1. Introduction
The United States (U.S.) is currently facing significant outbreaks of the H5N1 strain of hghly pathogenic avian influenza (HPAI). Since February 2022, HPAI has been detected in both wild and domestic bird populations across all 50 states, leading to substantial poultry losses and economic impacts [
1]. Notably, in 2024, the B3.13 subtype of H5N1 HPAI was identified in U.S. dairy cattle for the first time [
2], and subsequent detection of subtype D1.1 [
3] raises concerns about cross-species transmission [
4]. Additionally, human cases have emerged, including 70 cases at the time of this manuscript preparation and the first death reported in Louisiana in January 2025 [
5]. These concurrent outbreaks underscore the critical need for vigilant surveillance and comprehensive response strategies to mitigate the spread and impact of these viruses on both animal and public health.
African swine fever, classical swine fever, and foot-and-mouth disease are significant foreign animal diseases (FADs) to the United States, with severe economic consequences to the livestock industries [
6]. The spread of African swine fever virus (ASFV) to the Dominican Republic in 2021 heightens the potential risk to U.S. domestic and feral swine populations [
7]. Classical swine fever virus (CSFV) remains a global concern, with outbreaks reported in regions of Asia, parts of Europe, and South America [
8]. The recent foot-and-mouth disease virus (FMDV) detection in a
water buffalo farm in Germany—the first detection in Germany since 1988 and the first in the European Union since 2011 [
9]
—indicates the need for continued surveillance, early detection, and reliable diagnostic methods to prevent outbreaks.
The continued HPAI outbreak underscores the excellent efforts of the National Animal Health Laboratory Network (NAHLN) (established in the U.S. in 2002) to coordinate diagnostic testing to detect and respond to animal disease outbreaks in the United States. The NAHLN’s mission is to enhance the nation’s animal health infrastructure by providing reliable, rapid diagnostic testing, ensuring early detection of significant animal diseases, and supporting emergency response efforts to protect animal and public health [
10]. High-throughput (96-well format) molecular testing solutions are approved by the NAHLN and are the backbone of high volume diagnostic and surveillance testing. In addition to high-throughput platforms, low- and medium-throughput equipment are required when testing a new suspected outbreak, as only a few samples are submitted, and these samples require immediate results. The magnetic particle processors, such as the KingFisher Duo Prime (KF Duo Prime), IndiMag 48s (IM48s), and IndiMag 2 (IM2) enhance flexibility in laboratory workflows for PCR testing, particularly for low number of samples for NAHLN assays. Several magnetic-based extraction kits are approved for sample extraction for the NAHLN assays, requiring the reagents are manually pipetted into the extraction plates for each equipment. The IndiMag Pathogen prefilled extraction available for the KingFisher 96-well family of extractors, the IM48s, and the IM2 offers improved workflow efficiency, reduced technical errors, and provides time and cost savings by eliminating reagent dispensing steps.
At the start of this study in 2022, the approved extraction protocols for Influenza A virus (IAV) and avian paramyxovirus Type-1 (APMV-1) include Trizol LS reagents, RNeasy spin column-based extraction kit, and MagMAX Viral RNA Isolation Kit, and MagMAX Pathogen RNA/DNA Kit on Biosprint 96, MagMAX Express 96, KingFisher 96, and KingFisher Flex magnetic particle processors. The approved extraction protocols for FADs are MagMAX Pathogen RNA/DNA Kit, MagMAX CORE Nucleic Acid Purification Kit, and IndiMag Pathogen Kit on Biosprint 96, MagMAX Express 96, KingFisher 96, and KingFisher Flex magnetic particle processors. The IndiMag Pathogen Kit is NAHLN-approved for FADs detection assays, but lacks NAHLN approval for IAV and APMV-1 surveillance testing and is evaluated in this study. Aside from the approved KingFisher 96-well family of extractors, additional low- and medium-throughput nucleic acid extractors and reagents should be suggested to provide reliable equipment and reagents alternatives, minimizing the risk of supply chain issue and monopolization of governmental testing. In this study, the KF Duo Prime, IM48s, IM2, IndiMag Pathogen kits, are investigated as alternate solutions to NAHLN IAV, APMV-1, ASFV, CSFV, and FMDV testing.
2. Materials and Methods
Field and Reference Samples
For diagnostic sensitivity evaluation, a total of 31 previously positive field samples containing low pathogenic influenza A (LPAI) or APMV-1 viruses were used. These consisted of three tracheal swabs from domestic turkey, four oropharyngeal swabs from pigeons, and 24 pooled oropharyngeal or cloacal swabs from waterfowl. Ten of the positive pooled oropharyngeal or cloacal swabs were spiked into 10 negative IAV tissue homogenates from turkeys for testing. Thirty previously IAV and APMV-1 negative field samples (7 tissue homogenates and 23 trachea, oropharyngeal, or cloacal swabs) were used for diagnostic specificity evaluation. All samples were dispensed and stored in an ultralow freezer until ready for use. Field samples were not evaluated for the three FADs due to the lack of availability.
Three LPAI and three APMV-1 reference strains were kindly provided by the National Veterinary Service Laboratories (NVSL), while the Foreign Animal Disease Diagnostic Laboratory (FADDL) provided a single synthetic phage construct each for the ASFV, CSFV, and FMDV (Appendix Table A1) for the analytical sensitivity evaluation. Ten-fold serial dilutions of the references were prepared in preferred media (BHI or TBTB), and multiple aliquots were made and stored in an ultralow freezer until ready for use.
Extraction Chemistries and Equipment
The ThermoFisher MagMAX Viral RNA Kit (MagMAX Viral) and the MagMAX CORE Nucleic Acid Purification Kit (MagMAX CORE) were the approved extraction chemistries for the IAV and APMV-1 assays at the time of this study (NVSL-SOP-0068, “NAHLN Real-time RT-PCR Detection of Influenza A and Avian Paramyxovirus Type-1”). The MagMAX CORE and the INDICAL IndiMag Pathogen Kits were approved for FADs testing (NVSL-SOP-0643, “Nucleic Acid Extraction Using the MagMAX Core Nucleic Acid Purification kit on a Magnetic Particle Processor” and NVSL-SOP-0645, “Nucleic Acid Extraction Using the INDICAL BIOSCIENCE IndiMag Pathogen Kit on a Magnetic Particle Processor”). The study involved comparing the manual fill and prefilled versions of the IndiMag Pathogen kits to the MagMAX Viral or MagMAX CORE extraction kits.
The Kingfisher Flex (KF Flex) (ThermoFisher) is NAHLN-approved for the extraction of IAV and APMV-1 (NVSL-SOP-0068) and FADs (NVSL-SOP-0643; NVSL-SOP-0644, “Nucleic Acid Extraction Using the MagMAX Pathogen RNA/DNA Kit on a Magnetic Particle Processor”; NVSL-SOP-0645). The KF Duo Prime (ThermoFisher, 5400110), IndiMag 48s (INDICAL, IN943048s) and IndiMag 2 (INDICAL, IN950048) were not NAHLN-approved equipment in the previous SOPs at the time of this study and were evaluated as alternative extraction platforms.
