Submitted:
07 October 2025
Posted:
08 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
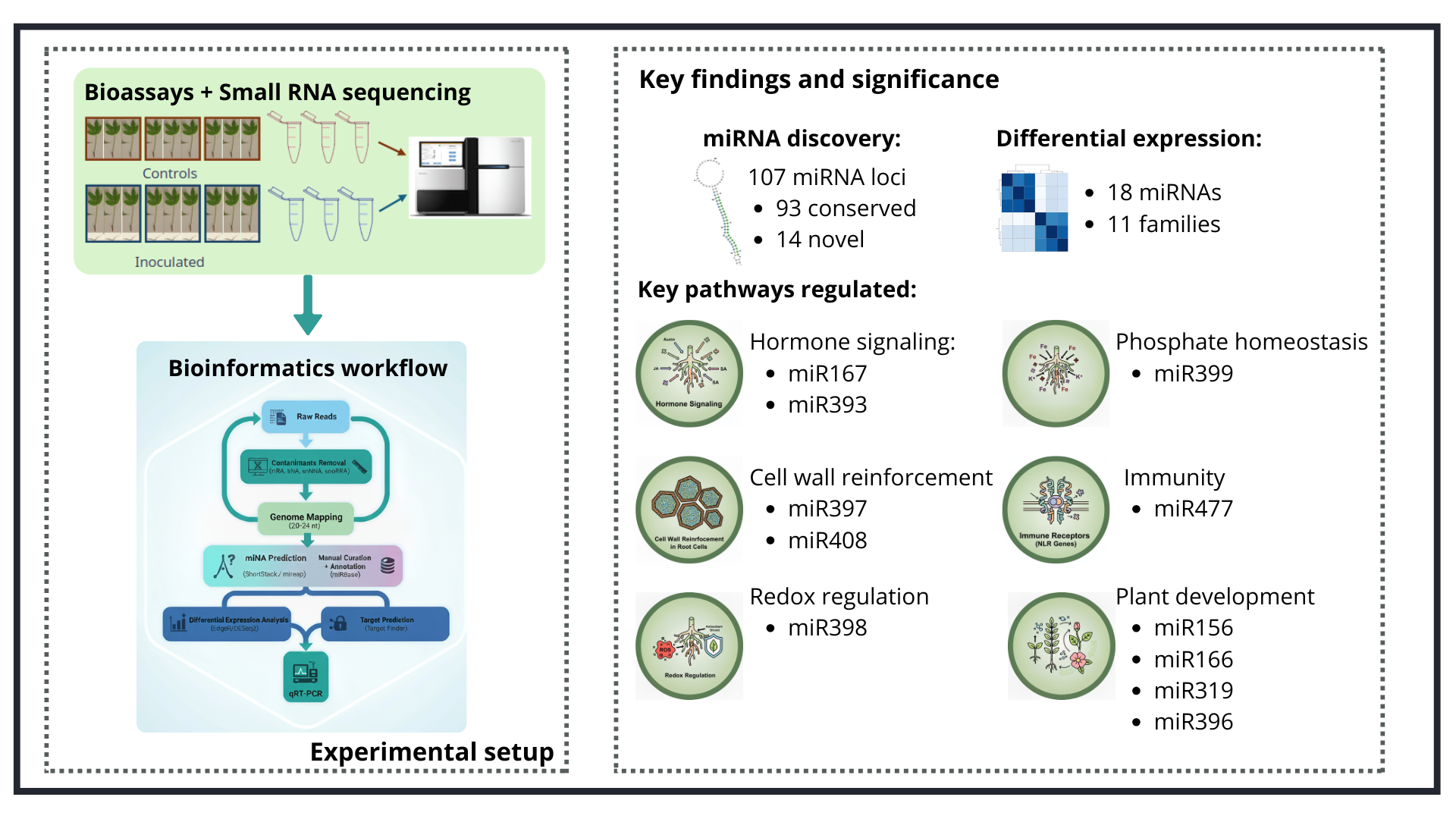
2. Results
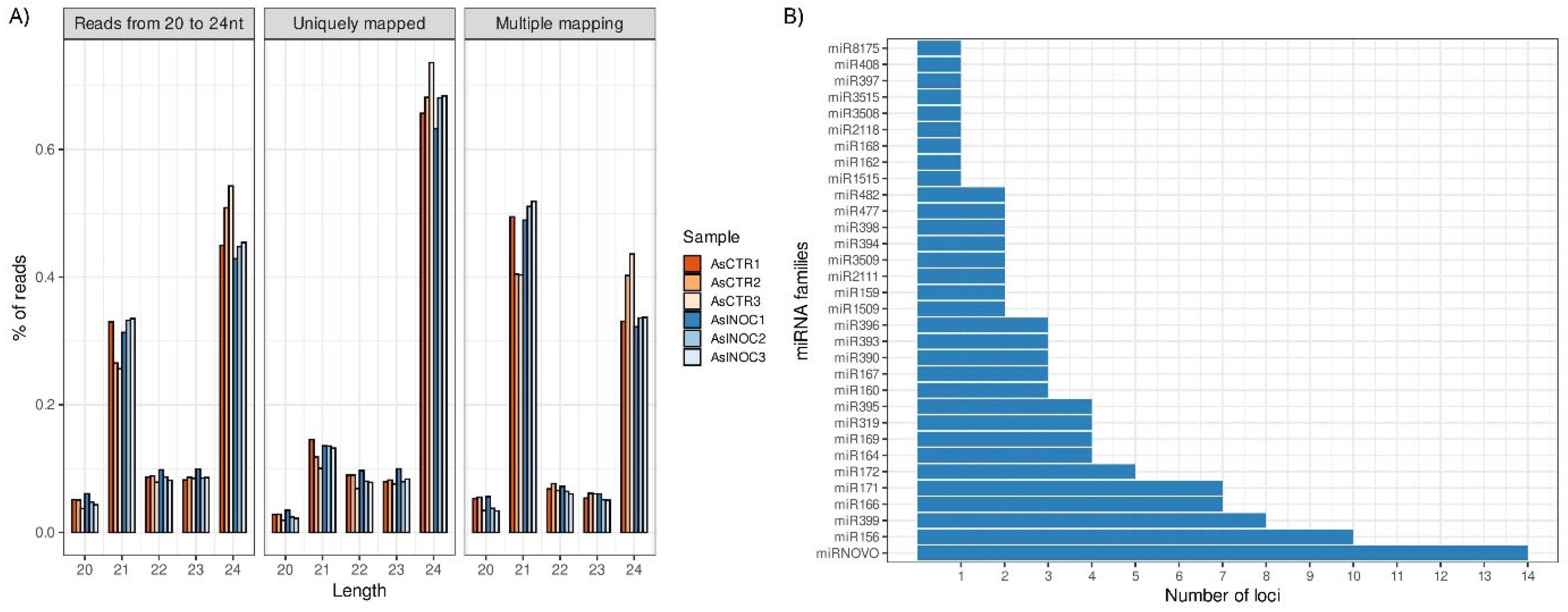
2.1. miRNA Sequencing and Analysis
2.2. A. stenosperma miRNAs Expression During M. arenaria Infection
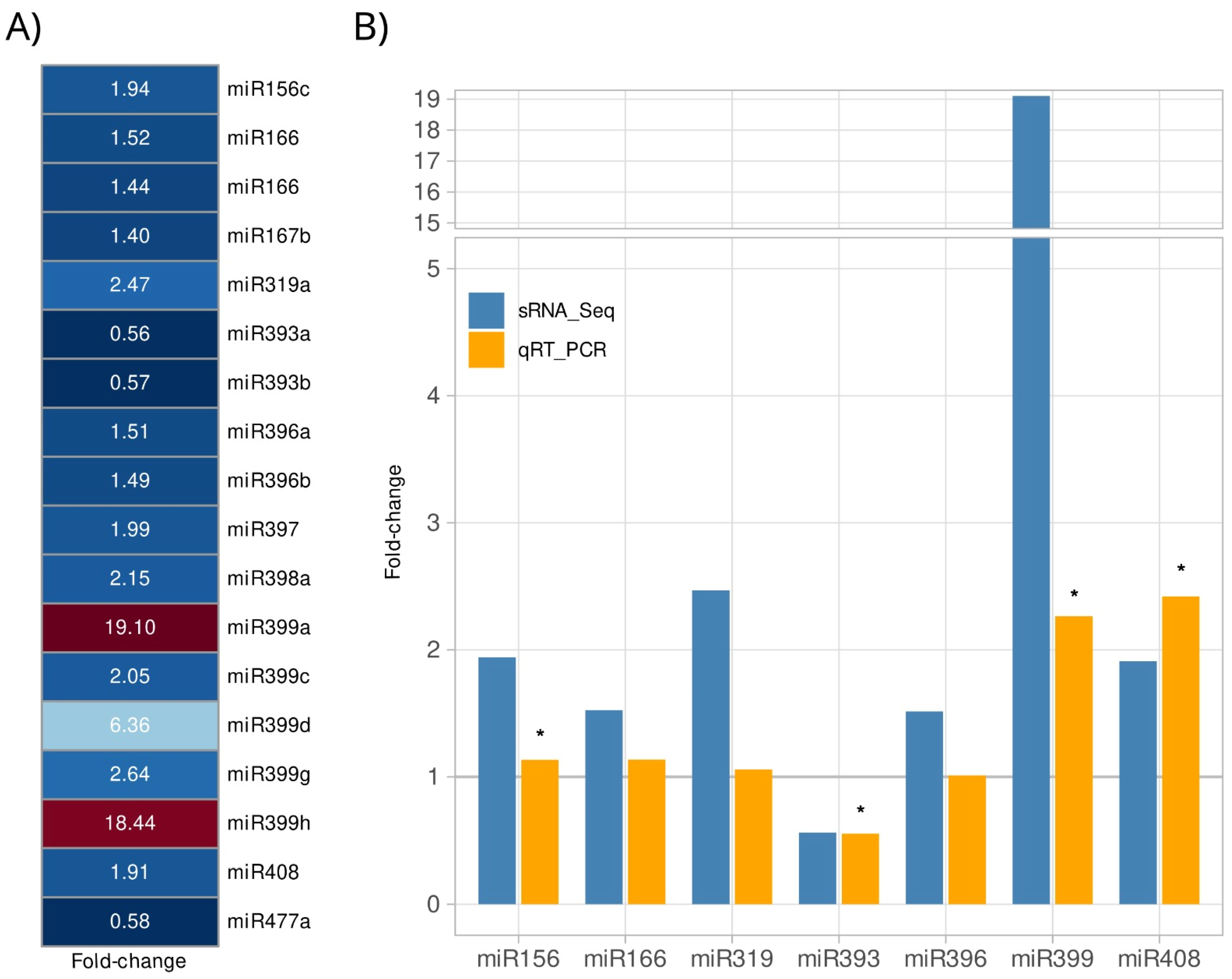
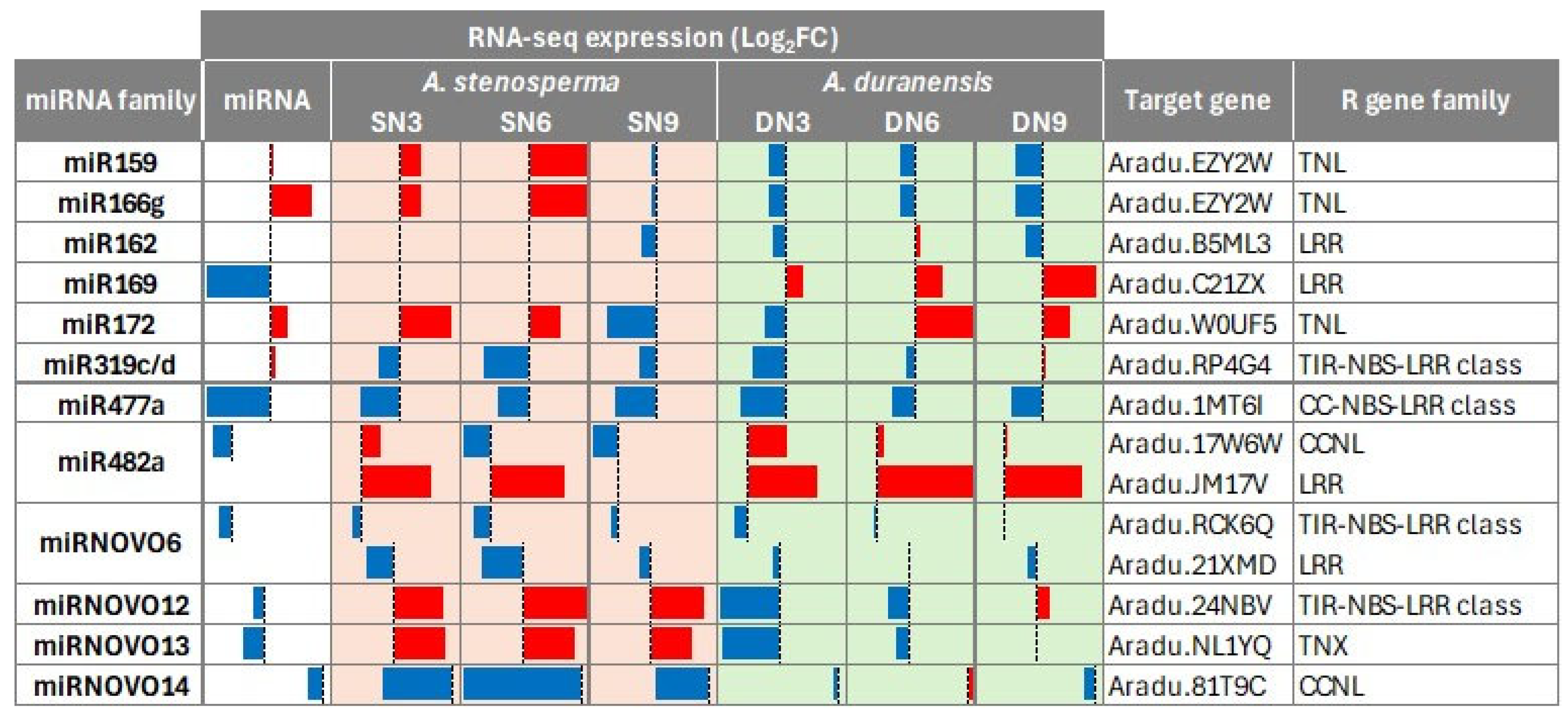
2.3. A. stenosperma Differentially Expressed miRNAs (DEMs) in Response to M. arenaria Infection
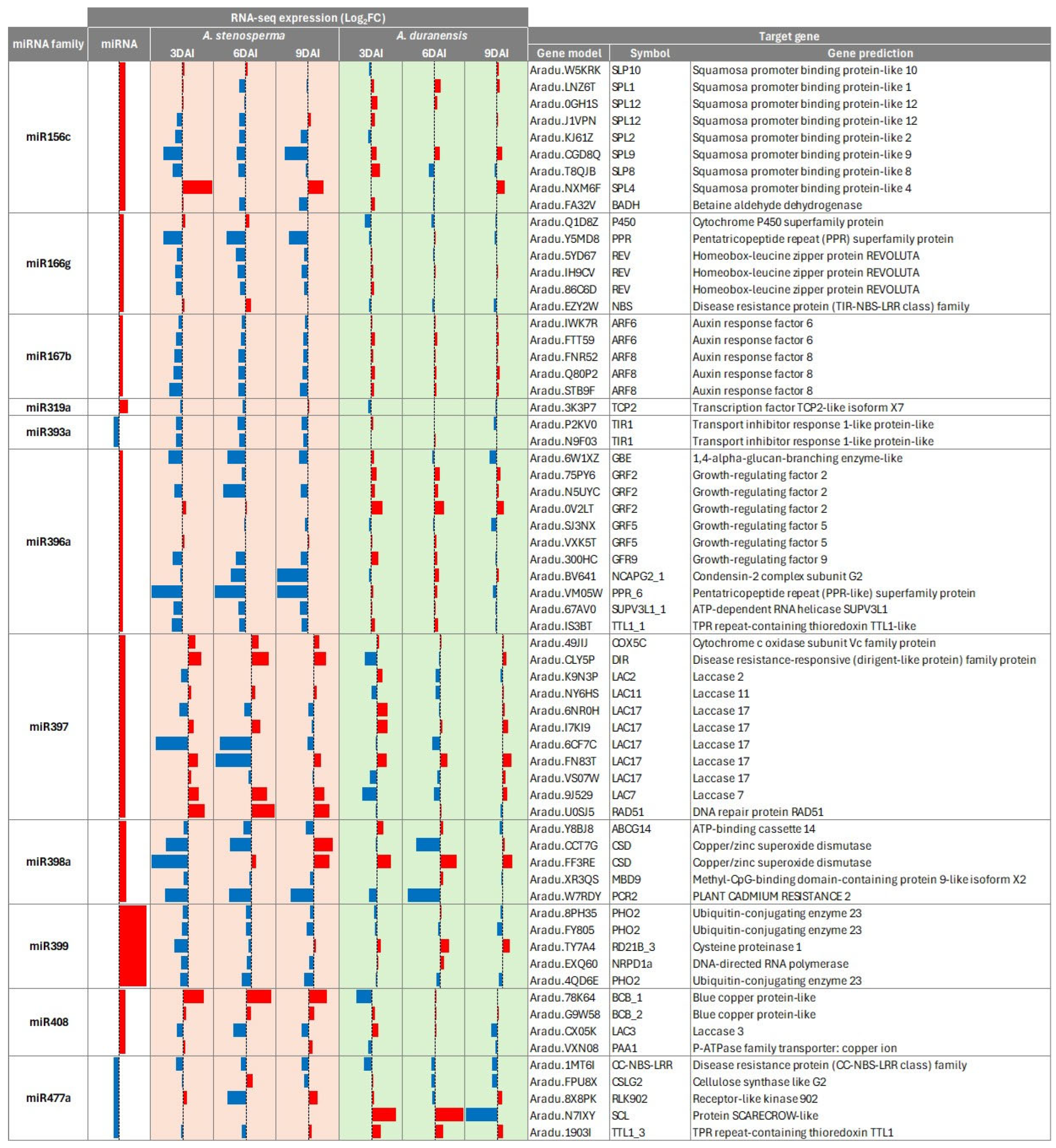
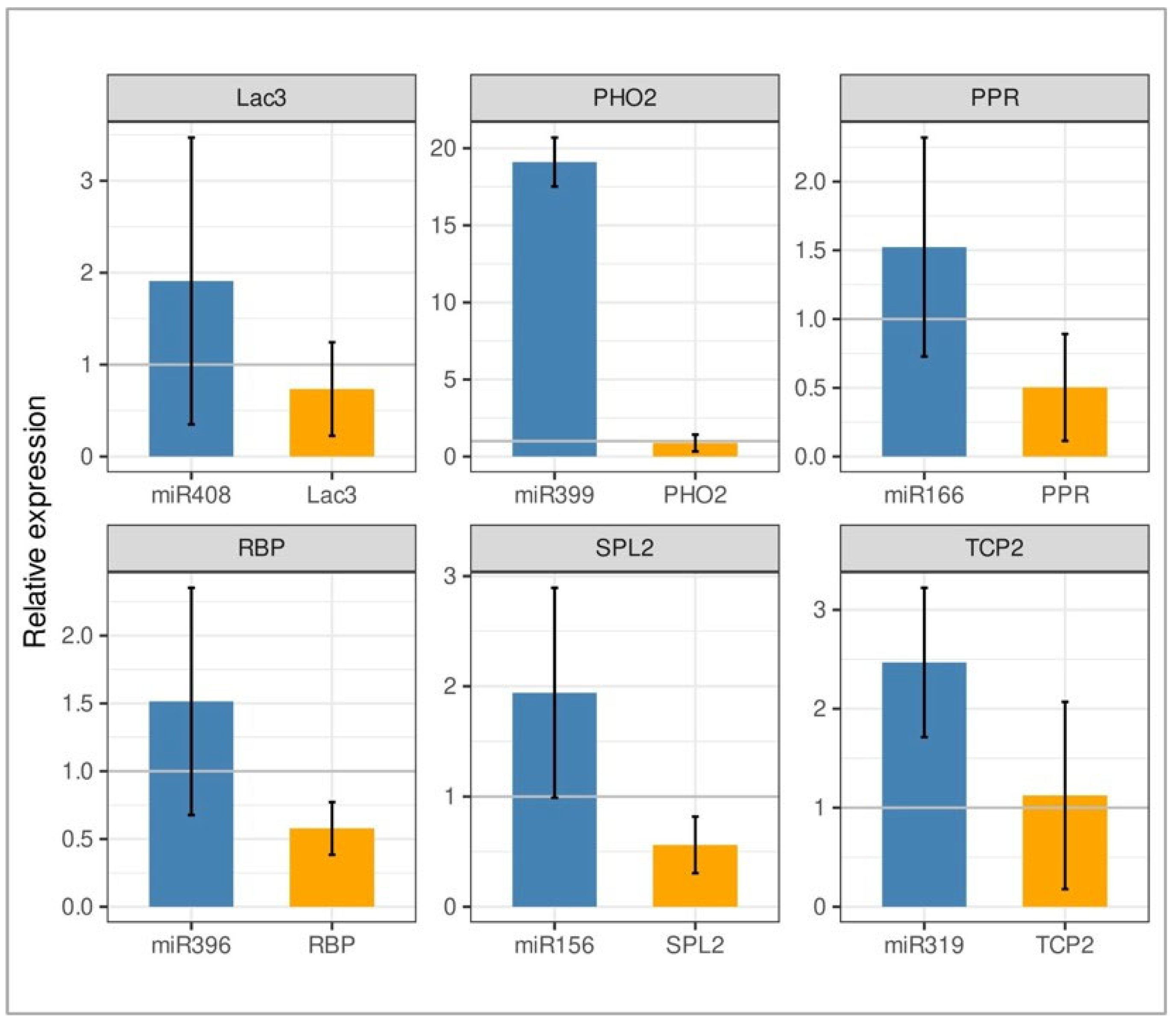
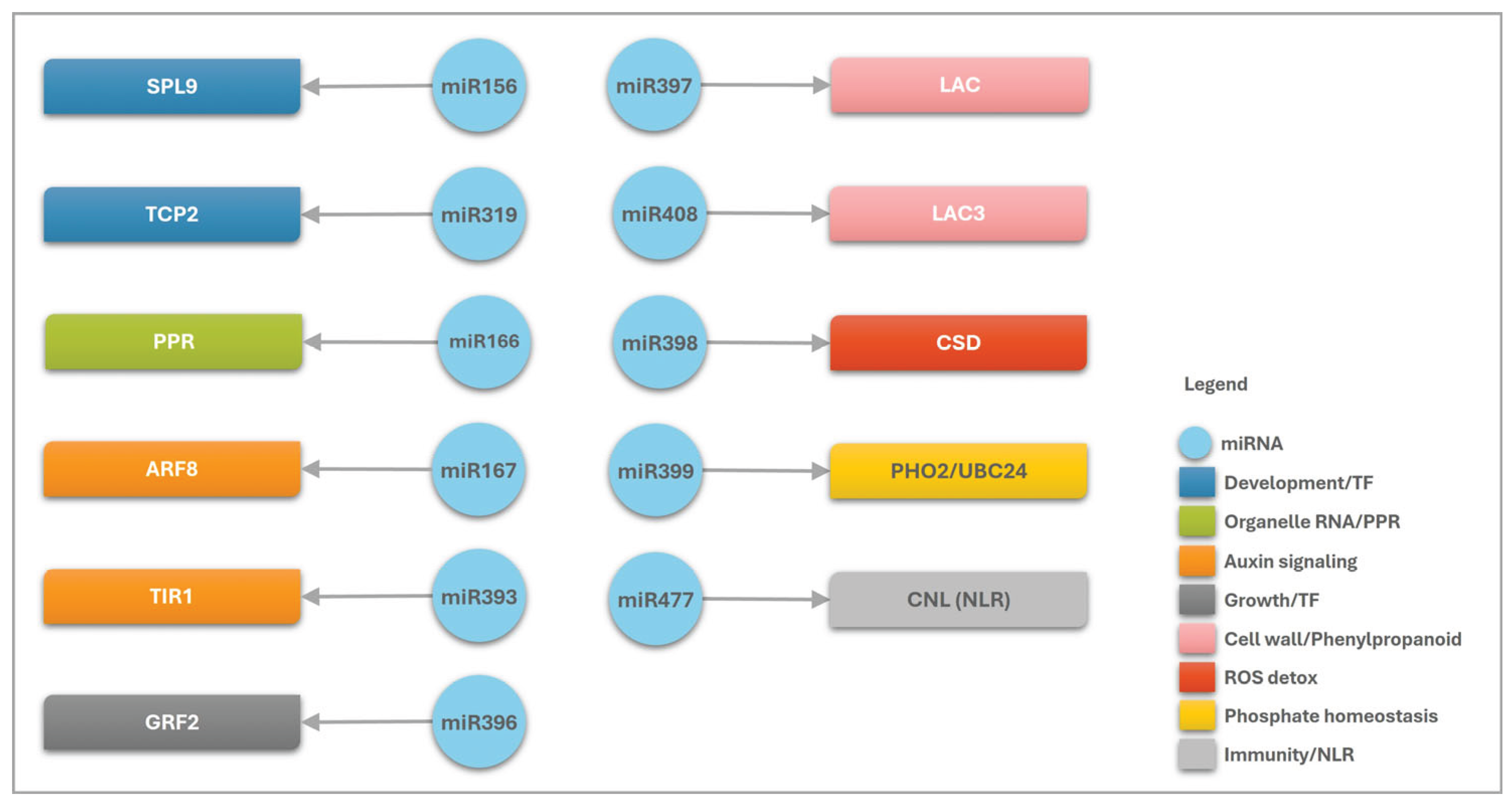
2.4. Correlation of Expression Profiles Between miRNAs and Their Target Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Nematode Bioassays
4.2. miRNA Extraction and Sequencing
4.3. miRNA Identification and Target Prediction
4.4. Differentially Expressed miRNAs (DEMs)
4.5. Expression Analysis by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNAs | MicroRNAs |
| DEMs | Differentially Expressed MicroRNAs |
| NLR | Nucleotide-binding site Leucine-Rich |
| RKN | Root-Knot Nematode |
| mRNA | Messenger RNA |
| PR | Pathogenesis-related genes |
| R | Resistance genes |
| AsCTR | Control samples (not inoculated) of Arachis stenosperma |
| AsINOC | Inoculated samples of Arachis stenosperma |
| Mare | Sample of Meloidogyne arenaria |
| Mb | Million base pairs |
| nt | Nucleotides |
| miRNOVO | New miRNA candidates |
| qRT-PCR | Quantitative Reverse Transcription-Polymerase Chain Reaction |
| RNA-Seq | RNA sequencing |
| DAI | Days After Inoculation |
| MLP | Major Latex Protein |
| MLO | Mildew Locus O |
| TF | Transcription Factor |
| SPL | Squamosa Promoter-Binding Protein-Like |
| HD-ZIP | Homeodomain-leucine Zipper |
| FC | Fold-change |
| TCP2 | Teosinte branched1/Cycloidea/Proliferating cell factor |
| PHO2 | Phosphatase2 |
| DEG | Differentially Expressed Gene |
| UV | Ultraviolet |
| TIR-NBS-LRR | Toll-Interleukin Receptor /Nucleotide-Binding Site/Leucine-Rich Repeat |
| PPR | Pentatricopeptide Repeat |
| CN | Cyst Nematode |
| GRF | Growth Regulating Factor |
| Pi | Phosphate |
| ROS | Reactive Oxygen Species |
| PAMP | Pathogen-Associated Molecular Patterns |
| CCNL | Cyclin L |
| phasiRNA | Phased secondary small interfering RNAs |
| TNL | TIR-NBS-LRR |
| MYB | Myeloblastosis |
| TNX | Toll-Interleukin Receptor /Nucleotide-Binding Site without LRR domain |
| AP2 | APETALA |
| amiRNA | Artificial miRNA |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| cv | Cultivate |
References
- Islam, W.; Islam, S. ul; Qasim, M.; Wang, L. Host-Pathogen Interactions Modulated by Small RNAs. RNA Biol 2017, 14, 891–904. [Google Scholar] [CrossRef]
- Balmer, D.; Mauch-Mani, B. Small Yet Mighty - MicroRNAs in Plant-Microbe Interactions. MicroRNA 2013, 2, 73–80. [Google Scholar] [CrossRef]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; et al. MicroRNAs Inhibit the Translation of Target MRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell 2013, 153, 562–574. [Google Scholar] [CrossRef]
