1. Introduction
Screening bovine semen for pathogens is essential to safeguard animal health, ensure reproductive success, and maintain the integrity of national and international breeding programs. Bovine semen can be a vector for transmitting a variety of viral, bacterial, and protozoal pathogens, such as
Brucella abortus,
Campylobacter fetus,
Tritrichomonas foetus,
Mycoplasma bovis (M. bovis), bovine herpesvirus-1 (BHV-1), and bovine viral diarrhea virus (BVDV), especially when proper screening and biosecurity measures are not in place [
1]. Semen is believed to be the plausible source of the introduction of
M. bovis into New Zealand [
2,
3], illustrating the importance of screening semen as a biosecurity measure.
A foreign animal disease (FAD) in the United States, foot and mouth disease virus (FMDV) is detectable and can spread in semen [
1]. The potential shed of FMDV in semen could facilitate dissemination both within and between herds, including across geographic boundaries [
4,
5]. Another FAD, highly pathogenic avian influenza (HPAI) virus, was unexpectedly detected in U.S. dairy cattle, raising concerns about the virus’s capacity for cross-species transmission and spread [
6,
7]. While respiratory and oral routes remain the primary focus for transmission studies, the detection of HPAI RNA in non-traditional sample types such as raw milk has prompted new questions about alternative pathways for viral spread [
7,
8]. At present, confirmations of HPAI detection in bull semen are lacking, and FMDV is absent from the US [
8]. However, the topic is important for animal health surveillance and trade requirements.
Detection of pathogen DNA and RNA in semen presents unique technical challenges due to its viscosity and complex biochemical composition, with high concentrations of proteins, nucleases, and polysaccharides, as well as lipid-rich seminal plasma, all of which can interfere with nucleic acid extraction efficiency and downstream amplification, leading to decreased assay sensitivity and the potential for false negatives [
1,
9,
10]. Bovine semen extender is also added to raw bull semen to preserve sperm viability and create multiple doses [
11,
12]. Extenders contain key components such as egg yolk, milk proteins, soy lecithin, or other plant-based derivatives, which shield the sperm cell membrane from physical and chemical damage, maintain optimal pH (6.8 - 7.2), and supply metabolizable sugars like fructose or glucose to fuel sperm motility [
11,
12]. Antibiotics are also included to control bacterial contamination, which can compromise sperm quality and transmit venereal diseases [
12]. However, proteins and lipids from extender components can act as PCR inhibitors, potentially complicating nucleic acid extraction from semen and downstream assay performance.
In addition, various pathogens can be intercellular, intracellular, or both, thus requiring a total sample extraction method and avoiding centrifugation steps for pathogen screening [
1]. Modified extraction protocols, including the use of reagents capable of mitigating inhibitory effects, and inclusion of internal positive controls, are often required to overcome these numerous issues to obtain high-quality nucleic acid from semen. As a result, two National Animal Health Laboratory Network (NAHLN)-approved extraction platforms (MagMAX CORE Nucleic Acid Purification Kit and IndiMag Pathogen Kit) were targeted for method development to ensure reliable detection of various pathogens in semen matrices to streamline extraction workflows if semen were part of FAD surveillance.
2. Materials and Methods
2.1. Field and Reference Samples
Residual semen samples submitted to the Wisconsin Veterinary Diagnostic Laboratory (WVDL) for routine diagnostic testing from January to September 2024 were pooled by submission and extender type to provide sufficient material for method evaluation. Known negative residual straws from 3-4 animals per submission were combined to create 88 pooled field bovine semen samples for this study. The extended semen samples consisted of four categories: egg yolk-based extender (Yellow), milk-based extender sexed-semen (Green or Pink), and milk-based extender (White). All semen samples were stored in an ultralow freezer at −80 °C until ready for use with the various extraction protocols.
A total of 36 naturally infected semen samples, as determined previously (2008 –2022) by the WVDL, were used for diagnostic sensitivity evaluation. These consisted of eight M. bovis, five BVDV, and 23 bovine herpesvirus-1 (BHV-1) samples. Multiple frozen 0.25 cc semen straws from each of the infected animals were pooled into a homogenized mixture and stored in an ultralow freezer at −80 °C until ready for use in the study.
Nine HxNx IAV strains (
Appendix A,
Table A1) at various concentrations were spiked into twenty-seven negative semen for diagnostic sensitivity evaluation of the IAV assays. Thirty previously negative field residual semen samples were used to evaluate diagnostic specificity for the IAV assays. All samples were dispensed and stored in an ultralow freezer at −80 °C until ready for use.
To determine the limit of detection (LOD), a single strain of
M. bovis and three influenza A virus (IAV) reference strains were spiked in semen. The ATCC 25,523
M. bovis strain was acquired from the American Type Culture Collection (ATCC), while three IAV reference strains were kindly provided by the National Veterinary Service Laboratories (NVSL) Diagnostic Virology Laboratory (Ames, IA) for the analytical sensitivity evaluation (
Appendix A,
Table A1). Ten-fold serial dilutions of the references were prepared in negative semen pools and stored in an ultralow freezer at −80 °C until ready for use.
2.2. Extraction Chemistries and Equipment
The Thermo Fisher SCIENTIFIC MagMAX CORE Nucleic Acid Purification Kit (MagMAX CORE) (Thermo Fisher SCIENTIFIC, Waltham, MA, USA) and the INDICAL IndiMag Pathogen Kits (INDICAL BIOSCIENCE, Leipzig, Germany) approved by National Animal Health Laboratory Network (NAHLN) for IAV, African swine fever virus, classical swine fever virus, and FMDV testing were compared, with extractions done using the Kingfisher Flex (Thermo Fisher SCIENTIFIC, Waltham, MA, USA) [
13,
14,
15,
16].
For the MagMAX CORE extraction evaluation, each of the samples was extracted using the 200 µL sample input volume per semen import requirement by New Zealand Ministry for Primary Industries (CORE 200-na) [
17]; a reduced 50 µL sample input volume (CORE 50-na); a reduced 50 µL sample input volume with a pretreatment process using 10 µL of proteinase K, 5 µL of 1M DTT, and 35 µL of 2% SDS, with heating at 60 °C for 5 minutes (CORE 50-pretreatment); and a reduced 12.5 µL sample input volume with above pretreatment process (CORE 12.5-pretreatment). When the sample input was less than 200 µL, 1× phosphate-buffered saline (PBS) was added to achieve a total input volume of 200 µL.
For the IndiMag Pathogen Kit extraction evaluation, samples were extracted using the 200 µL sample input volume, mimicking CORE 200-na for semen import requirement by New Zealand Ministry for Primary Industries (Pathogen 200-na); 100 µL semen and 100 µL PBS (Pathogen 100-na); and a 100 µL semen with a pretreatment process using 20 µL of proteinase K, 90 µL of Buffer ATL (Qiagen, Germantown, MD, USA), and heating and mixing at 56 °C for 10 minutes (Pathogen 100-pretreament).
Two microliters of VetMAX Xeno Internal Positive Control RNA (Xeno RNA, Thermo Fisher SCIENTIFIC, Waltham, MA, USA) and 1 µL of the WVDL internal control (WVDL IC) were added to the lysis solution per sample for each extraction kit. The remainder of the extraction process was conducted according to the manufacturer’s instructions. Eluted RNA was used for Polymerase Chain Reactions (PCR) evaluation or stored in an ultralow freezer at −80 °C until ready for use.
2.3. Polymerase Chain Reactions (PCR)
The RNA extracted using the described methods was evaluated using the NAHLN IAV Matrix PCR assay (NAHLN assay) and the WVDL in-house influenza A Matrix PCR assay (WVDL assay). The NAHLN assay utilized AgPath-ID One Step PCR reagents (Thermo Fisher SCIENTIFIC, Waltham, MA, USA) and the VetMAX Xeno Internal Positive Control-VIC Assay for master mix per NVSL-SOP-0068 [
15]. The harmonized IAV thermocycling program was used on ABI 7500 (Thermo Fisher SCIENTIFIC, Waltham, MA, USA). The NAHLN-approved protocols are available online:
https://www.aphis.usda.gov/animal_health/lab_info_services/downloads/ApprovedSOPList.pdf. The NAHLN program office controls the distribution of protocols and the detailed information in the previously listed protocols; these can be requested by emailing NAHLN@usda.gov.
The WVDL IAV,
M. bovis, BVDV, and BHV-1 assays utilized the VetMAX Master Mix (Thermo Fisher SCIENTIFIC, Waltham, MA, USA). Each assay consisted of primers and probes for the pathogen and the WVDL IC (
Supplementary Table S1) and 5 µL of extracted nucleic acids in a 15 µL reaction. A thermocycling program of 50 °C for 5 minutes, 95 °C for 10 minutes, and 40 cycles of 95 °C for 3 seconds and 58 °C for 30 seconds was used on ABI 7500 (Thermo Fisher SCIENTIFIC, Waltham, MA, USA).
2.4. PCR and Statistical Analysis
All PCR analysis was performed at a 5% manual threshold value for the IAV, M. bovis, BVDV, BHV-1, Xeno RNA, and WVDL IC controls. A 5% manual threshold setting for Xeno RNA deviated from the auto-threshold setting in the NAHLN NSVL-SOP-0068. The auto-threshold algorithm was influenced by the samples within each run, resulting in fluctuations between runs. The 5% manual threshold value for Xeno RNA accurately positioned the threshold around the midpoint of the log-linear phase for the Xeno RNA analysis across all runs.