For the IAV and APMV-1 extraction evaluation, each of the serial diluted reference strains and samples were extracted using the MagMAX Viral on the KF Flex according to NVSL-SOP-0068, which was considered the gold standard for comparison of the performance of extraction chemistry and equipment. The 6 serial diluted reference strains and samples were each extracted on the KF Flex and IM48s using manual and/or prefilled versions of IndiMag Pathogen kits to evaluate reagents and IM48s equipment performance. Subsequently, the test samples and a single IAV and APMV-1 reference strain were extracted with the IndiMag Pathogen Kit on IM2 and the MagMAX CORE on Duo Prime to evaluate equipment performance. The manufacturer’s instructions for the MagMAX Viral, MagMAX CORE, and IndiMag Pathogen kits were followed except for a single modification. MagMAX Viral has a recommended 50 µL sample input while the MagMAX CORE and IndiMag Pathogen kits have a recommended 200 µL input sample volume. Thus, a mixture of 50 µL sample input with 150 µL of phosphate-buffered saline (PBS) was used as starting material for the MagMAX CORE and IndiMag Pathogen kits for equivalent input sample volume between the extraction kits.
For the FADs extraction evaluation, each reference construct (200 µL) was extracted using the MagMAX CORE on KF Flex according to NVSL-SOP-0643, which was the gold standard for comparison of the performance of equipment under evaluation. Each reference construct was also extracted with the IndiMag Pathogen Kit on the IM48s and IM2, and the MagMAX CORE on KF Duo Prime. Extraction was carried out per manufacturers’ instruction.
Polymerase Chain Reactions (PCR)
All IAV and APMV-1 extracts were evaluated using AgPath-ID One Step PCR reagents (ThermoFisher) in the ABI 7500 with version 2.3 software using the harmonized IAV protocol and the APMV-1 protocol per NAHLN SOP (NVSL-SOP-0068). Current harmonized IAV protocol indicates analysis at 5% manual threshold value and the APMV-1 protocol uses auto threshold setting for analysis of results. This study compared data across multiple PCR runs, and the auto threshold fluctuated from run to run as the algorithm was influenced by the samples in each run. We performed multiple analyses and found that the 5% manual threshold value for IAV runs was within the proximity of 0.2, and a threshold of 0.2 also landed accurately around the midpoint of the log linear phase for the APMV-1 assays. Based on this observation, all the IAV and APMV-1 PCR runs were normalized with a manual threshold at 0.2 (with auto baseline setting), to allow for ease of comparison of all data points across the runs.
All FADs extracts were evaluated using the TaqMan Fast Virus One-Step Master Mix (ThermoFisher) in the ABI 7500 with version 2.3 software using the protocol per NAHLN SOP (NVSL-SOP-0642, “Classical Swine Fever and Foot and Mouth Disease rRT-PCR using the Taqman Fast Virus 1-Step Master Mix on the Applied Biosystems 7500 Real-time PCR System” or NVSL-SOP-0646, “Preparation, Performance, and Interpretation of the African Swine Fever rPCR Assay on the Applied Biosystems 7500 Real-time PCR System”). FAD targets and VetMAX Xeno Internal Positive Control RNA (Xeno, ThermoFisher) exogenous control were analyzed at the set threshold per SOP (ASFV and FMDV at 0.2, CSFV at 0.05, and Xeno at 0.1).
Analytical Sensitivity