- Subramanian, S.; Fu, Y.; Sunkar, R.; Barbazuk, W.B.; Zhu, J.K.; Yu, O. Novel and Nodulation-Regulated MicroRNAs in Soybean Roots. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Chand Jha, U.; Nayyar, H.; Mantri, N.; Siddique, K.H.M. Non-Coding RNAs in Legumes: Their Emerging Roles in Regulating Biotic/Abiotic Stress Responses and Plant Growth and Development. Cells 2021, Vol. 10, Page 1674 2021, 10, 1674. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Fang, Y.; Chen, L.; Wang, J.; Chen, X. Role of Non-Coding RNAs in Plant Immunity. Plant Commun 2021, 2, 100180. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A New Target for Improving Plant Tolerance to Abiotic Stress. J Exp Bot 2015, 66, 1749. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Figueroa, B.E.; Gao, L.; Diop, N.N.; Wu, Z.; Ehlers, J.D.; Roberts, P.A.; Close, T.J.; Zhu, J.K.; Liu, R. Identification and Comparative Analysis of Drought-Associated MicroRNAs in Two Cowpea Genotypes. BMC Plant Biol 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Shi, Y.; Ntambiyukuri, A.; Li, X.; Zhan, J.; Wang, A.; Xiao, D.; He, L. Integration of Small RNA and Degradome Sequencing Reveals the Regulatory Network of Al-Induced Programmed Cell Death in Peanut. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Anco, D.J.; Thomas, J.S.; Jordan, D.L.; Shew, B.B.; Monfort, W.S.; Mehl, H.L.; Small, I.M.; Wright, D.L.; Tillman, B.L.; Dufault, N.S.; et al. Peanut Yield Loss in the Presence of Defoliation Caused by Late or Early Leaf Spot. Plant Dis 2020, 104, 1390–1399. [Google Scholar] [CrossRef]
- Biswal, A.K.; Ozias-Akins, P.; Holbrook, C.C. Recent Technological Advancements for Identifying and Exploiting Novel Sources of Pest and Disease Resistance for Peanut Improvement. Agronomy 2024, Vol. 14, Page 3071 2024, 14, 3071. [Google Scholar] [CrossRef]
- Guimaraes, P.M.; Guimaraes, L.A.; Morgante, C. V.; Silva, O.B.; Araujo, A.C.G.; Martins, A.C.Q.; Saraiva, M.A.P.; Oliveira, T.N.; Togawa, R.C.; Leal-Bertioli, S.C.M.; et al. Root Transcriptome Analysis of Wild Peanut Reveals Candidate Genes for Nematode Resistance. PLoS One 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Mota, A.P.Z.; Vidigal, B.; Danchin, E.G.J.; Togawa, R.C.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Araujo, A.C.G.; Brasileiro, A.C.M.; Guimaraes, P.M. Comparative Root Transcriptome of Wild Arachis Reveals NBS-LRR Genes Related to Nematode Resistance. BMC Plant Biol 2018, 18, 1–16. [Google Scholar] [CrossRef]
- Kumar, D.; Kirti, P.B. The Genus Arachis: An Excellent Resource for Studies on Differential Gene Expression for Stress Tolerance. Front Plant Sci 2023, 14, 1275854. [Google Scholar] [CrossRef]
- Brasileiro, A.C.M.; Lacorte, C.; Pereira, B.M.; Oliveira, T.N.; Ferreira, D.S.; Mota, A.P.Z.; Saraiva, M.A.P.; Araujo, A.C.G.; Silva, L.P.; Guimaraes, P.M. Ectopic Expression of an Expansin-like B Gene from Wild Arachis Enhances Tolerance to Both Abiotic and Biotic Stresses. Plant Journal 2021, 107, 1681–1696. [Google Scholar] [CrossRef]
- Leal-Bertioli, S.C.M.; Moretzsohn, M.C.; Roberts, P.A.; Ballén-Taborda, C.; Borba, T.C.O.; Valdisser, P.A.; Vianello, R.P.; Araújo, A.C.G.; Guimarães, P.M.; Bertioli, D.J. Genetic Mapping of Resistance to Meloidogyne Arenaria in Arachis Stenosperma: A New Source of Nematode Resistance for Peanut. G3: Genes, Genomes, Genetics 2016, 6, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, P.M.; Brasileiro, A.C.M.; Mehta, A.; Araujo, A.C.G. Functional Genomics in Peanut Wild Relatives. 2017, 149–164. [CrossRef]