The effectiveness of the extraction protocol was determined by PCR amplification of the Xeno RNA and WVDL IC. Both PCRs were performed to 40 cycles. In accordance with the NAHLN acceptance criteria, the passing CT for Xeno RNA and WVDL IC was set at a CT value of < 34.5. The various pathogen and internal positive control targets with no amplification were assigned a cycle threshold (CT) value of 40 (the maximum cycle of the assays) for statistical analysis.
The M. bovis and IAV assays’ performance was evaluated using LOD, the coefficient of correlation of the standard curve (R2), and PCR efficiency. The IAV assays were used to evaluate diagnostic sensitivity and diagnostic specificity. For the LOD, a standard curve was generated using ten-fold dilutions for each of the reference strains that was extracted in triplicate. The endpoint dilution of the reference strain determined the LOD, where all three replicates were detected. PCR efficiency and R2 were obtained from the standard curve generated by the Design and Analysis software (Thermo Fisher SCIENTIFIC, Waltham, MA, USA).
Repeatability within a single assay was assessed with the samples used for diagnostic sensitivity and specificity evaluation. Testing was performed on a second set of samples 4 months later to evaluate the inter-run repeatability and reproducibility.
Paired T-tests and Analysis of variance (ANOVA) tests were used to infer statistical significance among various methods. The statistical analysis was performed using Prism software version 10.6.0 (GraphPad, Boston, MA, USA). Figures were generated using Tableau software, Public Edition (Salesforce, San Francisco, CA, USA).
3. Results
3.1. Extraction Method Optimization
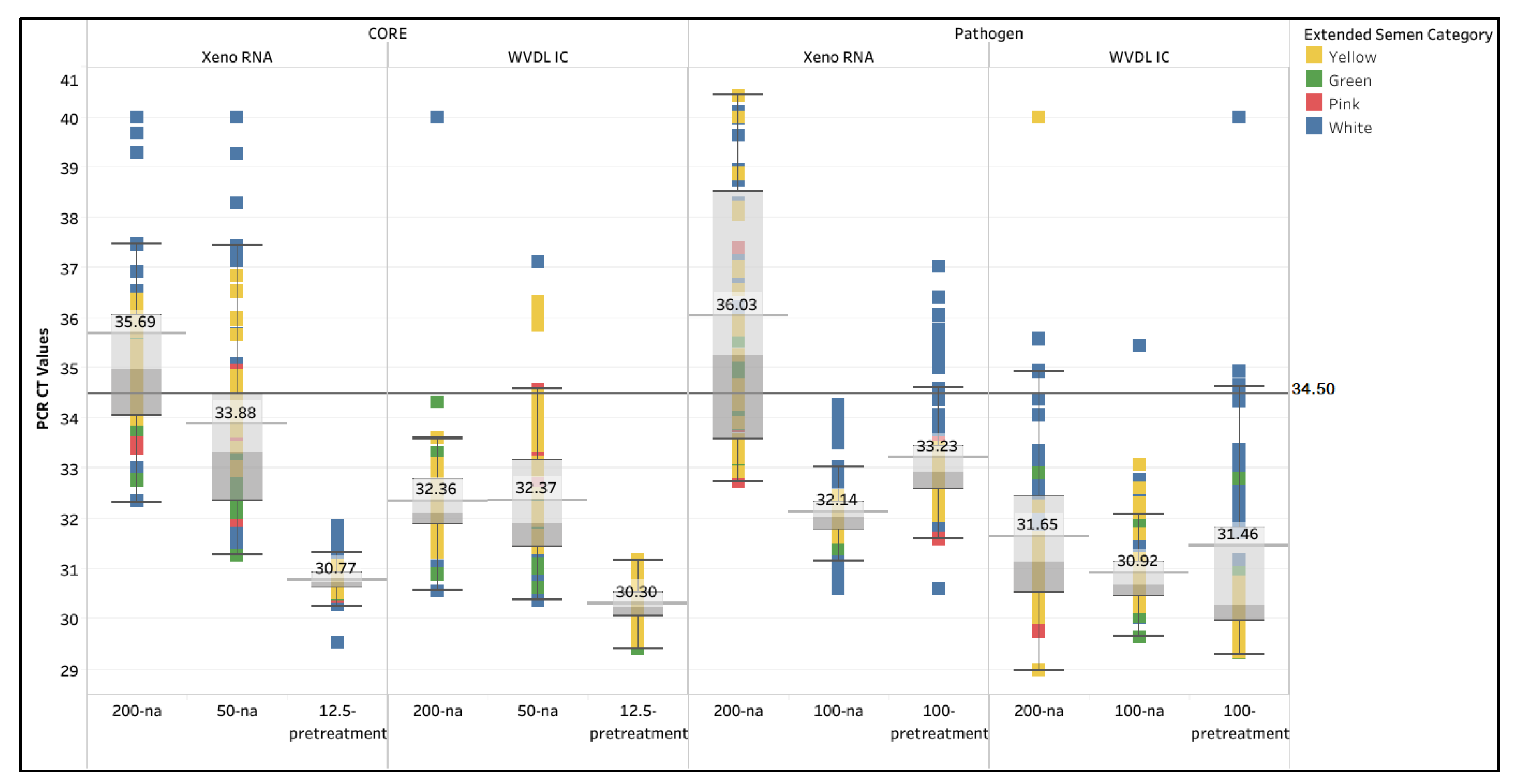
The first part of the study evaluated the performance of the current NALHN-approved extraction kits, the MagMAX CORE and IndiMag Pathogen kits, using 88 negative extended semen samples with the New Zealand required sample input of 200 µL. The Xeno RNA with the NAHLN IAV matrix PCR assay as a reference method and the WVDL IC (routinely used to screen semen for pathogens at the WVDL) with the WVDL in-house influenza A PCR assay were utilized to investigate the inhibitory effect. The overall passing rates for the MagMAX CORE (CORE 200-na) were 31.8% and 97.7% for Xeno RNA and WVDL IC, respectively. The overall passing rates for the IndiMag Pathogen (Pathogen 200-na) were 37.5% and 94.3% for Xeno RNA and WVDL IC, respectively (
Table 1). The passing rate varied significantly (3.1% to 100%) depending on the type of semen extender and internal positive control (
Table 1 and
Supplemental Table S2).
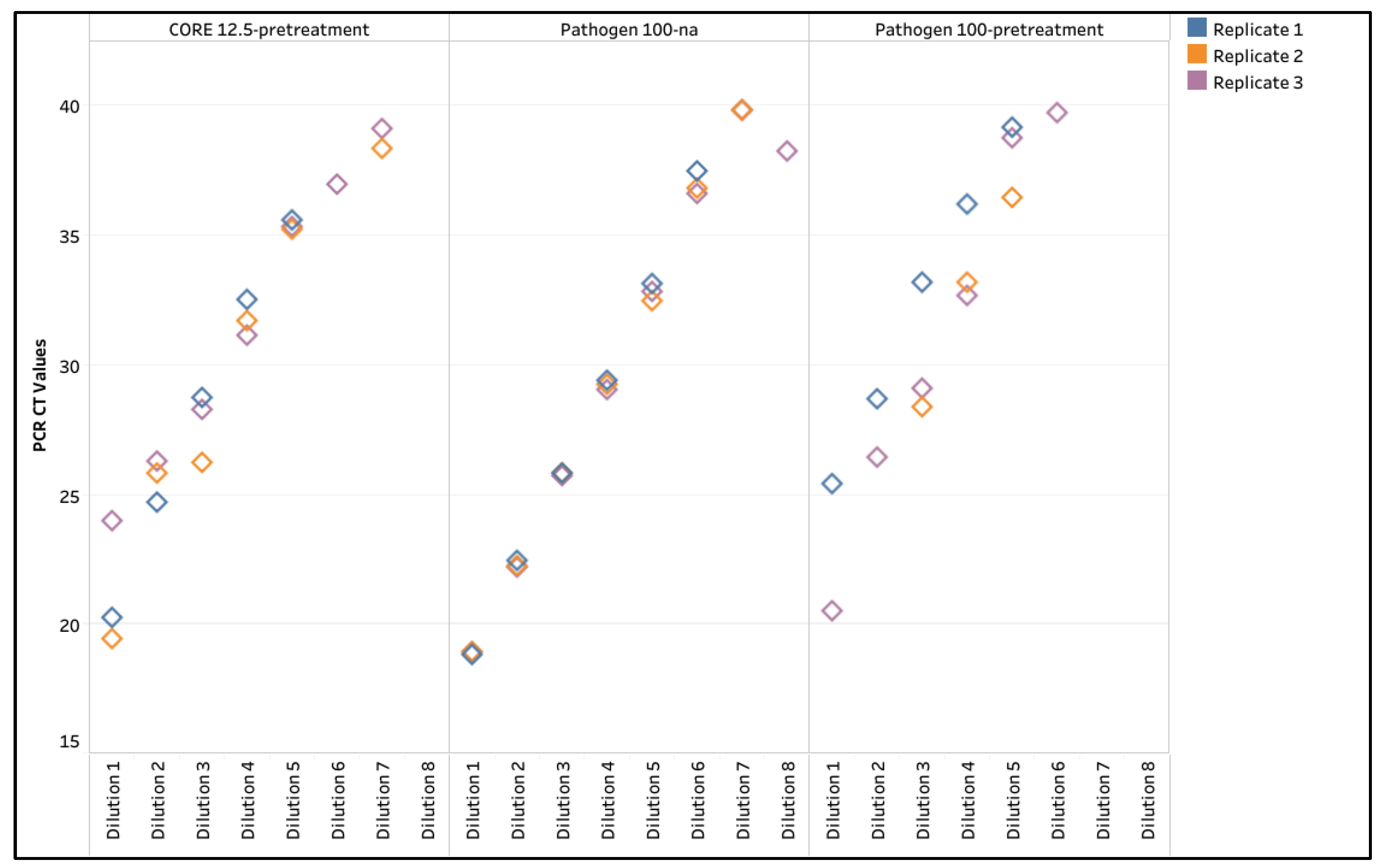
The manufacturer were contacted, and their suggested pretreatment protocols were attempted. With MagMAX CORE, a 50 µL input with pretreatment (CORE 50-pretreatment) was attempted, but the elution contained significant bead residue and could not be pipetted due to its clumpiness. Thus, 50 µL input without pretreatment (CORE 50-na) and a 12.5 µL input with pretreatment (CORE 12.5-pretreatment) were attempted. With the IndiMag Pathogen Kit, a 100 µL input without pretreatment (Pathogen 100-na) and a 100 µL input with pretreatment (Pathogen 100-pretreatment) were attempted. Upon testing with the CORE 50-na, overall passing rates increased to 73.9% and 93.2% for the Xeno RNA and the WVDL IC, respectively, while the CORE 12.5-pretreatment overall passing rate was 100% for the Xeno RNA and the WVDL IC. For the Pathogen 100-na, the overall passing rate increased to 100% for the Xeno RNA and the WVDL IC. The Pathogen 100-pretreatment reduced the overall passing rates to 88.6% and 85.2% on the Xeno RNA and the WVDL IC, respectively. The CT values for the non-passing samples moved into the passing range as the successful modifications were applied (
Figure 1).