A standard curve was generated using the ten-fold diluted samples for each of the references. The limit of detection (LOD) was determined by the end point dilution of the reference strains. The PCR efficiency and the R2 were obtained from the standard curve generated by the ABI 7500 software.
The repeatability within a single assay for IAV and APMV-1 was performed by running a high and a low concentration of the references. The low concentration was prepared to approximately ten-fold dilution more concentrated than the reported LOD endpoint, and the high concentration was a dilution with a 25-30 cycle threshold (CT) range. Each sample was extracted and PCR in 5 replicates to assess the single-run repeatability. This was repeated one more time (a total of 2 runs) to assess the inter-run repeatability.
Diagnostic Sensitivity and Diagnostic Specificity
For IAV and APMV-1 assays, diagnostic sensitivity was evaluated using 31 positive samples (20 LPAI and 11 APMV-1) from waterfowl or domestic poultry. Samples were tested as collected (oropharyngeal, trachea, or cloacal swabs) or spiked into pooled tissue matrix. Diagnostic specificity was evaluated using 30 negative diagnostic samples as collected (oropharyngeal, trachea, or cloacal swabs and tissues).
Statistical Analysis
The assay performance was compared using the LOD, R2 , PCR efficiency, diagnostic sensitivity, and diagnostic specificity. Standard deviation and coefficient of variant (CV) were calculated for assay precision. Standard F-test or ANOVA tests were used to infer statistical significance among various methods. Statistical analysis was computed using the Prism software (GraphPad). Figures were generated using Tableau software (Salesforce).
3. Results
The study was divided into sections due to availability of testing material and the launch of the IM2. The first study compared the manual fill and prefilled versions of the IndiMag Pathogen Kit on the KF Flex and IM48s to the MagMAX Viral on KF Flex. The LOD was the same across the extraction chemistries and instrumentation (
Table 1, supplemental data in
Table S1, LOD plots in Appendix Figure A1). When LOD differences occurred, the last CT value did not represent a ten-fold dilution and deemed non-repeatable was excluded. The R
2 ranged between 0.951 and 1.00 across the 6 references and instrumentation. The PCR efficiency ranged between 82.5% and 107.6%, with the lowest value occurring with prefilled IndiMag Pathogen kit on the IM48s and the highest value occurring with MagMAX Viral on the KF Flex.
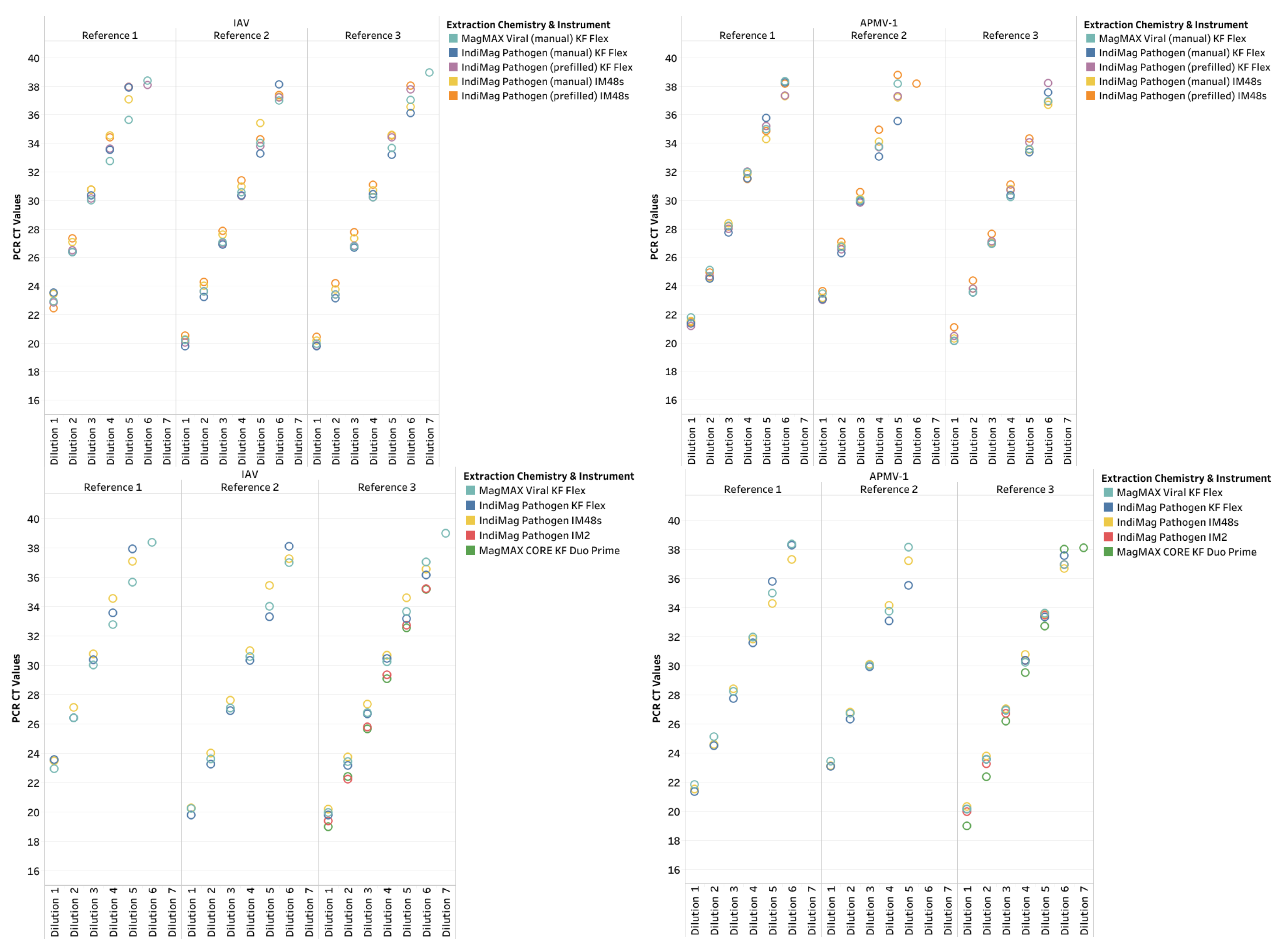
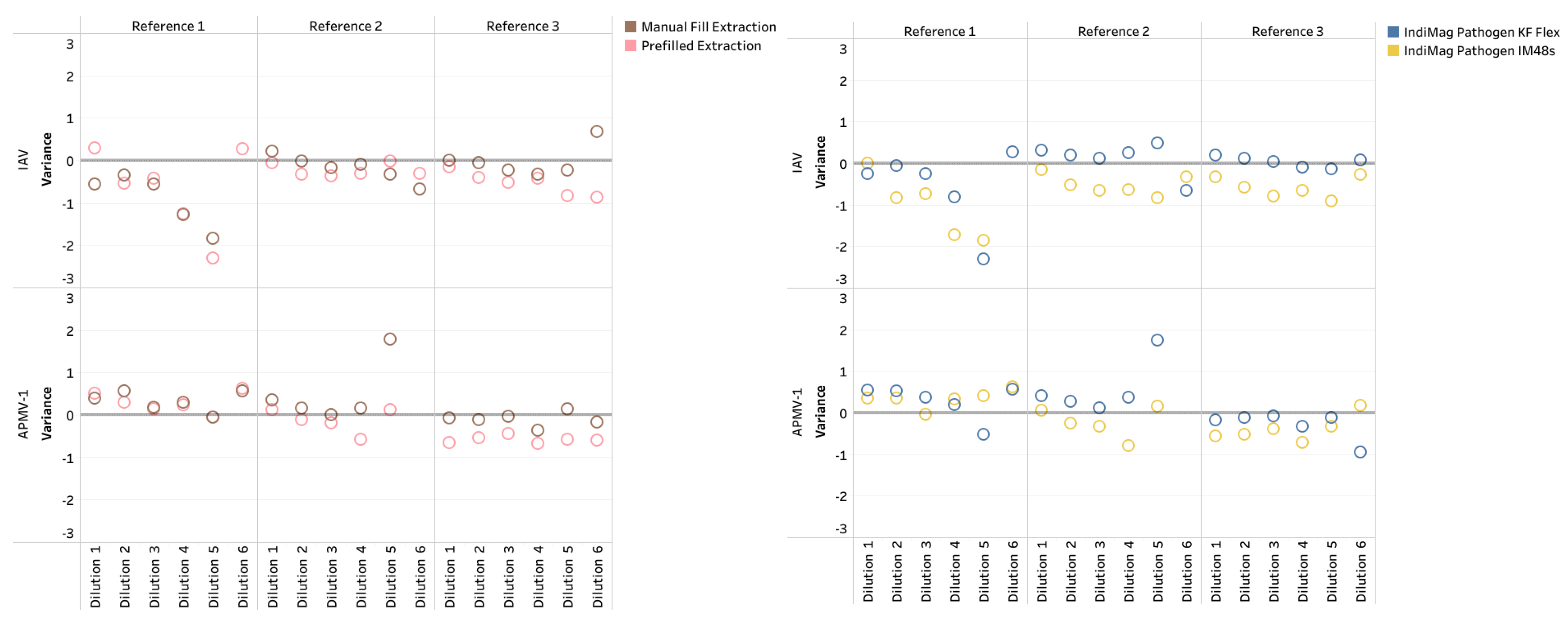
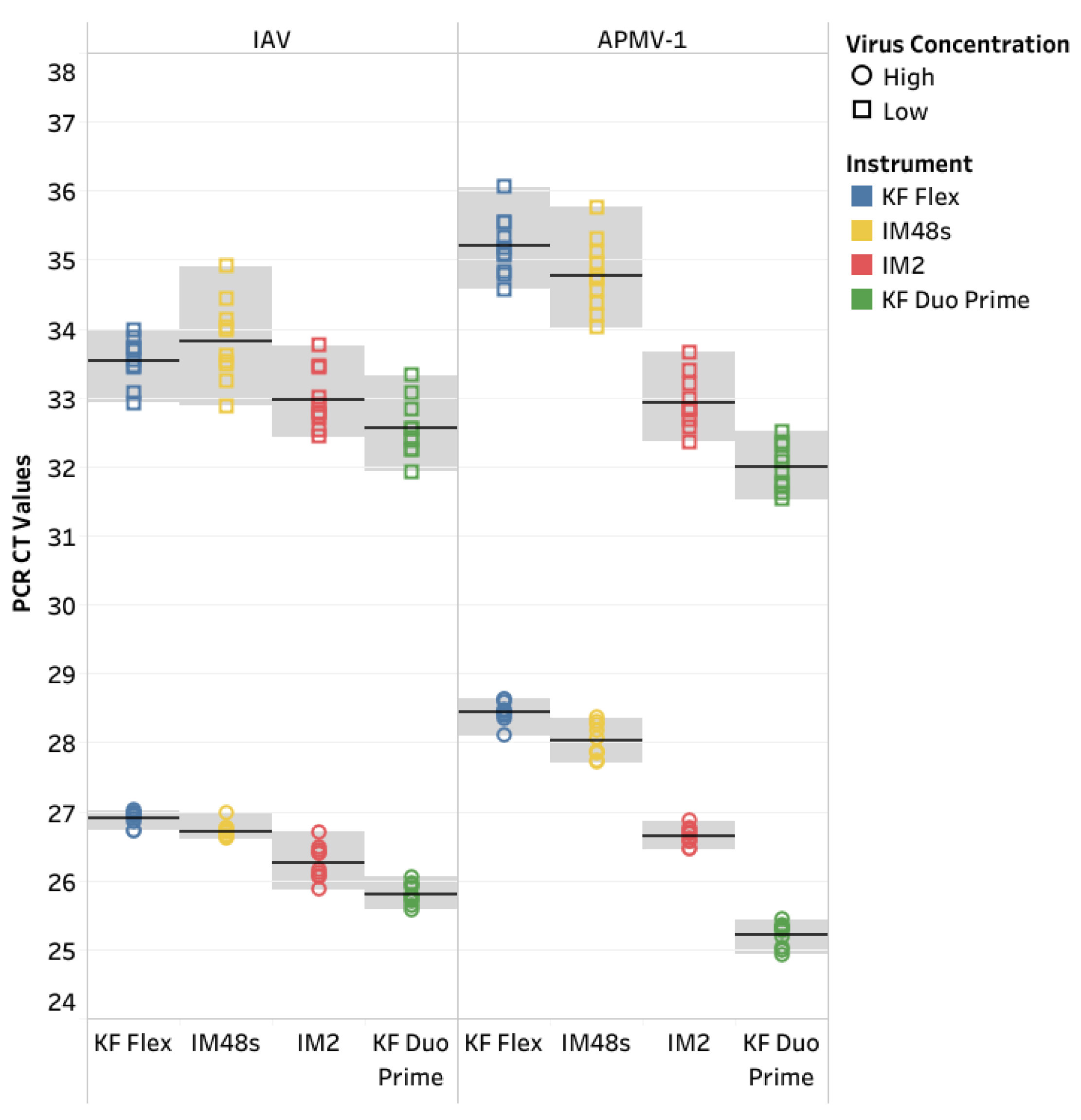
The CT values of manual fill and prefilled versions of the IndiMag Pathogen Kit on the KF Flex and IM48s for each dilution of the individual reference were averaged and compared to the MagMAX Viral on the KF Flex to investigate CT value differences in the format of the IndiMag Pathogen kit (
Figure 1a). Both the manual fill and prefilled versions of the IndiMag Pathogen Kit have the same dynamic range as the NAHLN-approved MagMAX Viral. The mean CT value difference between the manual fill and prefilled versions of the IndiMag Pathogen Kit to MagMAX Viral on the KF Flex was -0.06 and -0.33, respectively. The standard error of the means (SEM) was -0.2691 ± 1.391, with a lack of significance (F-test, p= 0.9061) between formats of the IndiMag Pathogen kit.
