | Ov-stdR | Wn-avPH | Ov-GmnR | Wn-rsWn | Wn-std | Ov-mnR | Ov-stdB |
| Whole | ||||||||||
| Acidity | Gl-cont | Ov-skR | Ov-skG | Ov-skB | Rn-mn | Ov-medR | Ov-stdR | Ov-stdB | VEG | GCC |
| Antioxidants | Gl-cont | Ov-GmnB | Gl-IDM | L-QmnB | L-mnB | L-skB | L-kuB | L-stdB | Wn-avVD | RGBVI |
| Carbohydrates | Rn-mxPH | Rn-mx | Gl-IDM | Wn-mi | Wn-mxVD | Wn-avVD | Wn-amWn | Rn-amRn | L-GmnG | L-GmnGr |
| Carotenes | Wn-xpvh | Ov-mnG | Ov-skB | Wn-rsWn | Wn-std | Ov-GmnB | Wn-amWn | Rn-amRn | Wn-mx | Wn-avVD |
| Crude fat | L-GmnG | L-GmnGr | Gl-corr | Rn-med | Wn-mn | Wn-mx | Wn-Gmn | Gl-ASM | L-GmnR | L-GmnB |
| Flavonoids | Rn-rsRn | Rn-std | Rn-med | Rn-mxPH | Rn-mx | Rn-avVD | Gl-cont | Ov-medR | Wn-avPH | Wn-mn |
| MC | Gl-cont | Ov-skR | Ov-medR | Ov-stdB | Ov-stdR | Ov-GmnR | Ov-mnR | GCC | ExG2 | VDVI |
| pH | Ov-medG | Ov-mnB | CIVE | ExG | Ov-GmnB | WI | RBDR | COMB1 | COMB2 | RGDR |
| Phenolics | Ov-skB | Ov-mnG | Ov-GmnG | Wn-avPH | Ov-GmnB | Ov-stdR | Wn-rsWn | Wn-std | Wn-amWn | Rn-amRn |
| Proteins | Gl-cont | Ov-mnG | Ov-GmnG | Ov-GmnB | Rn-mxPH | Rn-mx | Ovr-medR | Rn-avVD | Rn-std | Rn-rsRn |
| Reducing sugars | Wn-xpvh | Ov-GmnB | Wn-mxVD | Wn-mx | Ov-skG | Ov-mnG | Ov-mnB | CIVE | ExG | Wn-rsWn |
| Tannins | Wn-xpvh | Rn-mx | Rn-mxPH | Wn-mxVD | Ov-GmnB | Wn-mx | Ov-medB | CIVE | Ov-mnB | ExG |
| Total Sugars | Ov-GmnB | Ov-mnB | CIVE | ExG | L-kuB | Ov-medG | COMB2 | COMB1 | L-skB | VDVI |
| TSS | Ov-skB | Ov-skG | Wn-xpvh | Wn-rsWn | Wn-std | Wn-avVD | Wn-amWn | Rn-amRn | Ov-mnG | Wn-mx |
| Vitamin C | Ov-skR | Rn-mn | Ov-stdR | Ov-skB | Rn-mi | Rn-mxVD | Ov-GmnR | Ov-mnR | Ov-medR | Ov-stdB |
| Wet chem. (y) | x | Model () | x | Model () | Wet chem. (y) | x | Model () | x | Model () |
|---|---|---|---|---|---|---|---|---|---|
| Slice | |||||||||
| Stage = 817 | Stage = 819 | Stage = 817 | Stage = 819 | ||||||
| Acidity | Ov-medR | WI | Antioxidant | VDVI | VEG | ||||
| Ov-mnR | RBDR | GCC | Ov-mnG | ||||||
| Ov-GmnR | Ov-medG | ExG2 | COMB2 | ||||||
| Carbohydrates | Ov-stdR | Ov-skG | Carotenoids | VEG | Rn-xpH | ||||
| Ov-GmnB | Ov-kuB | COMB1 | Rn-mx | ||||||