The CT values for the various methods were further analyzed. The internal positive control CT values ranged from 29.51 to 40 with the NAHLN assay and from 29.40 to 40 with the WVDL assay (
Figure 1 and
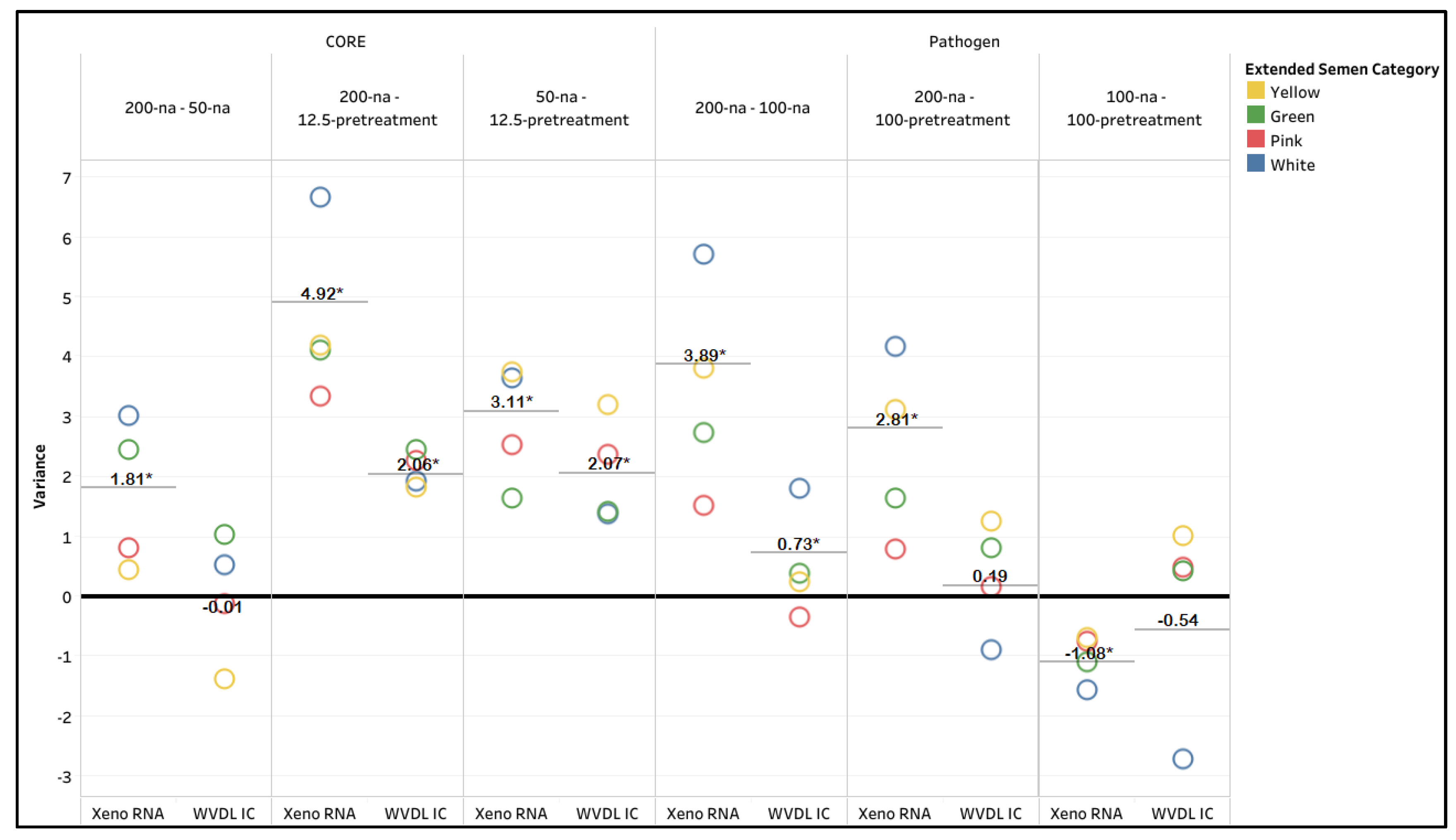
Supplemental Table S2). For MaxMAX CORE, a significant difference in CT values was observed for improvement from CORE 200-na to CORE 50-na on the NAHLN assay and to CORE 12.5-pretreatment for both the NAHLN and WVDL assays (Paired ANOVA test, p < 0.0001). Reduced sample input alone (CORE 50-na) had a minimal effect on the mean variance compared to CORE 200-na with the NAHLN and WVDL assays (
Figure 2,
Supplemental Table S2). However, the CORE 12.5-pretreatment drastically improved the CT values, with the lowest and tightest CT range (29.51 - 31.85 and 29.40 - 31.17 for the NAHLN and WVDL assays, respectively). The high variance demonstrates successful modification of the protocol for minimizing PCR inhibitory effects (
Figure 2).
For the Pathogen Kit, major improvements were achieved by reducing the sample input volume (Pathogen 100-na) or by incorporating additional pretreatment steps at the reduced volume (Pathogen 100-pretreatment) (
Figure 1 and
Figure 2). The pretreatment provided no further improvement to the reduced volume samples (
Figure 2 and
Supplemental Table S2). The CT value for Pathogen 100-na ranged from 30.61 to 34.28 and 29.64 to 35.43 for the NAHLN and WVDL assays, respectively. The CT value for Pathogen 100-pretreatment ranged from 30.60 to 37.06 for the NAHLN assay and 29.30 to 40 for the WVDL assay. A significant difference in CT values and high variance (
Figure 2) were observed for improvement from Pathogen 200-na to Pathogen 100-na for both the NAHLN and WVDL assays and to Pathogen 100-pretreatment on the NAHLN assay (Paired ANOVA test, p < 0.0001)
3.2. Evaluation of Diagnostic Sensitivity for the Selected Extraction Protocols
A total of 36 extended semen samples naturally infected with
M. bovis (n=8), BVDV (n=5), or BHV-1 (n=23) were extracted in duplicate using separate extractions for the CORE 12.5-pretreatment, Pathogen 100-na, and Pathogen 100-pretreatment protocols with the WVDL pathogen-specific assays (
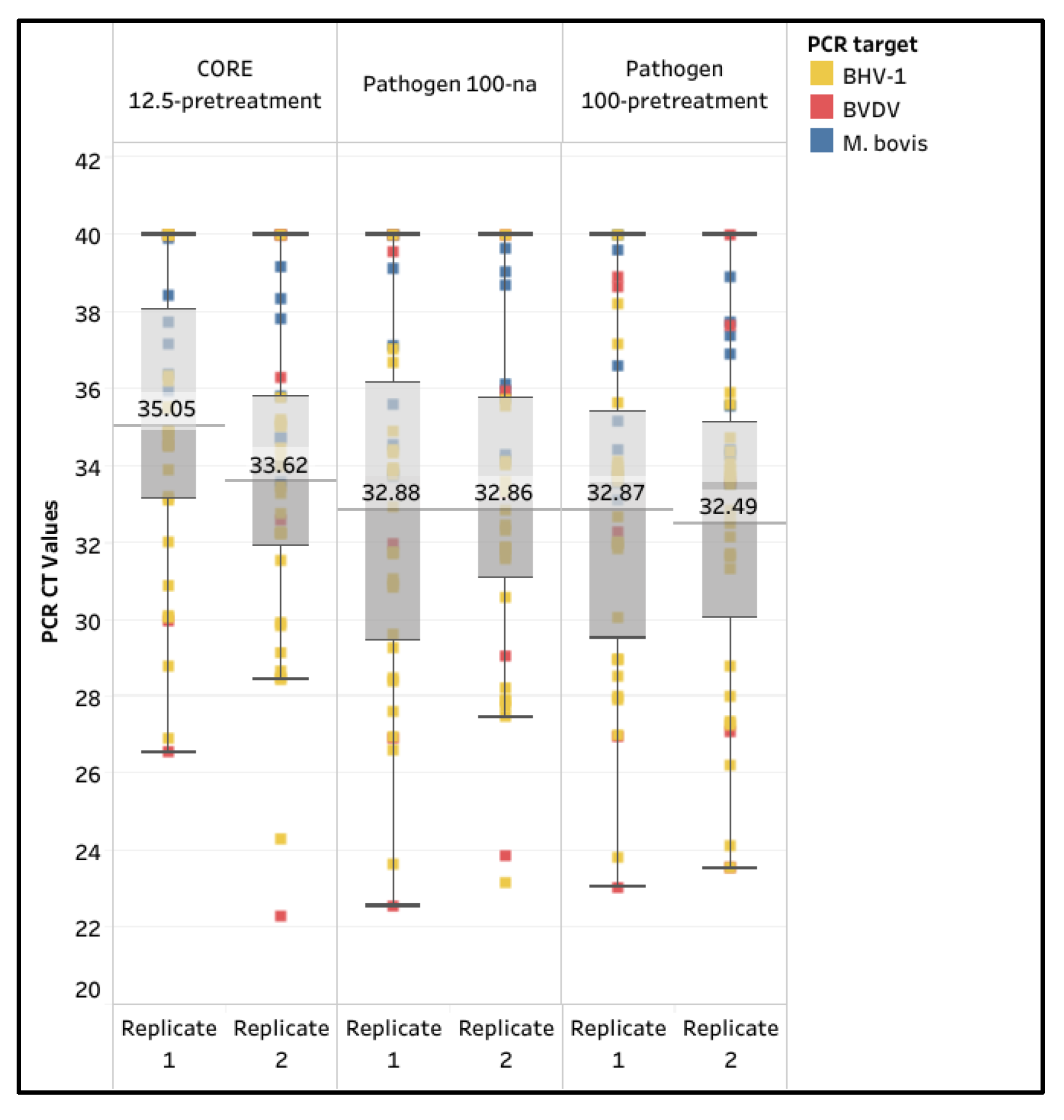
Table 2). The Core 12.5-pretreatment and the Pathogen 100-na were selected based on the 100% passing rate for internal positive controls; the Pathogen 100-pretreatment was also included to investigate the effect of pretreatment in naturally infected samples. Samples with a pathogen CT value less than 40 were considered positive, regardless of the CT value of the internal positive controls.