- Shen, Y.; Liu, Y.H.; Zhang, X.J.; Sha, Q.; Chen, Z.D. Gynophore MiRNA Analysis at Different Developmental Stages in Arachis Duranensis. Genetics and Molecular Research 2016, 15. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Q.; Chen, K.; Zhao, S.; Zhang, C.; Pan, R.; Cai, T.; Deng, Y.; Wang, X.; Chen, Y.; et al. Integrated MicroRNA and Transcriptome Profiling Reveals a MiRNA-Mediated Regulatory Network of Embryo Abortion under Calcium Deficiency in Peanut (Arachis Hypogaea L.). BMC Genomics 2019 20:1 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Gao, C.; Wang, P.; Zhao, S.; Zhao, C.; Xia, H.; Hou, L.; Ju, Z.; Zhang, Y.; Li, C.; Wang, X. Small RNA Profiling and Degradome Analysis Reveal Regulation of MicroRNA in Peanut Embryogenesis and Early Pod Development. BMC Genomics 2017, 18, 1–18. [Google Scholar] [CrossRef]
- Li, J.; Ma, Y.; Hu, M.; Zhao, Y.; Liu, B.; Wang, C.; Zhang, M.; Zhang, L.; Yang, X.; Mu, G. Multi-Omics and MiRNA Interaction Joint Analysis Highlight New Insights Into Anthocyanin Biosynthesis in Peanuts (Arachis Hypogaea L.). Front Plant Sci 2022, 13. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Zhao, K.; Li, F.; Li, K.; Ning, L.; He, J.; Xin, Z.; Yin, D. Small RNA and Degradome Deep Sequencing Reveals the Roles of MicroRNAs in Seed Expansion in Peanut (Arachis Hypogaea L.). Front Plant Sci 2018, 9, 330144. [Google Scholar] [CrossRef]
- Zhao, C.; Xia, H.; Cao, T.; Yang, Y.; Zhao, S.; Hou, L.; Zhang, Y.; Li, C.; Zhang, X.; Wang, X. Small RNA and Degradome Deep Sequencing Reveals Peanut MicroRNA Roles in Response to Pathogen Infection. Plant Mol Biol Report 2015, 33, 1013–1029. [Google Scholar] [CrossRef]
- Zhao, C.; Li, T.; Zhao, Y.; Zhang, B.; Li, A.; Zhao, S.; Hou, L.; Xia, H.; Fan, S.; Qiu, J.; et al. Integrated Small RNA and MRNA Expression Profiles Reveal MiRNAs and Their Target Genes in Response to Aspergillus Flavus Growth in Peanut Seeds. BMC Plant Biol 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Joshi, P.; Sharma, V.; Pandey, A.K.; Nayak, S.N.; Bajaj, P.; Sudini, H.K.; Sharma, S.; Varshney, R.K.; Pandey, M.K. Identification of MiRNAs Associated with Aspergillus Flavus Infection and Their Targets in Groundnut (Arachis Hypogaea L.). BMC Plant Biol 2025, 25, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, X.; Hou, R.; Zhang, X.; Li, S.; Yue, F.; Zhang, X.; Zhu, X. Multiple MicroRNAs Are Involved in Regulating Peanut (Arachis Hypogaea L.) Resistance to Sclerotium Rolfsii at the Early Stage. Trop Plant Biol 2022, 15, 276–287. [Google Scholar] [CrossRef]
- Xu, P.; Li, H.; Wang, X.; Zhao, G.; Lu, X.; Dai, S.; Cui, X.; Yuan, M.; Liu, Z. Integrated Analysis of the LncRNA/CircRNA-MiRNA-MRNA Expression Profiles Reveals Novel Insights into Potential Mechanisms in Response to Root-Knot Nematodes in Peanut. BMC Genomics 2022, 23, 1–12. [Google Scholar] [CrossRef]