Next the CT values of the KF Flex and the IM48s for the two versions of the IndiMag Pathogen Kit for each dilution of the individual reference were averaged and compared to the MagMAX Viral on the KF Flex to investigate differences in instrumentation (
Figure 1b). The mean CT value difference between the KF Flex and IM48s with the two versions of the IndiMag Pathogen kit to MagMAX Viral on KF Flex was 0.00 and -0.39, respectively. The SEM was -0.3932 ± 1.391, with a lack of significant difference (F-test, p= 0.9269) using the IndiMag Pathogen Kit on the KF Flex and IM48s.
Subsequently, both versions of the IndiMag Pathogen Kit using the KF Flex and IM48s became approved by NAHLN for IAV and APMV-1 during the study. Thus, only a single IAV and APMV-1 reference strain (per NAHLN requirements) was evaluated using the IndiMag Pathogen kit on the IM2 and using MagMAX CORE on the KF Duo Prime as a verification of these instruments. Similar to the previous LOD results, the last CT value did not represent a ten-fold dilution and was deemed non-repeatable (
Table 1, also supplemental data in
Table S2, LOD plots in Appendix Figure A1). The R
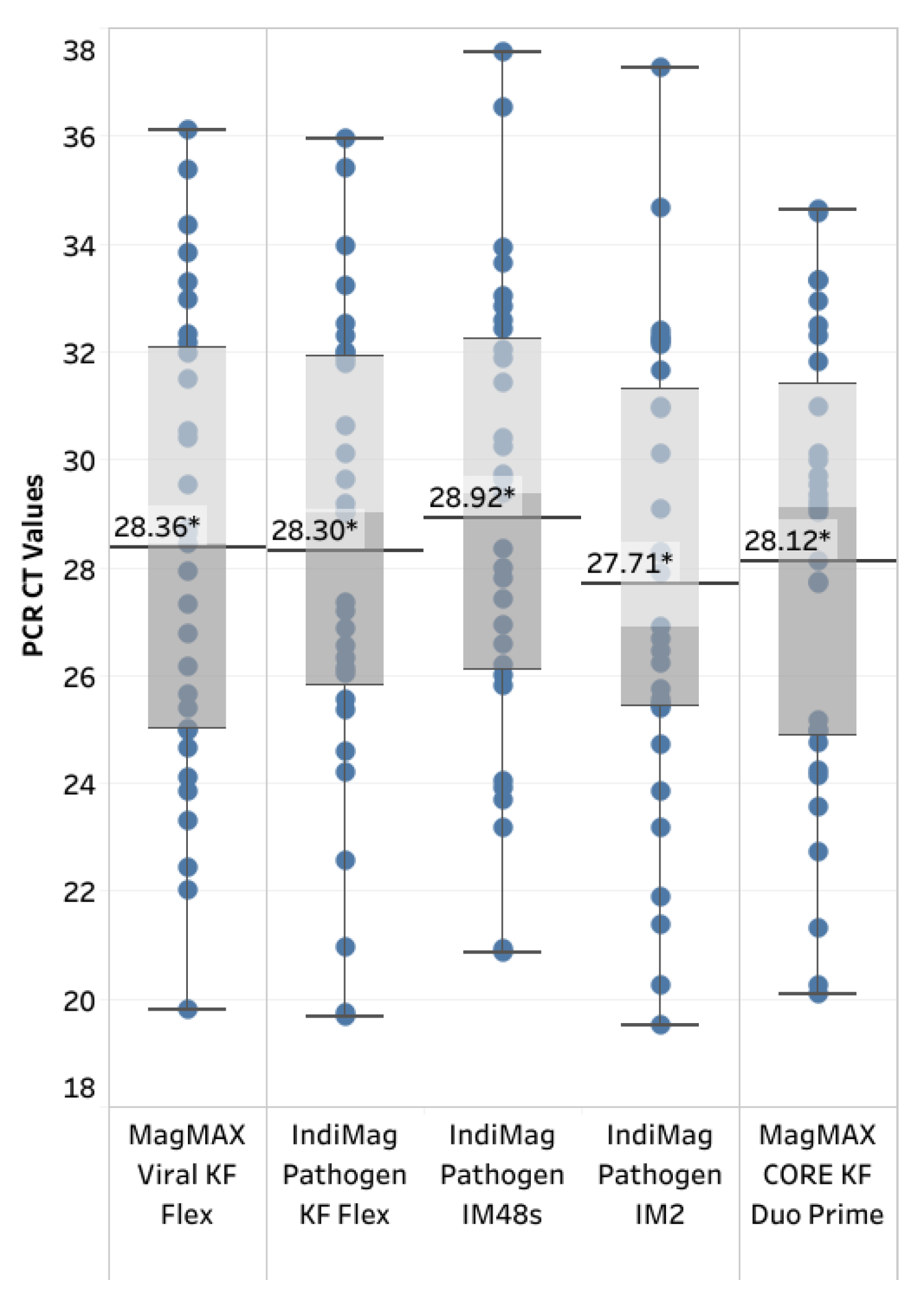
2 ranged between 0.995 and 1.00 across the two reference strains and two instruments. The PCR efficiency ranged between 96.62% and 104.4%, with the lowest value occurring with the IndiMag Pathogen kit on the IM2 and the highest value occurring with MagMAX CORE on the KF Duo Prime. All three of the low- and medium-throughput instruments have the same dynamic range as the NAHLN-approved high-throughput KF Flex. The CT value difference of the IM2 using the IndiMag Pathogen kit and the KF Duo Prime using MagMAX CORE compared to MagMAX Viral on KF Flex was 0.91 and 0.60, respectively. When the average CT values for all four new chemistry/instrument combinations were compared to MagMAX Viral on KF Flex (
Figure 2), the SEM was 0.6249 ± 1.405, 0.2958 ± 1.405, 1.852 ± 1.958 and 1.361 ± 1.901 for IndiMag Pathogen kit on KF Flex, on IM48s, on IM2 and MagMAXCORE on KF Duo Prime respectively, with a lack of significant difference (ANOVA test, p= 0.8738) observed.