The diagnostic sensitivity of the CORE 12.5-pretreatment was the lowest, with the replicates at 80.6% and 88.9%, while the Pathogen 100-pretreatment had the highest sensitivity, with the replicates at 94.9% and 97.2%. The overall mean CT values for the CORE 12.5-pretreatment were 35.05 (95% CI: 33.76 - 36.35) and 33.62 (95% CI: 32.2 - 35.04), while the Pathogen 100-na CT values were 32.88 (95% CI: 31.29 - 34.46) and 32.86 (95% CI: 31.45 - 34.27), and the Pathogen 100-pretreatment CT values were 32.87 (95% CI: 31.41 - 33.34) and 32.49 (95% CI: 31.05 - 33.94), respectively. Overall, the Pathogen 100-na and Pathogen 100-pretreatment protocols provided higher detection rates and lower mean CT values for
M. bovis, BVDV, and BHV-1, indicating significantly better diagnostic sensitivity compared to the CORE 12.5-pretreatment protocol (ANOVA test,
p < 0.0001), while a lack of significant difference existed between Pathogen 100-na and Pathogen 100-pretreatment protocols (Paired T test, p = 0.3849) (
Figure 3 and
Supplemental Table S3).
3.3. Evaluation of Analytical Sensitivity
Understanding the importance of bioexclusion of
M. bovis for New Zealand, the reference strain ATCC 25,523 was diluted ten-fold in prepared negative semen (White) for extraction with CORE 12.5-pretreatment, Pathogen 100-na, and Pathogen 100-pretreatment to evaluate the analytical sensitivity. The LOD was dilution 6 with Pathogen 100-na—and two of the three replicates were detected for dilution 7—compared to dilution 5 with CORE 12.5-pretreatment and Pathogen 100-pretreatment (
Table 3,
Figure 4, and
Supplemental Table S4). The data suggested that the heat treatments were negatively affecting the artificially spiked samples, so these protocols were not further evaluated. For Pathogen 100-na, the R
2values ranged from 0.999 to 1.000, with the percent PCR efficiency ranging between 90.4 and 94.1.
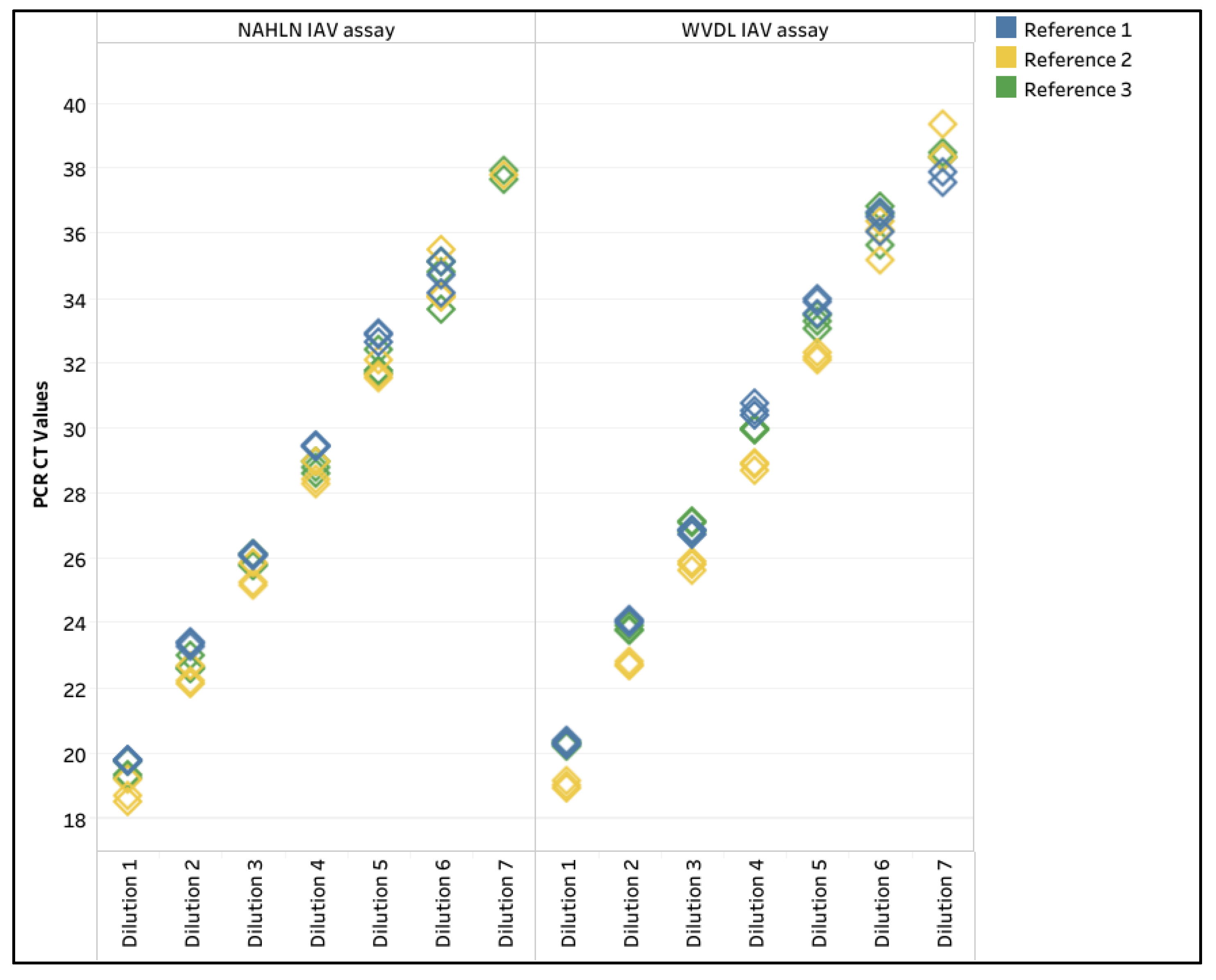
Given the epidemic of IAV in US dairy herds and concerns of pathogens in semen, three low pathogenic avian influenza (LPAI) virus reference strains were diluted tenfold in negative semen pools (White, Yellow, and Green). The LOD for the three reference strains was dilution 6 for both the NAHLN and WVDL PCR assays (
Table A2,
Figure A1, and
Supplemental Table S5). Dilution 7 was detected once or twice out of the three replicates for reference strains 2 and 3 for the NAHLN and WVDL PCR assays (
Table A2). The R
2values ranged from 0.990 to 0.999, with the percent PCR efficiency ranging between 97.8 and 120.7. In addition, Xeno RNA in the NAHLN IAV assay and the WVDL IC in the WVDL IAV assay were evaluated to investigate IC differences. The Xeno RNA CT value range was 29.49 to 31.27, while the WVDL IC CT value range was 28.87 to 32.14.
3.4. Evaluation of Pathogen 100-Na Protocol for the IAV Assays
Since the diagnostic sensitivity of naturally infected M. bovis samples was previously investigated, and due to a lack of naturally IAV-infected semen, diagnostic sensitivity and specificity were assessed using 27 IAV spiked-positive and 30 negative semen samples using the NAHLN and WVDL PCR assays. The inter-run repeatability was assessed by extracting the same set of samples four months after the initial extraction.