- Li, F.; Han, Y.; Zhang, Z.; Zhang, B.; Zhou, Y.; Li, L.; Li, Q.; Davis, K.E.; Patterson, C.; Oo, S.; et al. Response of Root Growth and Development to Nitrogen and Potassium Deficiency as Well as MicroRNA-Mediated Mechanism in Peanut (Arachis Hypogaea L.). Front Plant Sci 2021, 12, 695234. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Dhingra, A.; Dawar, P.; Payton, P.; Rock, C.D. The Role of MicroRNAs in Responses to Drought and Heat Stress in Peanut (Arachis Hypogaea). Plant Genome 2023, 16. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, C.; Xue, Y.; Tian, Y.; Zhang, H.; Li, N.; Sheng, C.; Jiang, H.; Bai, D. Small RNA and Degradome Deep Sequencing Reveals the Roles of MicroRNAs in Peanut (Arachis Hypogaea L.) Cold Response. Front Plant Sci 2022, 13, 920195. [Google Scholar] [CrossRef]
- Garg, V.; Agarwal, G.; Pazhamala, L.T.; Nayak, S.N.; Kudapa, H.; Khan, A.W.; Doddamani, D.; Sharma, M.; Kavi Kishor, P.B.; Varshney, R.K. Genome-Wide Identification, Characterization, and Expression Analysis of Small RNA Biogenesis Purveyors Reveal Their Role in Regulation of Biotic Stress Responses in Three Legume Crops. Front Plant Sci 2017, 8, 238864. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nature Genetics 2016 48:4 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yang, Q.; Chen, X.; Wang, J.; Pan, L.; Chen, M.; Yang, Z.; He, Y.; Liang, X.; Yu, S. Identification and Characterization of MicroRNAs from Peanut (Arachis Hypogaea L.) by High-Throughput Sequencing. PLoS One 2011, 6, e27530. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A Highly Sensitive RT-PCR Method for Detection and Quantification of MicroRNAs. Plant Methods 2007, 3, 1–12. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, N.; Tian, C.; Wen, S.; Zhang, C.; Zheng, A.; Hu, X.; Fang, J.; Zhang, Z.; Lai, Z.; et al. The MiR166 Targets CsHDZ3 Genes to Negatively Regulate Drought Tolerance in Tea Plant (Camellia Sinensis). Int J Biol Macromol 2024, 264, 130735. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, MicroRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants. Plant Physiol 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Huang, D.; Guo, Z.; Kuang, Z.; Zhang, H.; Xie, X.; Ma, Z.; Gao, S.; Lerdau, M.T.; Chu, C.; et al. Overexpression of MicroRNA408 Enhances Photosynthesis, Growth, and Seed Yield in Diverse Plants. J Integr Plant Biol 2018, 60, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Mota, A.P.Z.; Brasileiro, A.C.M.; Vidigal, B.; Oliveira, T.N.; da Cunha Quintana Martins, A.; Saraiva, M.A. de P.; de Araújo, A.C.G.; Togawa, R.C.; Grossi-de-Sá, M.F.; Guimaraes, P.M. Defining the Combined Stress Response in Wild Arachis. Scientific Reports 2021 11:1 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Samynathan, R.; Venkidasamy, B.; Shanmugam, A.; Ramalingam, S.; Thiruvengadam, M. Functional Role of MicroRNA in the Regulation of Biotic and Abiotic Stress in Agronomic Plants. Front Genet 2023, 14. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of MiR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis Thaliana. PLoS Genet 2016, 12. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhang, Y.; Wu, C.; Wang, S.; Hao, L.; Wang, S.; Li, T. Md-MiR156ab and Md-Mir395 Target WRKY Transcription Factors to Influence Apple Resistance to Leaf Spot Disease. Front Plant Sci 2017, 8, 256407. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-Wide Analysis of the Rice PPR Gene Family and Their Expression Profiles under Different Stress Treatments. BMC Genomics 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial Control of Gene Expression by MiR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol 2018, 176, 1694–1708. [Google Scholar] [CrossRef]