The precision was evaluated using the IndiMag Pathogen kit on the KF Flex, IM48s, and IM2 and using MagMAX CORE on the KF Duo Prime. Five replicates at a high and a low concentration for both IAV and APMV-1 were run on different days, totaling 10 replicates at a high and a low concentration for IAV and APMV-1 for each instrument (
Figure 3, supplemental data
Table S3). The average mean CT value for the high and low concentrations of IAV was 26.42 and 33.22, respectively. The average inter-run standard deviation for the high and low concentrations of IAV was 0.16 and 0.45, respectively. The coefficient of variation (CV) ranged between 0.16% to 1.76% for IAV. The average mean CT value for the high and low concentrations of APMV-1 was 27.08 and 33.72, respectively. The average inter-run standard deviation for the high and low concentrations of APMV-1 was 0.18 and 0.42, respectively. The CV ranged between 0.50% to 1.52% for APMV-1.
A total of 32 positive and 30 negative samples were tested to evaluate diagnostic sensitivity and specificity for the IndiMag Pathogen kit on the KF Flex, IM48s, and IM2 and the MagMAX CORE on the KF Duo Prime compared to the MagMAX Viral on the KF Flex. The Kappa coefficient was k= 1.0, with 100 % in agreement for diagnostic sensitivity and specificity for the extraction chemistry on its respective instrumentation. The average CT value was slightly lower, with a positive shew using the IndiMag Pathogen kit on the IM2. However, a significant difference was lacking between the average CT values regardless of the extraction chemistry and instrumentation(ANOVA test, p= 0.8602) .
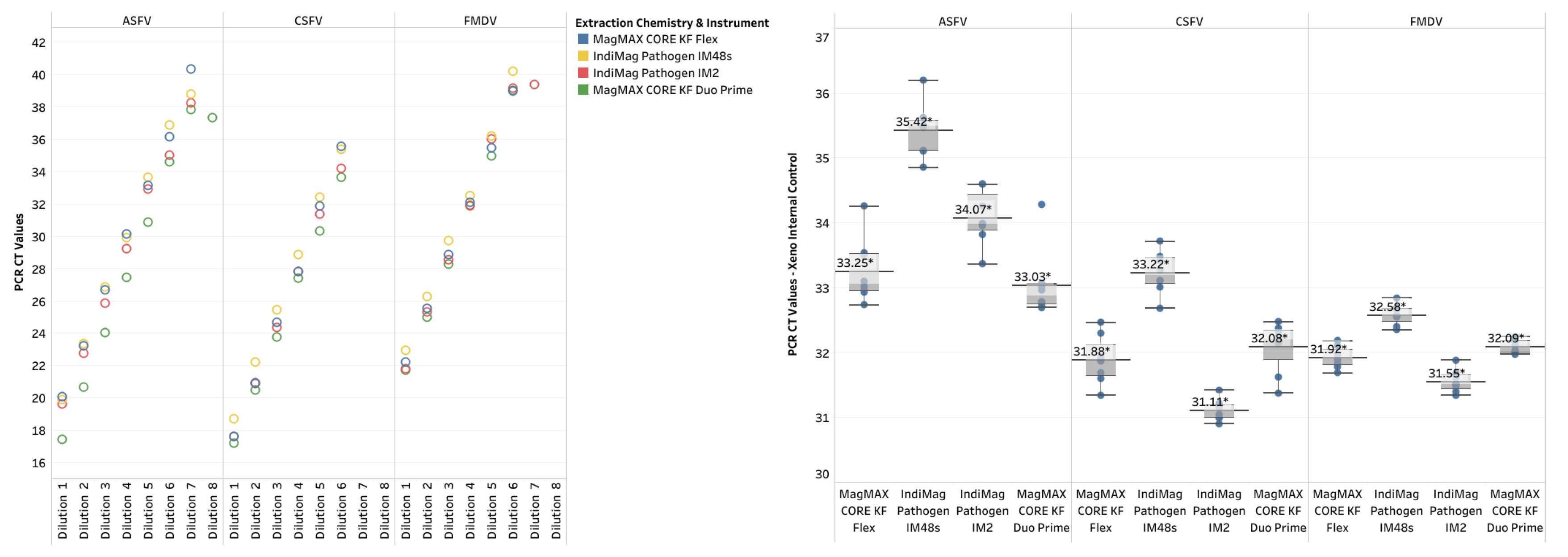
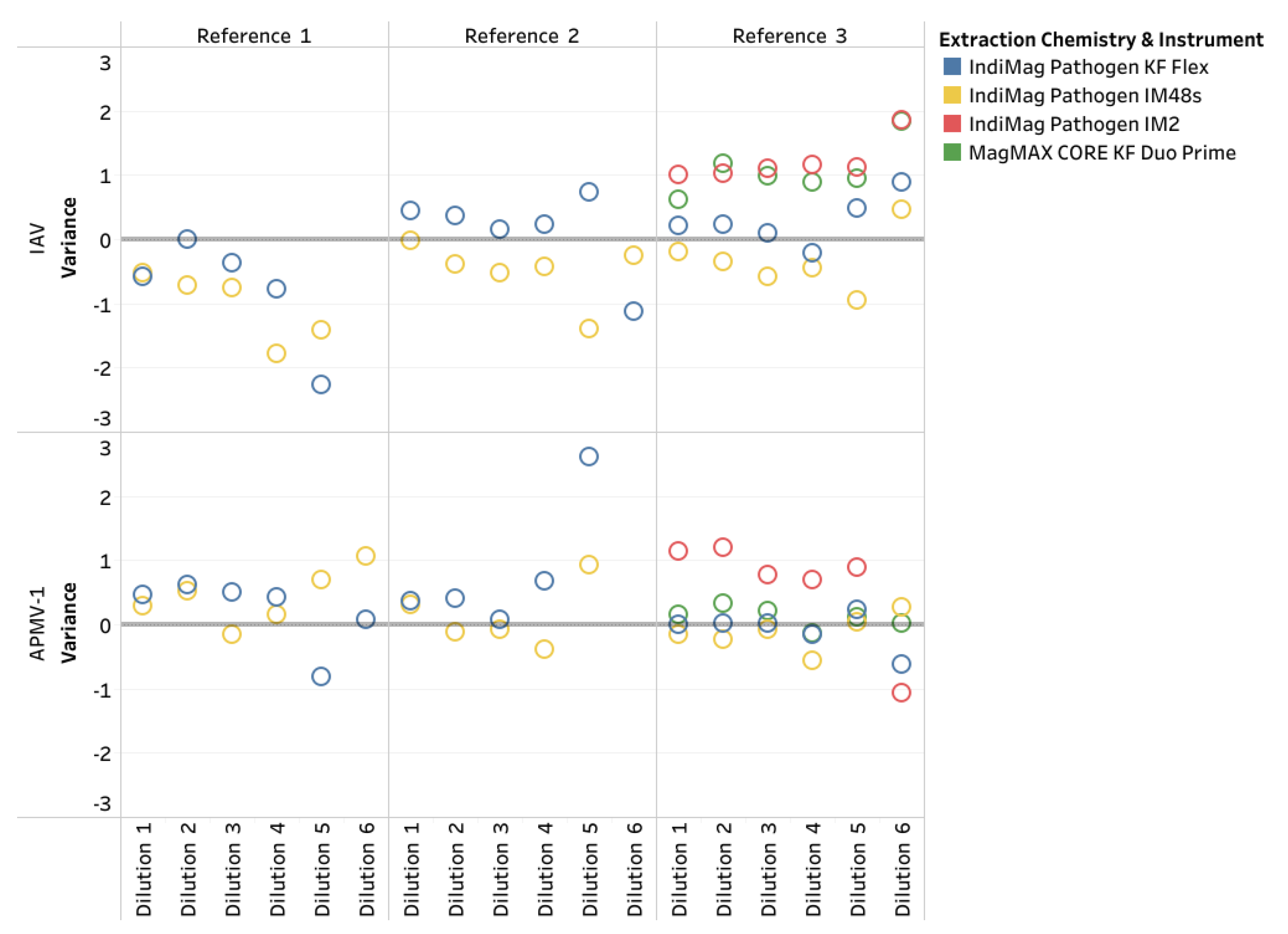
Finally, single references for ASFV, CSFV, and FMDV were evaluated using the IndiMag Pathogen Kit on the IM48s and IM2 and the MagMAX CORE on the KF Duo Prime compared to the MagMAX CORE on the KF Flex. Both extraction chemistries were NAHLN-approved for ASFV, CSFV, and FMDV. Similar to the IAV and APMV-1 LOD results, the CT value did not represent a ten-fold dilution when a difference in dilution level occurred, and was deemed non-repeatable (
Table 2, also supplemental material of CT values, LOD plots in Appendix A1). The R
2 ranged between 0.986 and 1.00 across the 3 references and instrumentation. The PCR efficiency ranged between 95.60% and 104.50%. All three of the low- and medium-throughput instruments have the same dynamic range as the NAHLN-approved high-throughput KF Flex. The CT value difference using the IndiMag Pathogen kit on the IM48s and IM2 and MagMAX CORE on the KF Duo Prime compared to MagMAX CORE on KF Flex was -0.4, 0.44, and 1.32, respectively (