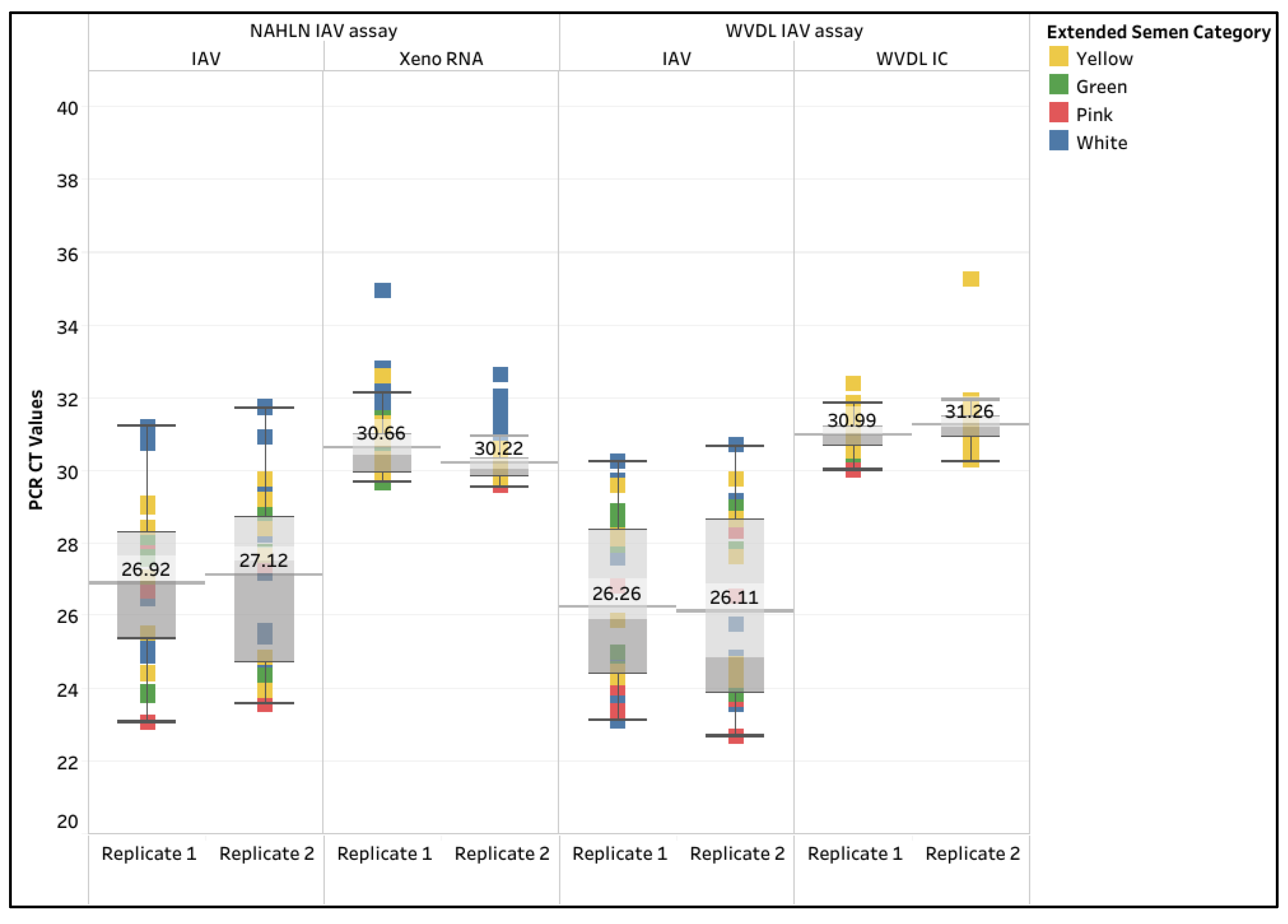
The overall mean CT values for IAV using the NAHLN assay were 26.92 (95% CI: 26.04 - 27.79) and 27.12 (95% CI: 26.20 - 28.04), while the mean IAV CT values using the WVDL assay were 26.26 (95% CI: 25.37 - 27.16) and 26.11 (95% CI: 25.14 - 27.09), respectively (Supplemental Tabel S6). The 30 negative samples tested negative by the NAHLN and WVDL IAV PCR assays. The overall mean CT values for the Xeno RNA were 30.66 (95% CI: 30.39 - 30.92) and 30.22 (95% CI: 30.05 - 30.39), while the WVDL IC CT values were 30.99 (95% CI: 30.88 - 31.11) and 31.26 (95% CI: 31.08 - 31.43), respectively (
Figure 5). One hundred percent diagnostic sensitivity and specificity were achieved for the evaluated samples.
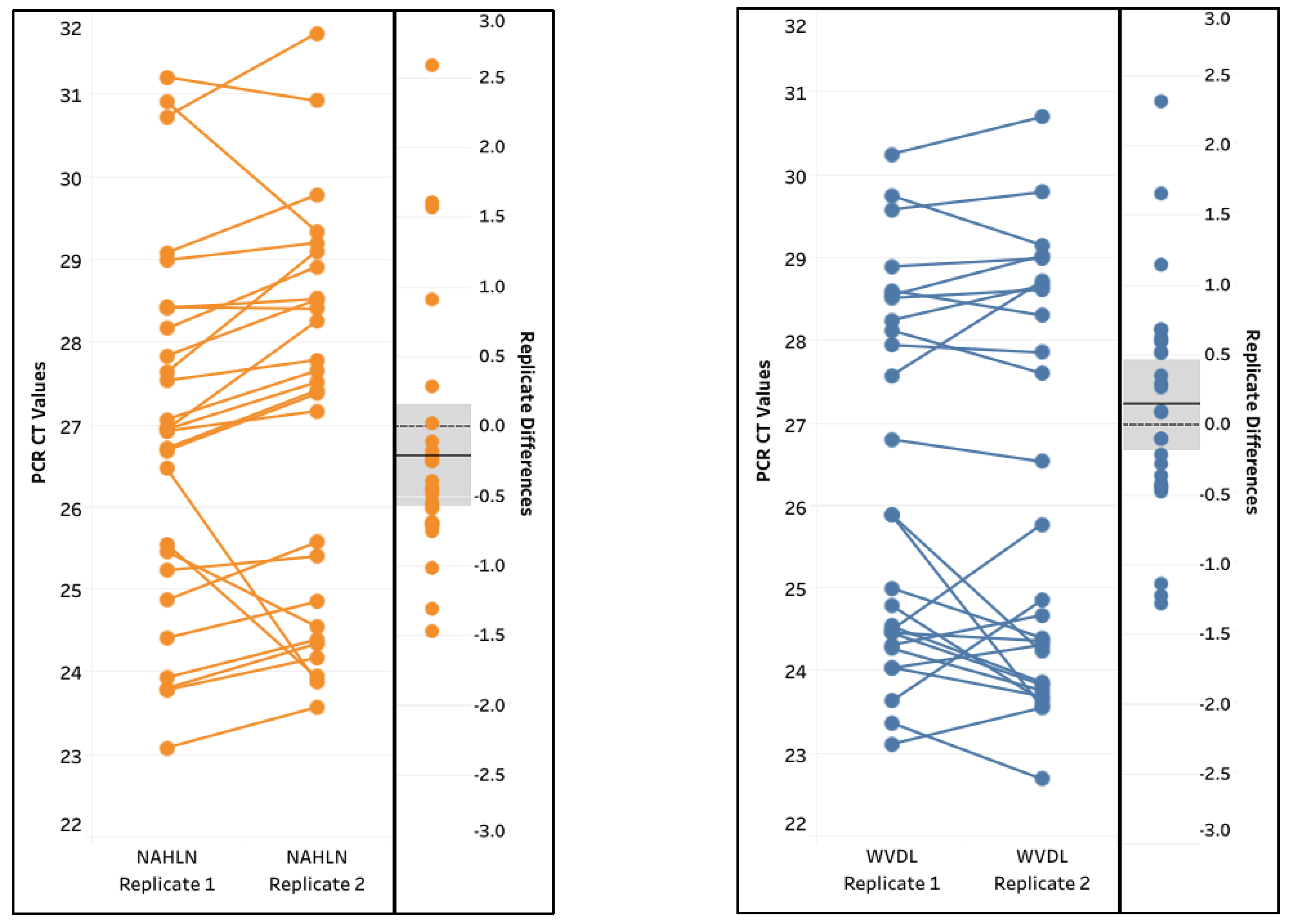
The repeatability and reproducibility of the method were assessed through a pairwise comparison of the two sets of replicated testing for the 27 IAV-spiked samples. Overall, excellent correlations were observed among the positive replicates, with Pearson correlation coefficients r = 0.9215 and r = 0.9448 for the NAHLN and WVDL assays, respectively (
Figure 6). There was a lack of significant difference between the IAV CT values of the two replicates (Paired T test, p = 0.2556 and p = 0.3557 for the NAHLN and WVDL assays, respectively). The cross-classification of the positive results was 100% with agreement. The data suggested excellent repeatability and reproducibility of the Pathogen 100-na extraction method.
Lastly, the 36 M. bovis, BVDV, or BHV-1 naturally infected samples were tested by the NAHLN and WVDL IAV PCR assays to investigate the absences of IAV in historical semen samples. As expected, the samples were negative, with mean CT values of 29.46 (95% CI: 28.95 - 2.97) and 29.61 (95% CI: 29.50 - 29.71) for the Xeno RNA and WVDL IC, respectively.