- Jeyaraj, A.; Elango, T.; Li, X.; Guo, G. Utilization of MicroRNAs and Their Regulatory Functions for Improving Biotic Stress Tolerance in Tea Plant [Camellia Sinensis (L.) O. Kuntze]. RNA Biol 2020, 17, 1365–1382. [Google Scholar] [CrossRef]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 Homeostasis Entails Coexpression of MIR168 and AGO1 and Preferential Stabilization of MiR168 by AGO1. Mol Cell 2006, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. Pho2, a Phosphate Overaccumulator, Is Caused by a Nonsense Mutation in a MicroRNA399 Target Gene. Plant Physiol 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, Y.; Wang, S.; Hao, L.; Wang, S.; Xu, C.; Jiang, F.; Li, T. Characterization of Genome-Wide MicroRNAs and Their Roles in Development and Biotic Stress in Pear. Planta 2019, 249, 693–707. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-Mediated Systemic down-Regulation of Copper Protein Expression in Response to Low Copper Availability in Arabidopsis. Journal of Biological Chemistry 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Martins, A.C.Q.; Mota, A.P.Z.; Carvalho, P.A.S.V.; Passos, M.A.S.; Gimenes, M.A.; Guimaraes, P.M.; Brasileiro, A.C.M. Transcriptome Responses of Wild Arachis to UV-C Exposure Reveal Genes Involved in General Plant Defense and Priming. Plants 2022, 11. [Google Scholar] [CrossRef]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis MicroRNA396-GRF1/GRF3 Regulatory Module Acts as a Developmental Regulator in the Reprogramming of Root Cells during Cyst Nematode Infection. Plant Physiol 2012, 159, 321–335. [Google Scholar] [CrossRef]
- Medina, C.; Da Rocha, M.; Magliano, M.; Ratpopoulo, A.; Revel, B.; Marteu, N.; Magnone, V.; Lebrigand, K.; Cabrera, J.; Barcala, M.; et al. Characterization of MicroRNAs from Arabidopsis Galls Highlights a Role for MiR159 in the Plant Response to the Root-Knot Nematode Meloidogyne Incognita. New Phytologist 2017, 216, 882–896. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Cabrera, J.; Martinez-Argudo, I.; Artaza, H.; Fenoll, C.; Escobar, C. Silenced Retrotransposons Are Major RasiRNAs Targets in Arabidopsis Galls Induced by Meloidogyne Javanica. Mol Plant Pathol 2018, 19, 2431–2445. [Google Scholar] [CrossRef]
- Cabrera, J.; Barcala, M.; García, A.; Rio-Machín, A.; Medina, C.; Jaubert-Possamai, S.; Favery, B.; Maizel, A.; Ruiz-Ferrer, V.; Fenoll, C.; et al. Differentially Expressed Small RNAs in Arabidopsis Galls Formed by Meloidogyne Javanica: A Functional Role for MiR390 and Its TAS3-Derived TasiRNAs. New Phytologist 2016, 209, 1625–1640. [Google Scholar] [CrossRef]
- Hewezi, T.; Howe, P.; Maier, T.R.; Baum, T.J. Arabidopsis Small RNAs and Their Targets during Cyst Nematode Parasitism. Molecular Plant-Microbe Interactions 2008, 21, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Guleria, P.; Mahajan, M.; Bhardwaj, J.; Yadav, S.K. Plant Small RNAs: Biogenesis, Mode of Action and Their Roles in Abiotic Stresses. Genomics Proteomics Bioinformatics 2012, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Shukla, N.; Joshi, G.; VijayaKumar, C.; Jagannath, A.; Agarwal, M.; Goel, S.; Kumar, A. Genome-Wide Identification and Characterization of MiRNAome from Tomato (Solanum Lycopersicum) Roots and Root-Knot Nematode (Meloidogyne Incognita) during Susceptible Interaction. PLoS One 2017, 12, e0175178. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate Response Decay and Defense Metabolite Accumulation Contributes to Age-Regulated Dynamics of Plant Insect Resistance. Nat Commun 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Kyndt, T.; Goverse, A.; Haegeman, A.; Warmerdam, S.; Wanjau, C.; Jahani, M.; Engler, G.; De Almeida Engler, J.; Gheysen, G. Redirection of Auxin Flow in Arabidopsis Thaliana Roots after Infection by Root-Knot Nematodes. J Exp Bot 2016, 67, 4559–4570. [Google Scholar] [CrossRef]
- Suzuki, R.; Kanno, Y.; Abril-Urias, P.; Seo, M.; Escobar, C.; Tsai, A.Y.L.; Sawa, S. Local Auxin Synthesis Mediated by YUCCA4 Induced during Root-Knot Nematode Infection Positively Regulates Gall Growth and Nematode Development. Front Plant Sci 2022, 13, 1019427. [Google Scholar] [CrossRef]
- Noureddine, Y.; Da Rocha, M.; An, J.; Médina, C.; Mejias, J.; Mulet, K.; Quentin, M.; Abad, P.; Zouine, M.; Favery, B.; et al. AUXIN RESPONSIVE FACTOR8 Regulates Development of the Feeding Site Induced by Root-Knot Nematodes in Tomato. J Exp Bot 2023, 74, 5752–5766. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. The Arabidopsis F-Box Protein TIR1 Is an Auxin Receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Noon, J.B.; Hewezi, T.; Baum, T.J. Homeostasis in the Soybean MiRNA396–GRF Network Is Essential for Productive Soybean Cyst Nematode Infections. J Exp Bot 2019, 70, 1653. [Google Scholar] [CrossRef]
- Ye, D.; Jiang, Y.; Wang, C.; Roberts, P.A. Expression Analysis of MicroRNAs and Their Target Genes in Cucumis Metuliferus Infected by the Root-Knot Nematode Meloidogyne Incognita. Physiol Mol Plant Pathol 2020, 111, 101491. [Google Scholar] [CrossRef]
- Ye, D.; Qi, Y.; Cao, S.; Duan, Y.; Huynh, B.L. Identification and Characterization of MicroRNA396 and Its Targets in Cucumis Metuliferus Infected with Meloidogyne Incognita. South African Journal of Botany 2024, 165, 417–427. [Google Scholar] [CrossRef]
- Kim, W.; Ahn, H.J.; Chiou, T.J.; Ahn, J.H. The Role of the MiR399-PHO2 Module in the Regulation of Flowering Time in Response to Different Ambient Temperatures in Arabidopsis Thaliana. Mol Cells 2011, 32, 83–88. [Google Scholar] [CrossRef]
- Tian, B.; Wang, S.; Todd, T.C.; Johnson, C.D.; Tang, G.; Trick, H.N. Genome-Wide Identification of Soybean MicroRNA Responsive to Soybean Cyst Nematodes Infection by Deep Sequencing. BMC Genomics 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Val-Torregrosa, B.; Bundó, M.; San Segundo, B. Crosstalk between Nutrient Signalling Pathways and Immune Responses in Rice. Agriculture 2021, Vol. 11, Page 747 2021, 11, 747. [Google Scholar] [CrossRef]