| Ov-stdB | Ov-stdR | COMB2 | Rn-mpvh | ||||||
| Crude fat | VEG | Gl-cont | Flavonoids | ExmR | VEG | ||||
| COMB2 | Rn-mx | TRI | COMB2 | ||||||
| COMB1 | Rn-xpH | RBD | COMB1 | ||||||
| MC | Ov-kuB | Rn-mx | pH | RBD | Ov-GmnB | ||||
| RBD | Rn-mxPH | TRI | CIVE | ||||||
| TRI | Rn-mpvh | ExmR | ExG | ||||||
| Phenolics | GRD | VEG | Proteins | Ov-GmnB | Ov-kuR | ||||
| RGD | COMB2 | Ov-stdB | Ov-kuG | ||||||
| ExmR | COMB1 | Ov-mnB | Ov-medR | ||||||
| R-sugars | Ov-stdR | Ov-skG | Tannins | VDVI | VEG | ||||
| Ov-stdB | Ov-stdR | GCC | COMB2 | ||||||
| Ov-GmnG | Ov-stdB | ExG2 | COMB1 | ||||||
| T-sugars | Ov-stdR | Ov-skG | TSS | VEG | Rn-mx | ||||
| Ov-stdB | Ov-medR | COMB1 | Rn-mxPH | ||||||
| Ov-GmnG | Ov-stdB | COMB2 | Rn-mpvh | ||||||
| Vitamin C | L-GmnR | Rn-mpvh | |||||||
| L-GmnGr | Rn-mx | ||||||||
| L-GmnB | Rn-mxPH | ||||||||
| Whole | |||||||||
| Stage = 817 | Stage = 819 | Stage = 817 | Stage = 819 | ||||||
| Acidity | WI | Ov-medG | Antioxidant | Ov-skG | Ov-skG | ||||
| RBDR | Ov-GmnB | Wn-mxPH | Wn-avVD | ||||||
| RGDR | Ov-mnB | Wn-mx | Wn-mxVD | ||||||
| Carbohydrates | Ov-skG | Gl-cont | Carotenoids | Ov-GmnG | Ov-skG | ||||
| Ov-kuB | COMB1 | Ov-stdR | Wn-mxVD | ||||||
| Ov-stdR | COMB2 | Ov-stdG | Wn-xpvh | ||||||
| Crude fat | Ov-mnG | VEG | Flavonoids | Wn-xpvh | Ov-kuB | ||||
| Ov-GmnG | L-GmnG | Ov-kuR | VEG | ||||||
| Gl-cont | L-GmnGr | WI | Wn-med | ||||||
| MC | Gl-cont | RGBVI | pH | WI | Ov-GmnB | ||||
| Ov-kuB | Gl-cont | RBDR | Ov-medG | ||||||
| Ov-medG | Ov-GmnB | RGDR | Ov-mnB | ||||||
| Phenolics | Wn-mx | Ov-skG | Proteins | Gl-cont | Ov-kuB | ||||
| Wn-xpvh | Ov-kuB | Ov-stdR | Ov-kuR | ||||||
| Ov-kuR | Wn-mxVD | Ov-stdB | WI | ||||||
| R-sugars | Ov-skG | Ov-stdR | Tannins | Wn-mx | Ov-skG | ||||
| Ov-stdR | RBRI | Wn-mxPH | Wn-avVD | ||||||
| Ov-medR | Ov-medG | Ov-mnG | Wn-mxVD | ||||||
| T-sugars | Ov-skG | VEG | TSS | Gl-cont | Gl-cont | ||||
| Ov-stdR | COMB1 | Ov-mnG | Ov-skG | ||||||
| Ov-medR | COMB2 | Ov-GmnG | Ov-medR | ||||||
| Vitamin C | Rn-avVD | Wn-Gmn | |||||||
| Wn-mn | Wn-mn | ||||||||
| Wn-Gmn | Wn-mxVD | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