4. Discussion
Effective isolation of pathogen nucleic acids is necessary in veterinary diagnostics because PCR inhibitors, such as proteins, lipids, and other sample compounds, can be problematic. In addition, improper storage of samples before testing can further interfere and reduce assay sensitivity, potentially generating false negatives [
18]. In this study, the two NAHLN-approved extraction kits (MagMAX CORE and IndiMag Pathogen) for detecting high-consequence diseases in various sample types, including swabs, tissues, milk, and whole blood, were evaluated [
13,
14,
15,
16]. These kits were rapidly adapted for IAV surveillance in milk, enabling reliable detection and streamlining the laboratory’s surveillance testing. Optimizing the semen extraction protocols for these kits can streamline the detection and surveillance of common pathogens and high-consequence diseases, such as FMDV and potentially HPAI, during a US outbreak.
The high occurrence of inhibitors in animal samples, particularly in semen with disulfide-linked nuclear proteins and lipid-rich seminal plasma, can be addressed by optimizing lysis chemistries using Proteinase K, detergents, reducing agents, and heat treatments to disrupt protein complexes and enhance nucleic acid recovery [
19,
20,
21,
22]. The MagMAX CORE protocol (CORE 200-na), IndiMag Pathogen Kit protocol (Pathogen 200-na), and reduced-input modification (CORE 50-na), did not sufficiently remove inhibition, consistent with the challenges described for other veterinary matrices [
18,
19].
Exogenous ICs are beneficial to identify inhibitor effects in the extracted nucleic acid. In a previous study using extended semen and the MagMAX CORE protocol (CORE 200-na), an exogenous IC was lacking to monitor PCR inhibition [
23], the semen was diluted in PBS (instead of diluting in negative semen), and assay performance was evaluated based on the detection of spiked
M. bovis target. These factors call into question the validity of using the MagMAX CORE protocol with 200 µL of semen.
Since the recommendation of the pretreatment protocols used in this study, the MagMAX CORE protocol was updated to include total semen and 300 µL input. A new version of complex semen extraction protocol for the MagMAX CORE Kit was subsequently released in April 2024, using the higher volume of semen and extended heat and proteinase K treatment [
24]. Unfortunately, the authors were not notified nor aware of the procedural change. Hence, this study does not command the complex semen protocol to investigate the effect on inhibition removal.
Using half the semen input volume with the IndiMag Pathogen Kit (Pathogen 100-na) reduced inhibition slightly better compared to adding Buffer ATL and heat pretreatment (Pathogen 100-pretreatment). While reducing sample input can help minimize PCR inhibition, lower sample volume may also decrease the sensitivity of pathogen detection. Thus, subsequent testing of naturally infected field samples containing
M. bovis, BVDV, or BHV-1 was conducted to verify that both the modified CORE (CORE 12.5-pretreatment) and Pathogen Kit (Pathogen 100-na and Pathogen 100-pretreatment) protocols achieved high sensitivity for pathogen detection, with a slight performance advantage for the Pathogen 100-pretreatment. Similarly, reducing milk input volume was found to reduce inhibition when optimizing extractions for the surveillance of IAV in milk samples [
25]. In veterinary diagnostics, careful monitoring of inhibition is critical to avoid reporting of false-negative results, leading to the spread of disease [
18,
26].
Due to bioexclusion of M. bovis in semen for New Zealand, the reference strain ATCC 25,523 was used to evaluate the analytical sensitivity for extraction with CORE 12.5-pretreatment, Pathogen 100-na, and Pathogen 100-pretreatment, with results illustrating that heating negatively impacted extracellularly-spiked target. Thus, only the Pathogen 100-na was evaluated with spiked IAV semen samples, which achieved high sensitivity for speculative IAV detection and provides the framework for validating this protocol for FMDV detection, if a US outbreak were to occur. While all the archived semen samples were negative for IAV (as expected), a well-developed semen extraction protocol is important for the detection of pathogens, specifically M. bovis for New Zealand import requirements. Subsequently, the Pathogen 100-na protocol was approved as a semen extraction method for New Zealand import requirements (D. Jaramillo, personal communication, October 16, 2025).
Establishing standardized nucleic acid extraction and pretreatment strategies that capture both intercellular and intracellular pathogens while providing a simple and high-throughput method for removal of inhibitors inherent to semen is crucial to ensure sensitive and specific detection, limit the spread of endemic and exotic pathogens through germplasm exchange, and support international biosecurity measures [
27].
5. Conclusions
In conclusion, this study highlights the critical role of optimizing nucleic acid extraction protocols to overcome PCR inhibition in milk-based and egg-based extended semen samples. While reducing sample input volumes and incorporating pretreatments effectively minimized inhibition, a balance must be struck between inhibitor removal and preserving viral integrity to maintain assay sensitivity. Validation with both naturally infected and spiked samples demonstrated that refined extraction protocols can support the reliable detection of viruses and bacteria.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: Primers and probes information for the influenza A virus,
Mycoplams bovis, bovine viral diarrhea virus, and bovine herpesvirus-1 PCR assays. Table S2: CT values and variance for the NAHLN IAV Matrix and the WVDL in-house influenza A PCR assays for the 88 negative semen samples using the various methodologies with the MagMAX CORE (CORE) and the IndiMag Pathogen (Pathogen) kits. Table S3: CT values for
M. bovis, BVDV, or BHV-1 specific PCR assays to assess the extraction method sensitivity for the modified MagMAX CORE (CORE) and the modified IndiMag Pathogen (Pathogen) kits extraction protocols using 36 naturally infected semen samples. Table S4: CT values for
M. bovis PCR assay to assess the modified MagMAX CORE (CORE 12.5-pretreatment), IndiMag Pathogen (Pathogen 100-na), and modified IndiMag Pathogen (Pathogen 100-pretreatment) extraction protocols with the
M. bovis-spiked samples. Table S5: CT values for NAHLN IAV and WVDL in-house influenza A PCR assays to assess the reduced input (100 µL) IndiMag Pathogen Kit extraction protocol with the influenza A virus-spiked standard curves. Table S6: CT values for the NAHLN IAV and WVDL in-house influenza A PCR assays to assess the sensitivity and specificity for the reduced input (100 µL) IndiMag Pathogen Kit extraction protocol with the influenza A virus-spiked samples. Table S7: CT values for the NAHLN IAV and WVDL in-house influenza A PCR assays to assess the influenza A status in
M. bovis, BVDV, or BHV-1 naturally infected semen samples.
Author Contributions
Conceptualization, D.G.M. and A.L.; methodology, A.Z., D.G.M., and A.L.; software, A.V.C. and A.L.; validation, A.Z., A.V.C., D.G.M., and A.L.; formal analysis, A.Z., D.G.M., and A.L.; investigation, A.Z., A.V.C., D.G.M., and A.L.; resources, A.L.; data curation, A.Z., A.V.C., and A.L.; writing—original draft preparation, A.Z., A.V.C., D.G.M., and A.L.; writing—review and editing, A.Z., A.V.C., D.G.M., and A.L.; visualization, A.V.C. and A.L.; supervision, A.L.; project administration, A.L.; funding acquisition, A.L. All authors have read and agreed to the published version of the manuscript.
Funding
WVDL funded the personnel and reagents for the study. INDICAL BIOSCIENCE provided some of the IndiMag Pathogen Kits for the study. This research received no extramural funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank the U.S. Department of Agriculture National Veterinary Services Laboratory for providing reference materials for this study; ThermoFisher SCIENTIFIC and INDICAL BIOSCIENCE Research & Development teams for their technical assistance.
Conflicts of Interest
The authors (A.Z., A.V.C., and A.L.) declare no conflicts of interest. D.M. is employed by INDICAL BIOSCIENCE, which provided some IndiMag Pathogen Kits for the study, participated in the design of the study, data analyses, and writing of the manuscript.
Abbreviations
The following abbreviations are used in this manuscript:
| ANOVA |
Analysis of Variance |
| BHV-1 |
Bovine herpesvirus-1 |
| BVDV |
Bovine viral diarrhea virus |
| CT |
Cycle threshold |
| FMDV |
Foot and mouth disease virus |
| HPAI |
Highly Pathogenic Avian Influenza |
| IAV |
Influenza A virus |
| IC |
Internal control |
| LOD |
Limit of detection |
| LPAI |
Low Pathogenic Avian Influenza |
| MagMAX CORE |
MagMAX CORE Nucleic Acid Purification Kit |
| M. bovis |
Mycoplasma bovis |
| NAHLN |
National Animal Health Laboratory Network |
| NVSL |
National Veterinary Service Laboratories |
| PBS |
Phosphate-buffered saline |
| PCR |
Polymerase chain reactions |
| R2
|
Coefficient of correlation of the standard curve |
| Xeno RNA |
VetMAX Xeno Internal Positive Control RNA |
| WVDL |
Wisconsin Veterinary Diagnostic Laboratory |
| WVDL IC |
WVDL internal control |
Appendix A
Table A1.
List of low pathogenic influenza A viruses used as reference strains and for generation of positive IAV samples for diagnostic sensitivity evaluation.
Table A1.