- Val-Torregrosa, B.; Bundó, M.; Martín-Cardoso, H.; Bach-Pages, M.; Chiou, T.J.; Flors, V.; Segundo, B.S. Phosphate-Induced Resistance to Pathogen Infection in Arabidopsis. Plant J 2022, 110, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, Y.; Mejias, J.; da Rocha, M.; Thomine, S.; Quentin, M.; Abad, P.; Favery, B.; Jaubert-Possamai, S. Copper MicroRNAs Modulate the Formation of Giant Feeding Cells Induced by the Root Knot Nematode Meloidogyne Incognita in Arabidopsis Thaliana. New Phytologist 2022, 236, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, Y.; Zhang, Q.; Xu, T.; Qiu, L.; Fan, Y.; Wang, L. Novel MiRNA and PhasiRNA Biogenesis Networks in Soybean Roots from Two Sister Lines That Are Resistant and Susceptible to SCN Race 4. PLoS One 2014, 9, e110051. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.W.; Lin, J.S.; Li, Y.C.; Jhu, M.Y.; King, Y.C.; Jeng, S.T. MicroR408 Regulates Defense Response upon Wounding in Sweet Potato. J Exp Bot 2019, 70, 469–483. [Google Scholar] [CrossRef]
- Shahbaz, M.; Pilon, M. Conserved Cu-MicroRNAs in Arabidopsis Thaliana Function in Copper Economy under Deficiency. Plants 2019, Vol. 8, Page 141 2019, 8, 141. [Google Scholar] [CrossRef]
- Klink, V.P.; Overall, C.C.; Alkharouf, N.W.; MacDonald, M.H.; Matthews, B.F. A Time-Course Comparative Microarray Analysis of an Incompatible and Compatible Response by Glycine Max (Soybean) to Heterodera Glycines (Soybean Cyst Nematode) Infection. Planta 2007, 226, 1423–1447. [Google Scholar] [CrossRef]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The Role of MicroRNAs and Other Endogenous Small RNAs in Plant Stress Responses. Biochim Biophys Acta 2008, 1779, 743–748. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants (Basel) 2020, 9. [Google Scholar] [CrossRef]
- Zsigmond, L.; Rigó, G.; Szarka, A.; Székely, G.; Ötvös, K.; Darula, Z.; Medzihradszky, K.F.; Koncz, C.; Koncz, Z.; Szabados, L. Arabidopsis PPR40 Connects Abiotic Stress Responses to Mitochondrial Electron Transport. Plant Physiol 2008, 146, 1721–1737. [Google Scholar] [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis Mitochondria-Localized Pentatricopeptide Repeat Protein PGN Functions in Defense against Necrotrophic Fungi and Abiotic Stress Tolerance. Plant Physiol 2011, 156, 2053–2068. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; Lou, Y. PRRs and NB-LRRs: From Signal Perception to Activation of Plant Innate Immunity. Int J Mol Sci 2019, 20, 1882. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P. V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.C.M.; Baulcombe, D.C. A MicroRNA Superfamily Regulates Nucleotide Binding Site-Leucine-Rich Repeats and Other MRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.M. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol 2010, 152, 2222–2231. [Google Scholar] [CrossRef]
- Zhai, J.; Jeong, D.H.; de Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as Master Regulators of the Plant NB-LRR Defense Gene Family via the Production of Phased, Trans-Acting SiRNAs. Genes Dev 2011, 25, 2540–2553. [Google Scholar] [CrossRef]
- De Vries, S.; Kloesges, T.; Rose, L.E. Evolutionarily Dynamic, but Robust, Targeting of Resistance Genes by the MiR482/2118 Gene Family in the Solanaceae. Genome Biol Evol 2015, 7, 3307–3321. [Google Scholar] [CrossRef]
- Zhang, Y.; Waseem, M.; Zeng, Z.; Xu, J.; Chen, C.; Liu, Y.; Zhai, J.; Xia, R. MicroRNA482/2118, a MiRNA Superfamily Essential for Both Disease Resistance and Plant Development. New Phytologist 2022, 233, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Xie, B.; Guan, P.; Jiang, N.; Cui, J. New Insight into the Molecular Mechanism of MiR482/2118 during Plant Resistance to Pathogens. Front Plant Sci 2022, 13, 1026762. [Google Scholar] [CrossRef] [PubMed]