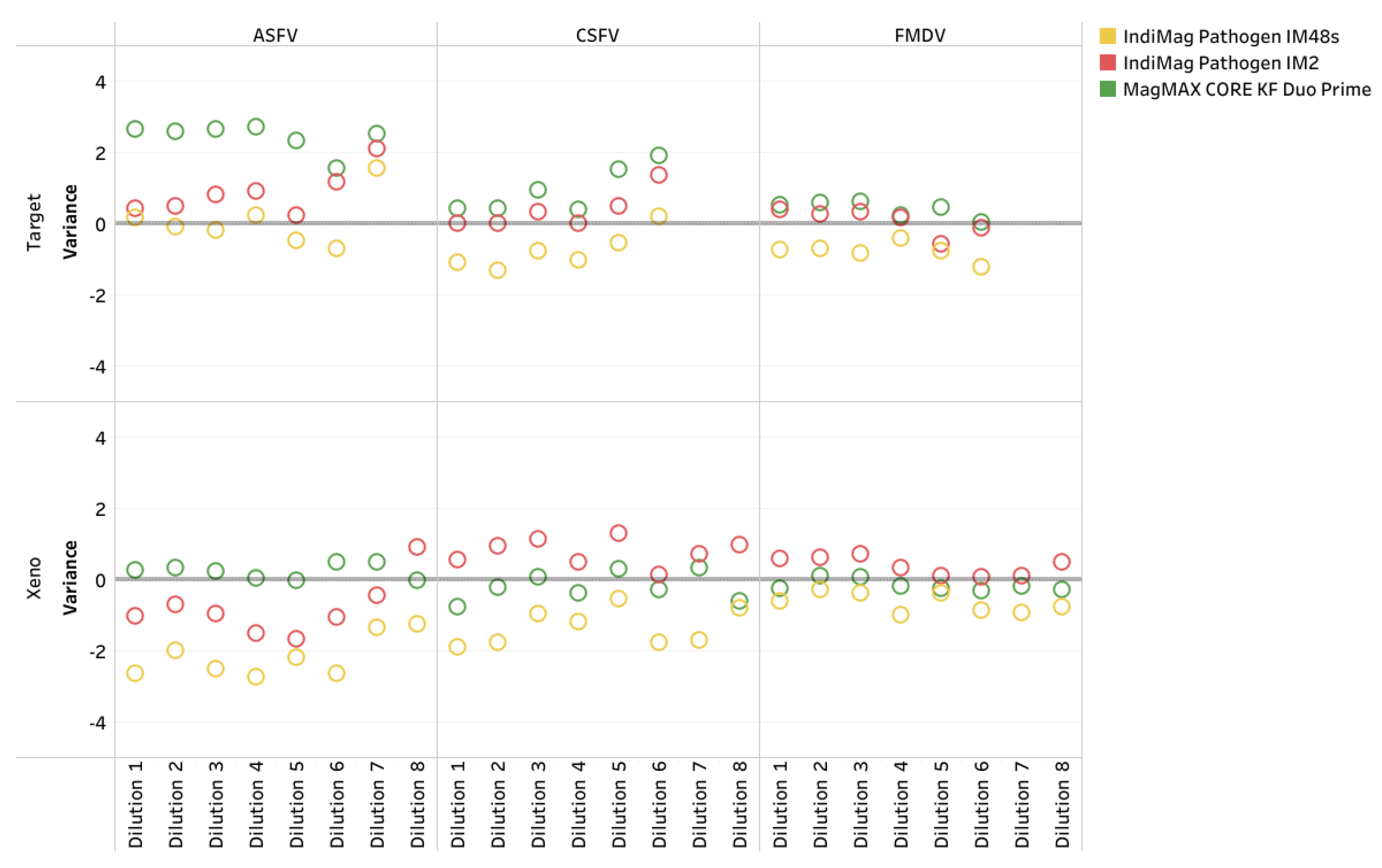
Figure 5). A significant difference was lacking between the average CT values for the FADs regardless of the extraction chemistry and instrumentation (ANOVA test, p= 0.8579). The VetMAX Xeno Internal Positive Control RNA was included in all of the ASFV, CSFV, and FMDV PCR assays; the CT values ranges are provided in
Figure A1. The CT value difference for Xeno RNA was -1.36, 0.09, and -0.05, respectively, with a lack of significant difference (ANOVA test, p= 0.7777).
4. Discussion
The ongoing H5N1 HPAI outbreak in wild and domestic bird populations across all 50 states in the United States and the detection of the B3.13 and D1.1 subtypes of H5N1 in U.S. dairy cattle pose substantial challenges to the U.S. agriculture industries. In addition to the HPAI outbreaks, the U.S. livestock industry faces ongoing threats from FADs, especially with ASFV in the Dominican Republic and the detection of FMDV in Germany, which highlights the critical need for vigilant surveillance, early detection, and robust diagnostic methods to prevent potential outbreaks in the United States, all of which is supported by NAHLN ensuring the nation’s animal health infrastructure and U.S. food supply through reliable and rapid diagnostic testing methods.
High-throughput molecular testing solutions have become essential to handle large volumes of samples during the HPAI outbreak. This study evaluated the performance of the IndiMag Pathogen Kit (manual fill and prefilled versions) on the KF Flex and IM48s compared to the NAHLN-approved MagMAX Viral extraction on KF Flex. Similar to previously reported [
11], there were no significant differences between manual fill versions of the IndiMag Pathogen kit and MagMAX Viral kit and the prefilled IndiMag Pathogen cartridges. The prefilled extraction significantly improves the laboratory workflow in multiple ways. First, the prefilled extraction eliminates numerous pipette steps of filling the extraction reagents into plastics, as only the sample and lysis buffer require pipetting. The reduced amount of manual handling leads to reduced risk of sample contaminations, failed extractions, and inaccurate and delayed results due to pipetting errors. Second, reducing the number of pipetting steps reduces the repetitive strain on the technician’s hands, providing overall a better working environment for the laboratory staff [
12]. Lastly, prefilled extraction streamlines the workflow and increases the efficiency, as the technician saves time from filling the extraction plates, leading to faster results.