List of low pathogenic influenza A viruses used as reference strains and for generation of positive IAV samples for diagnostic sensitivity evaluation.
| IAV |
Subtype |
Strain ID |
| IAV reference 1 |
H9N2 |
Influenza A Virus A/Turkey/CA/6889/1980 |
| IAV reference 2 |
H5N9 |
Influenza A Virus A/Turkey/Wisconsin/1968 |
| IAV reference 3 |
H7N3 |
Influenza A Virus A/Turkey/Oregon/1977 |
| LPAI 1 |
H3N8 |
Influenza A Virus A/Equine/Miami/1/63 |
| LPAI 2 |
H1N7 |
Influenza A Virus A/NJ/8/76/EQ-1 |
| LPAI 3 |
HON3 |
Influenza A Virus A/NWS-NOV2 |
| LPAI 4 |
H10N7 |
Influenza A Virus A/CK/GERM/49 |
| LPAI 5 |
H2N3 |
Influenza A Virus A/Mallard/A16/77 |
| LPAI 6 |
H4N8 |
Influenza A Virus A/MYNAH/Mass/71 |
| LPAI 7 |
H7N3 |
Influenza A Virus A/TY/ORE |
| LPAI 8 |
H4N8 |
Influenza A Virus A/DK/England/62 |
| LPAI 9 |
H3N8 |
Influenza A Virus A/DK/Ukraine/1/63 |
Table A2.
The limit of detection, coefficient of correlation of the standard curve (R2), and percent PCR efficiency for the three reference strains extracted using the IndiMag Pathogen Kit with 100 µL semen input and evaluated on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) assays.
Table A2.
The limit of detection, coefficient of correlation of the standard curve (R2), and percent PCR efficiency for the three reference strains extracted using the IndiMag Pathogen Kit with 100 µL semen input and evaluated on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) assays.
| |
Reference
Strain
|
NAHLN IAV assay |
WVDL IAV assay |
| |
Replicate 1 |
Replicate 2 |
Replicate 3 |
Replicate 1 |
Replicate 2 |
Replicate 3 |
| Limit of Detection |
1 |
6 |
6 |
6 |
6 |
6 |
6 |
| 2 |
7 |
6 |
6 |
6 |
7 |
7 |
| 3 |
7 |
6 |
7 |
6 |
7 |
7 |
| R2 value |
1 |
0.997 |
0.99 |
0.994 |
0.997 |
0.995 |
0.999 |
| 2 |
0.999 |
0.998 |
0.998 |
0.999 |
0.999 |
0.996 |
| 3 |
0.997 |
0.995 |
0.999 |
0.997 |
0.995 |
0.996 |
| (%) PCR Efficiency |
1 |
110.6 |
118.0 |
113.3 |
102.1 |
104.8 |
103.8 |
| 2 |
98.5 |
109.0 |
101.3 |
106.2 |
97.8 |
101.5 |
| 3 |
100.6 |
120.7 |
110.9 |
116.4 |
112.2 |
109.9 |
Figure A1.
Standard curve for the three influenza A (IAV) reference strains in different extended semen extracted using the IndiMag Pathogen Kit with 100 µL semen input and evaluated on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays. Reference 1 was spiked into negative semen with milk-based extender, Reference 2 was spiked into negative semen with egg yolk-based extender, and Reference 3 was spiked into negative sexed-semen with milk-based extender.
Figure A1.
Standard curve for the three influenza A (IAV) reference strains in different extended semen extracted using the IndiMag Pathogen Kit with 100 µL semen input and evaluated on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays. Reference 1 was spiked into negative semen with milk-based extender, Reference 2 was spiked into negative semen with egg yolk-based extender, and Reference 3 was spiked into negative sexed-semen with milk-based extender.
References
- Givens, M. D. Review: Risks of Disease Transmission through Semen in Cattle. Animal 2018, 12, s165–s171. [Google Scholar] [CrossRef]
- Jordan, A.; Sadler, R. J.; Sawford, K.; van Andel, M.; Ward, M.; Cowled, B. Mycoplasma Bovis Outbreak in New Zealand Cattle: An Assessment of Transmission Trends Using Surveillance Data. Transbound Emerg Dis 2021, 68, 3381–3395. [Google Scholar] [CrossRef]
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a Source of Mycoplasma Bovis Mastitis in Dairy Herds. Vet Microbiol 2018, 216, 60–66. [Google Scholar] [CrossRef]
- Sharma, G.; Subramaniam, S.; De, A.; Das, B.; Dash, B.; Sanyal, A.; Misra, A.; Pattnaik, B. Detection of Foot-and-Mouth Disease Virus in Semen of Infected Cattle Bulls. Indian Journal of Animal Sciences 2012, 82, 1472–1476. [Google Scholar] [CrossRef]
- Alexandersen, S.; Zhang, Z.; Donaldson, A. I.; Garland, A. J. M. The Pathogenesis and Diagnosis of Foot-and-Mouth Disease. Journal of Comparative Pathology 2003, 129, 1–36. [Google Scholar] [CrossRef]
- Burrough, E. R.; Magstadt, D. R.; Petersen, B.; Timmermans, S. J.; Gauger, P. C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A. C.; Gorden, P. J.; Plummer, P. J.; Main, R. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerg Infect Dis 2024, 30, 1335–1343. [Google Scholar] [CrossRef]
- Caserta, L. C.; Frye, E. A.; Butt, S. L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L. M.; Thompson, A. C.; Koscielny, M. P.; Cronk, B.; Johnson, A.; Kleinhenz, K.; Edwards, E. E.; Gomez, G.; Hitchener, G.; Martins, M.; Kapczynski, D. R.; Suarez, D. L.; Alexander Morris, E. R.; Hensley, T.; Beeby, J. S.; Lejeune, M.; Swinford, A. K.; Elvinger, F.; Dimitrov, K. M.; Diel, D. G. Spillover of Highly Pathogenic Avian Influenza H5N1 Virus to Dairy Cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef]
- Authority (EFSA), E. F. S.; Alvarez, J.; Bortolami, A.; Ducatez, M.; Guinat, C.; Stegeman, J. A.; Broglia, A.; Jensen, H.; Kryemadhi, K.; Gervelmeyer, A. Risk Posed by the HPAI Virus H5N1, Eurasian Lineage Goose/Guangdong Clade 2.3.4.4b. Genotype B3.13, Currently Circulating in the US. EFSA Journal 2025, 23, e9508. [Google Scholar] [CrossRef]
- Samanta, L.; Parida, R.; Dias, T. R.; Agarwal, A. The Enigmatic Seminal Plasma: A Proteomics Insight from Ejaculation to Fertilization. Reproductive Biology and Endocrinology 2018, 16, 41. [Google Scholar] [CrossRef]
- Gautier, C.; Aurich, C. “Fine Feathers Make Fine Birds” – The Mammalian Sperm Plasma Membrane Lipid Composition and Effects on Assisted Reproduction. Animal Reproduction Science 2022, 246, 106884. [Google Scholar] [CrossRef]
- Raheja, N.; Choudhary, S.; Grewal, S.; Sharma, N.; Kumar. , N. A Review on Semen Extenders and Additives Used in Cattle and Buffalo Bull Semen Preservation. Journal of Entomology and Zoology Studies 2018, 6, 293–245. [Google Scholar]
- Bodu, M.; Hitit, M.; Greenwood, O. C.; Murray, R. D.; Memili, E. Extender Development for Optimal Cryopreservation of Buck Sperm to Increase Reproductive Efficiency of Goats. Front. Vet. Sci. 2025, 12. [Google Scholar] [CrossRef]
- National Animal Health Laboratory Network. Nucleic Acid Extraction Using the INDICAL BIOSCIENCE IndiMag Pathogen Kit on a Magnetic Particle Processor (NVSL-SOP-0645). National Veterinary Services Laboratories NAHLN Document List. https://www.aphis.usda.gov/media/document/15399/file (accessed 2025-09-26).
- National Animal Health Laboratory Network. Standard operating procedure for Nucleic Acid Extraction Using the MagMAX Core Nucleic Acid Purification Kit on a Magnetic Particle Processor. (NVSL-SOP-0643. National Veterinary Services Laboratories NAHLN Document List. https://www.aphis.usda.gov/media/document/15399/file (accessed 2025-09-26).
- National Animal Health Laboratory Network. Real-time RT-PCR Detection of Influenza A and Avian Paramyxovirus Type-1. (NVSL-SOP-0068), /: Veterinary Services Laboratories NAHLN Document List. https, 1539.
- Vandenburg-Carroll, A.; Marthaler, D. G.; Lim, A. Enhancing Diagnostic Resilience: Evaluation of Extraction Platforms and IndiMag Pathogen Kits for Rapid Animal Disease Detection. Microbiology Research 2025, 16, 80. [Google Scholar] [CrossRef]
- Ministry for Primary Industries, N. Z. Import Health Standard: Bovine Germplasm. www.mpi.govt.nz/dmsdocument/46537-Bovine-Germplasm-2025-Import-Health-Standard/(accessed 2025-10-18).