- Morgante, C. V.; Brasileiro, A.C.M.; Roberts, P.A.; Guimaraes, L.A.; Araujo, A.C.G.; Fonseca, L.N.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Guimaraes, P.M. A Survey of Genes Involved in Arachis Stenosperma Resistance to Meloidogyne Arenaria Race 1. Functional Plant Biology 2013, 40, 1298–1309. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.M.; Arraes, F.; Quintana Martins, A.C.; Freitas Alves, N.S.; Melo, B.P.; Morgante, C.V.; Passos Saraiva, M.A.; Grossi-De-Sá, M.F.; Guimaraes, P.M.; Miranda Brasileiro, A.C. A Novel Soybean Hairy Root System for Gene Functional Validation. PLoS One 2023, 18, e0285504. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat Biotechnol 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol 2009, 10, 1–10. [Google Scholar] [CrossRef]
- Johnson, N.R.; Yeoh, J.M.; Coruh, C.; Axtell, M.J. Improved Placement of Multi-Mapping Small RNAs. G3: Genes, Genomes, Genetics 2016, 6, 2103–2111. [Google Scholar] [CrossRef]
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA Web Services. Methods in Molecular Biology 2015, 1269, 307–326. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2. 2016. [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem-Loop RT-PCR. Nucleic Acids Res 2005, 33. [Google Scholar] [CrossRef]
- Zhao, S.; Fernald, R.D. Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. Journal of Computational Biology 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res 2002, 30. [Google Scholar] [CrossRef]
- Santos, L.S.; Maximiano, M.R.; Megias, E.; Pappas, M.; Ribeiro, S.G.; Mehta, A. Quantitative Expression of MicroRNAs in Brassica Oleracea Infected with Xanthomonas Campestris Pv. Campestris. Mol Biol Rep 2019, 46, 3523–3529. [Google Scholar] [CrossRef] [PubMed]
- Morgante, C. V.; Guimarães, P.M.; Martins, A.C.Q.; Araújo, A.C.G.; Leal-Bertioli, S.C.M.; Bertioli, D.J.; Brasileiro, A.C.M. Reference Genes for Quantitative Reverse Transcription-Polymerase Chain Reaction Expression Studies in Wild and Cultivated Peanut. BMC Res Notes 2011, 4, 1–11. [Google Scholar] [CrossRef]
- Teotia, S.; Wang, X.; Zhou, N.; Wang, M.; Liu, H.; Qin, J.; Han, D.; Li, C.; Li, C.E.; Pan, S.; et al. A High-Efficiency Gene Silencing in Plants Using Two-Hit Asymmetrical Artificial MicroRNAs. Plant Biotechnol J 2023, 21, 1799–1811. [Google Scholar] [CrossRef]
- Zhou, Z.; Cao, Y.; Li, T.; Wang, X.; Chen, J.; He, H.; Yao, W.; Wu, J.; Zhang, H. MicroRNAs Are Involved in Maize Immunity Against Fusarium Verticillioides Ear Rot. Genomics Proteomics Bioinformatics 2020, 18, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene Silencing in Plants Using Artificial MicroRNAs and Other Small RNAs. Plant Journal 2008, 53, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Vázquez, L.A.; Castro-Pacheco, A.M.; Pérez-Vargas, R.; Velázquez-Jiménez, J.F.; Paul, S. The Emerging Applications of Artificial MicroRNA-Mediated Gene Silencing in Plant Biotechnology. Non-Coding RNA 2025, Vol. 11, Page 19 2025, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, J.; Zhang, N.; Wu, J.; Si, H. Roles of MicroRNAs in Abiotic Stress Response and Characteristics Regulation of Plant. Front Plant Sci 2022, 13, 919243. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