Low- and medium-throughput equipment like the IM48s, IM2, and KF Duo Prime provide flexibility for time and cost saving when testing a small number of samples. Previous studies reported excellent performances of several low- and medium throughput equipment including Indimag 48, IM48s and KF Duo Prime [
13,
14,
15,
16] . This study evaluated the performance of the IM48s, IM2, and KF Duo Prime— which are very useful for providing immediate results with a suspected farm break—compared to the NAHLN-approved KF Flex. Our results illustrate that these instruments generate comparable results to the KF Flex. Samples from farms suspected of HPAI, APMV-1, and FADs are routinely driven to the laboratory for immediate testing. In HPAI-suspected infections, up to 11 swabs are collected and pooled into a single sample for testing. With ASFV- or CSFV-suspected infections, tissue samples from a few field necropsy animals or a pool of up to 5 whole blood samples may be submitted, while lesion swabs from a few animals suspected of FMDV are submitted. Testing these small numbers of individual or pooled samples on a 96-well extraction instrument is a waste of resources due to the cost associated with the unused plastics in the 96-well extraction. In addition, the prefilled IndiMag Pathogen 8-sample cartridge is perfect for up to 6 samples and 2 extraction controls. The prefilled IndiMag Pathogen 8-sample cartridge provides time savings from the elimination of aliquoting four extraction buffers per each sample into extraction plates, also reducing possible technical errors while improving workflow efficiency, making it a valuable addition to NAHLN-approved assays.
Diagnostic sensitivity and specificity assessments using 32 positive and 30 negative samples demonstrated 100% agreement across all extraction chemistries and instruments, with a Kappa coefficient of 1.0. Precision evaluations revealed consistent CT values across instruments and extraction methods, with less than 3% of coefficients of variation across different concentrations evaluated for IAV and APMV-1. The average CT values were slightly lower using the IndiMag Pathogen kit on IM2, though not statistically significant. Similar to previous reports, our data confirmed the reliability of these methods [
13,
14,
15,
16]. At the time of manuscription preparation, the IndiMag Pathogen kits and the IndiMag 48s, IndiMag 2 and MagMAX Duo Prime platforms were approved by the NALHN program for use in the testing of IAV and APMV-1, and have been utilized for research and testing of the current HPAI outbreak [
17,
18].
One limitation of this study was the inability to use true samples for assessment of FADs which is limited to only reference laboratories and select agent-registered entities. Our study of analytical sensitivity was limited to the use of a phage clone of the PCR target segment of the FADs (provided by the FADDL reference laboratory), and diagnostic sensitivity and specificity was not conducted using field samples. Currently, the IndiMag 48s, IndiMag 2 and MagMAX Duo Prime platforms have not been accepted for use in FADs detection protocols. Our preliminary data are suggestive of the suitability of these platforms for consideration, and further validation should be done using the true virus and field samples to complete the validation process.
5. Conclusions
Overall, the study supports the adoption of IndiMag Pathogen kits and the IndiMag 48s, IndiMag 2 and MagMAX Duo Prime platforms as reliable alternatives for NAHLN diagnostic assays, enhancing flexibility, efficiency, and preparedness for managing current and emerging animal health threats.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org Table S1. Raw data and statistical analysis for manual and prefilled IndiMag Pathogen kits versus the MagMAX Viral RNA kit. The IndiMag Pathogen kit utilizing the KingFisher Flex and IndiMag 48s versus the MagMAX Viral RNA kit on the KingFisher Flex. Table S2. Raw data and statistical analysis for the IndiMag Pathogen kit utilizing the KingFisher Flex, IndiMag 48s, IndiMag 2 and MagAX CORE kit utilizing the KingFisher Duo Prime versus MagMAX Viral RNA kit utilizing the KingFisher Flex. Table S3. Raw data and statistical analysis for precision for the IAV and APMV-1 reference strains for the IndiMag Pathogen kit on the KF Flex, IM48s, and IM2 and MagMAX CORE on the KF Duo Prime. Table S4. Raw data and statistical analysis for the diagnostic sensitivity for the IndiMag Pathogen kit on the KF Flex, IM48s, and IM2 and using MagMAX CORE on the KF Duo Prime. Table S5. Raw data and statistical analysis for the IndiMag Pathogen kit utilizing the IndiMag 48s, IndiMag 2 and MagMAX CORE kit utilizing the KingFisher Duo Prime versus MagMAX CORE kit utilizing the KingFisher Flex.
Author Contributions
Conceptualization, D.M. and A.L.; methodology, A.L.; software, A.VC.; validation, A.VC., D.M. and A.L.; formal analysis, A.VC., D.M. and A.L.; investigation, D.M. and A.L.; resources, D.M. and A.L.; data curation, D.M. and A.L.; writing—original draft preparation, A.VC., D.M. and A.L.; writing—review and editing, A.VC., D.M. and A.L; visualization, A.VC.; supervision, A.L.; project administration, A.L.; funding acquisition, A.L.; All authors have read and agreed to the published version of the manuscript.
Funding
The portion of the research for KingFisher Duo Prime evaluation was funded by NAHLN Farm Bill, grant number AP21VSD&B000C029 and the portion for the validation of IndiMag pathogen kit, IndiMag 48s and IndiMag 2 were funded by INDICAL BIOSCIENCE.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank the U.S. Department of Agriculture National Veterinary Services Laboratory for providing reference materials for this study (Diagnostic Virology Laboratory for the IAV and APMV-1 reference strains, and the Foreign Animal Disease Diagnostic Laboratory for the phage constructs for ASFV, CSFV and FMDV); the U.S. Geological Survey National Wildlife Health Center for providing positive field samples; Joanna Colovas and Eryn Opgenorth for their technical assistance.
Conflicts of Interest
The authors (AVC and AL) declare no conflicts of interest. DM is employed by INDICAL BIOSCIENCE which partially funded the project, and participates in the design of the study; in the analyses of data; and in the writing of the manuscript.
Abbreviations
The following abbreviations are used in this manuscript:
| HPAI |
Highly Pathogenic Avian Influenza |
| ASFV |
African swine fever virus |
| CSFV |
Classical swine fever virus |
| FMDV |
Foot-and-mouth disease virus |
| FADs |
Foreign animal diseases |
| NAHLN |
National Animal Health Laboratory Network |
| PCR |
Polymerase chain reactions |
| KF Duo Prime |
KingFisher Duo Prime |
| IM48s |
IndiMag 48s |
| IM2 |
IndiMag 2 |
| KF Flex |
KingFisher Flex |
| IAV |
Influenza A virus |
| APMV-1 |
Avian paramyxovirus Type-1 |
| LPAI |
Low Pathogenic Avian Influenza |
| BHI |
Brain heart infusion |
| TBTB |
Tris-buffered tryptose broth |
| MagMAX Viral |
MagMAX Viral RNA Isolation kit |
| MagMAX CORE |
MagMAX CORE Nucleic Acid Purification kit |
| PBS |
Phosphate-buffered saline |
| LOD |
Limit of detection |
| CT |
Cycle threshold |
| CV |
Correlation of variance |
Appendix
Table A1.
Information of the reference viruses used for the analytical evaluation.