- Yan, L.; Toohey-Kurth, K. L.; Crossley, B. M.; Bai, J.; Glaser, A. L.; Tallmadge, R. L.; Goodman, L. B. Inhibition Monitoring in Veterinary Molecular Testing. J Vet Diagn Invest 2020, 32, 758–766. [Google Scholar] [CrossRef]
- Lee, S. M.; Balakrishnan, H. K.; Doeven, E. H.; Yuan, D.; Guijt, R. M. Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review. Biosensors (Basel) 2023, 13, 980. [Google Scholar] [CrossRef]
- Schellhammer, S. K.; Hudson, B. C.; Cox, J. O.; Dawson Green, T. Alternative Direct-to-amplification Sperm Cell Lysis Techniques for Sexual Assault Sample Processing. J Forensic Sci 2022, 67, 1668–1678. [Google Scholar] [CrossRef]
- Hennekens, C. M.; Cooper, E. S.; Cotton, R. W.; Grgicak, C. M. The Effects of Differential Extraction Conditions on the Premature Lysis of Spermatozoa. J Forensic Sci 2013, 58, 744–752. [Google Scholar] [CrossRef]
- Yoshida, K.; Sekiguchi, K.; Mizuno, N.; Kasai, K.; Sakai, I.; Sato, H.; Seta, S. The Modified Method of Two-Step Differential Extraction of Sperm and Vaginal Epithelial Cell DNA from Vaginal Fluid Mixed with Semen. Forensic Science International 1995, 72, 25–33. [Google Scholar] [CrossRef]
- Jaramillo, D.; Foxwell, J.; Burrows, L.; Snell, A. Mycoplasma Bovis Testing for the Screening of Semen Imported into New Zealand. New Zealand Veterinary Journal 2023, 71, 200–208. [Google Scholar] [CrossRef]
- ThermoFisher SCIENTIFIC. MagMAXTM CORE Nucleic Acid Purification Kit USER GUIDE, Revison D00, 2024. https://documents.thermofisher.com/TFS-Assets/LSG/manuals/MAN0015944_MagMAXCORE_NA_Kit_UG.pdf.
- Hsiao, C.-C.; Lin, C.-C.; Chen, Y.-M.; Cheng, M.-C.; Davison, S.; Ma, J.; Dai, H.-L. Quantitative Real-Time PCR Detection of Inactivated H5 Avian Influenza Virus in Raw Milk Samples by Miniaturized Instruments Designed for On-Site Testing. bioRxiv 2025, 2025.06.02.657307. [Google Scholar] [CrossRef]
- Miller, M. R.; Braun, E.; Ip, H. S.; Tyson, G. H. Domestic and Wild Animal Samples and Diagnostic Testing for SARS-CoV-2. Vet Q 2023, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Foot and Mouth Disease; OIE Terrestrial Manual 2009.
Figure 1.
Box and whisker plot illustrating the Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC) CT values for each semen sample using the MagMAX CORE (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment). The categories of semen extenders are egg yolk-based (Yellow), sexed milk-based (Green or Pink), and milk-based (White). The passing CT value criteria of 34.50 is represented by the black colored horizontal line.
Figure 1.
Box and whisker plot illustrating the Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC) CT values for each semen sample using the MagMAX CORE (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment). The categories of semen extenders are egg yolk-based (Yellow), sexed milk-based (Green or Pink), and milk-based (White). The passing CT value criteria of 34.50 is represented by the black colored horizontal line.
Figure 2.
Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC) CT value variance between the different extraction methods using MagMAX CORE (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment). The categories of semen extenders are egg yolk-based (Yellow), sexed milk-based (Green or Pink), and milk-based (White). The asterisks indicate significance with p < 0.0001.
Figure 2.
Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC) CT value variance between the different extraction methods using MagMAX CORE (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment). The categories of semen extenders are egg yolk-based (Yellow), sexed milk-based (Green or Pink), and milk-based (White). The asterisks indicate significance with p < 0.0001.
Figure 3.
Box and whisker plot illustrating the Mycoplasma bovis (M. bovis), bovine viral diarrhea virus (BVDV), and bovine herpesvirus-1 (BHV-1) positive extended semen samples with passing internal controls using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretreatment) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
Figure 3.
Box and whisker plot illustrating the Mycoplasma bovis (M. bovis), bovine viral diarrhea virus (BVDV), and bovine herpesvirus-1 (BHV-1) positive extended semen samples with passing internal controls using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretreatment) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
Figure 4.
Standard curve for M. bovis reference strain ATCC 25,523 using the MagMAX CORE with 12.5 μL semen input and pretreatment (CORE 12.5-pretreatment) and the IndiMag Pathogen Kit with 100 μL semen input (Pathogen 100-na) and pretreatment (Pathogen 100-pretreatment).
Figure 4.
Standard curve for M. bovis reference strain ATCC 25,523 using the MagMAX CORE with 12.5 μL semen input and pretreatment (CORE 12.5-pretreatment) and the IndiMag Pathogen Kit with 100 μL semen input (Pathogen 100-na) and pretreatment (Pathogen 100-pretreatment).
Figure 5.
Box and whisker plot of the CT values for the spiked influenza A virus (IAV) and the internal controls in semen extracted using the IndiMag Pathogen Kit with 100 µL semen input on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays.
Figure 5.
Box and whisker plot of the CT values for the spiked influenza A virus (IAV) and the internal controls in semen extracted using the IndiMag Pathogen Kit with 100 µL semen input on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays.
Figure 6.
Repeatability for the spiked influenza A virus (IAV) samples extracted using the IndiMag Pathogen Kit with 100 µL semen input on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays. The solid black line near zero represents mean variance.
Figure 6.
Repeatability for the spiked influenza A virus (IAV) samples extracted using the IndiMag Pathogen Kit with 100 µL semen input on the National Animal Health Laboratory Network (NAHLN) and Wisconsin Veterinary Diagnostic Laboratory (WVDL) IAV assays. The solid black line near zero represents mean variance.
Table 1.
Percentage of samples passing using the MagMAX CORE Kit (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment), using Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC). The categories of semen are egg yolk-based extender (Yellow), milk-based extender sexed-semen (Green or Pink), and milk-based extender (White).
Table 1.
Percentage of samples passing using the MagMAX CORE Kit (CORE) with 200 µL semen input (200-na), 50 µL semen input (50-na), and 12.5 µL semen input with pretreatment (12.5-pretreatment); and the IndiMag Pathogen Kit (Pathogen) with 200 µL semen input (200-na), 100 µL semen input (100-na), and 100 µL semen input with pretreatment (100-pretreatment), using Xeno RNA and Wisconsin Veterinary Diagnostic Laboratory internal control (WVDL IC). The categories of semen are egg yolk-based extender (Yellow), milk-based extender sexed-semen (Green or Pink), and milk-based extender (White).
| Extraction Kit |
CORE |
Pathogen |
|
| Internal Control |
Xeno RNA |
WVDL IC |
Xeno RNA |
WVDL IC |
| Semen input & modification |
200-na |
50-na |
12.5-pretreat-ment |
200-na |
50-na |
12.5-pretreat-ment |
200-na |
100-na |
100-pretreat-ment |
200-na |
100-na |
100-pretreat-ment |
| Yellow (n=24) |
25.0 |
62.5 |
100.0 |
100.0 |
83.3 |
100.0 |
41.7 |
100.0 |
100.0 |
95.8 |
100.0 |
100.0 |
| Green (n=16) |
12.5 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
50.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
| Pink (n=16) |
81.3 |
87.5 |
100.0 |
100.0 |
93.8 |
100.0 |
87.5 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
| White (n=32) |
21.9 |
62.5 |
100.0 |
93.8 |
96.9 |
100.0 |
3.1 |
100.0 |
59.4 |
83.3 |
100.0 |
68.8 |
| Overall % Passing rate (n=88) |
31.8 |
73.9 |
100.0 |
97.7 |
93.2 |
100.0 |
37.5 |
100.0 |
85.2 |
94.3 |
100.0 |
88.6 |
Table 2.
Percentage of pathogen detection for Mycoplasma bovis (M. bovis), bovine viral diarrhea virus (BVDV), and bovine herpesvirus-1 (BHV-1) positive extended semen samples using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretrement) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
Table 2.
Percentage of pathogen detection for Mycoplasma bovis (M. bovis), bovine viral diarrhea virus (BVDV), and bovine herpesvirus-1 (BHV-1) positive extended semen samples using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretrement) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
| |
CORE 12.5-pretreatment |
Pathogen
100-na |
Pathogen
100-pretreatment |
| Replicate |
1 |
2 |
1 |
2 |
1 |
2 |
|
M. bovis (n=8) |
75.0% |
87.5% |
75.0% |
100.0% |
87.5% |
100.0% |
| BVDV (n=5) |
60.0% |
60.0% |
80.0% |
80.0% |
100.0% |
80.0% |
| BHV-1 (n=23) |
87.0% |
95.7% |
95.7% |
95.7% |
95.7% |
100.0% |
| Sensitivity (n=36) |
80.6% |
88.9% |
88.9% |
94.4% |
94.4% |
97.2% |
Table 3.
The limit of detection, coefficient of correlation of the standard curve (R2), and percent PCR efficiency for M. bovis extracted using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretrement) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
Table 3.
The limit of detection, coefficient of correlation of the standard curve (R2), and percent PCR efficiency for M. bovis extracted using the MagMAX CORE Kit (CORE) extraction with 12.5 µL semen input and pretreatment (12.5-pretrement) and using the IndiMag Pathogen Kit (Pathogen) extraction with 100 µL semen input (100-na) and 100 µL semen input with pretreatment (100-pretreatment).
| |
CORE
12.5-pretreatment
|
Pathogen
100-na
|
Pathogen
100-pretreatment
|
| Replicate |
1 |
2 |
3 |
1 |
2 |
3 |
1 |
2 |
3 |
| Limit of Detection |
5 |
5 |
6 |
6 |
7 |
7 |
5 |
5 |
5 |
| R2 value |
0.996 |
0.956 |
0.986 |
1.000 |
1.000 |
0.999 |
0.994 |
0.988 |
0.981 |
| PCR Efficiency (%) |
81.6 |
84.7 |
133.5 |
90.4 |
93.2 |
94.1 |
92.8 |
76.8 |
71.3 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).