Table A1.
Information of the reference viruses used for the analytical evaluation.
| Reference strains |
Virus |
| IAV Reference Strain 1 |
Influenza A Virus A/Turkey/CA/6889/1980 (H9N2) LPAI |
| IAV Reference Strain 2 |
Influenza A Virus A/Turkey/Oregon/1971 (H7N3) LPAI |
| IAV Reference Strain 3 |
Influenza A Virus A/Turkey/Wisconsin/1968 (H5N9) LPAI |
| APMV-1 Reference Strain 1 |
Newcastle disease virus APMV-1 Texas GB vNDV |
| APMV-1 Reference Strain 2 |
Newcastle disease virus Pigeon-44407 strain (APMV-1) |
| APMV-1 Reference Strain 3 |
Newcastle disease virus LaSota strain (APMV-1) |
| ASFV Reference Construct |
Synthetic phage construct for ASFV PCR assay control |
| CSFV Reference Construct |
Synthetic phage construct for CSFV PCR assay control |
| FMDV Reference Construct |
Synthetic phage construct for FMDV PCR assay control |
Figure A1.
Standard curves for references and CT values for the Xeno RNA. (a) Standard curves for IAV reference strains extracted using manual and prefilled IndiMag Pathogen kits; (b) Standard curves for APMV-1 reference strains extracted using manual and prefilled IndiMag Pathogen kits; (c) Standard curves for IAV reference strains extracted using instrumentations under evaluation; (d) Standard curves for APMV-1 reference strains extracted using instrumentations under evaluation; (e) Standard curves for FADs reference constructs; and (f) CT values for Xeno RNA run with FADs assays. The * indicates no significant difference among the average CT values reported in the chart.
Figure A1.
Standard curves for references and CT values for the Xeno RNA. (a) Standard curves for IAV reference strains extracted using manual and prefilled IndiMag Pathogen kits; (b) Standard curves for APMV-1 reference strains extracted using manual and prefilled IndiMag Pathogen kits; (c) Standard curves for IAV reference strains extracted using instrumentations under evaluation; (d) Standard curves for APMV-1 reference strains extracted using instrumentations under evaluation; (e) Standard curves for FADs reference constructs; and (f) CT values for Xeno RNA run with FADs assays. The * indicates no significant difference among the average CT values reported in the chart.
References
- Detections of Highly Pathogenic Avian Influenza. Animal and Plant Health Inspection Service |U.S. Department of Agriculture. Available online: https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections?utm_source=chatgpt.com (accessed on 27 February 2025).
- Burrough, E. R.; Magstadt, D. R.; Petersen, B.; Timmermans, S. J.; Gauger, P. C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A. C.; Gorden, P. J.; Plummer, P. J.; Main, R. Early Release—Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerging Infectious Diseases 2024, 30. [Google Scholar] [CrossRef] [PubMed]
- APHIS Confirms D1.1 Genotype in Dairy Cattle in Nevada. Animal and Plant Health Inspection Service | U.S. Department of Agriculture. Available online: https://www.aphis.usda.gov/news/program-update/aphis-confirms-d11-genotype-dairy-cattle-nevada-0 (accessed on 27 February 2025).
- Avian influenza virus type A (H5N1) in, U.S.; dairy cattle. American Veterinary Medical Association. Available online: https://www.avma.org/resources-tools/animal-health-and-welfare/animal-health/ avian-influenza/avian-influenza-virus-type-h5n1-us-dairy-cattle?utm_source=chatgpt.com (accessed on 27 February 2025).
- H5N1 Bird Flu: Current Situation. Avian Influenza (Bird Flu) | CDC. Available online: https://www.cdc.gov/bird-flu/situation-summary/index.html (accessed on 27 February 2025).
- Brown, V. R.; Miller, R. S.; McKee, S. C.; Ernst, K. H.; Didero, N. M.; Maison, R. M.; Grady, M. J.; & Shwiff, S. A.; Shwiff, S. A. Risks of introduction and economic consequences associated with African swine fever, classical swine fever and foot-and-mouth disease: A review of the literature. Transboundary and emerging diseases 2021, 68, 1910–1965. [Google Scholar] [CrossRef] [PubMed]
- Disease Alert: African Swine Fever. Animal and Plant Health Inspection Service | U.S. Department of Agriculture. Available online: https://www.aphis.usda.gov/livestock-poultry-disease/swine/african-swine-fever (accessed on 27 February 2025).
- Classical swine fever. WOAH—World Organisation for Animal Health. Available online: https://www.woah.org/en/disease/classical-swine-fever/ (accessed on 27 February 2025).
- Statement on recent Foot and Mouth Disease (FMD) outbreak in Germany. WOAH—World Organisation for Animal Health. Available online: https://www.woah.org/en/statement-on-recent-foot-and-mouth-disease-fmd-outbreak-in-germany/ (accessed on 27 February 2025).
- National Animal Health Laboratory Network. Animal and Plant Health Inspection Service | U.S. Department of Agriculture. Available online: https://www.aphis.usda.gov/labs/nahln (accessed on 27 February 2025).
- Elnagar, A.; Pikalo, J.; Beer, M.; Blome, S.; Hoffmann, B. Swift and Reliable “Easy Lab” Methods for the Sensitive Molecular Detection of African Swine Fever Virus. International Journal of Molecular Sciences 2021, 22, 2307. [Google Scholar] [CrossRef] [PubMed]
- Caskey, C. R. Ergonomics in the clinical laboratory. Clinical laboratory science: journal of the American Society for Medical Technology 1999, 12, 140–11. [Google Scholar] [PubMed]
- Aebischer, A.; Beer, M.; Hoffmann, B. Development and Validation of Rapid Magnetic Particle Based Extraction Protocols. Virology Journal 2014, 11. [Google Scholar] [CrossRef]
- Korthase, C.; Elnagar, A.; Beer, M.; Hoffmann, B. Easy Express Extraction (TripleE)—a Universal, Electricity-Free Nucleic Acid Extraction System for the Lab and the Pen. Microorganisms 2022, 10, 1074. [Google Scholar] [CrossRef]
- Dei Giudici, S.; Loi, F.; Ghisu, S.; Angioi, P. P.; Zinellu, S.; Fiori, M. S.; Carusillo, F.; Brundu, D.; Franzoni, G.; Zidda, G. M.; Tolu, P.; Bandino, E.; Cappai, S.; Oggiano, A. The Long-Jumping of African Swine Fever: First Genotype II Notified in Sardinia, Italy. Viruses 2023, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Dziadek, K.; Świętoń, E.; Kozak, E.; Wyrostek, K.; Tarasiuk, K.; Styś-Fijoł, N.; Śmietanka, K. Phylogenetic and Molecular Characteristics of Wild Bird-Origin Avian Influenza Viruses Circulating in Poland in 2018−2022: Reassortment, Multiple Introductions, and Wild Bird–Poultry Epidemiological Links. Transboundary and Emerging Diseases 2024, 2024. [Google Scholar] [CrossRef]
- Caserta, L. C.; Frye, E. A.; Butt, S. L.; Laverack, M.; Nooruzzaman, M.; Covaleda, L. M.; Thompson, A. C.; Koscielny, M. P.; Cronk, B.; Johnson, A.; Kleinhenz, K.; Edwards, E. E.; Gomez, G.; Hitchener, G.; Martins, M.; Kapczynski, D. R.; Suarez, D. L.; Alexander Morris, E. R.; Hensley, T.; Beeby, J. S. Spillover of Highly Pathogenic Avian Influenza H5N1 Virus to Dairy Cattle. Nature 2024, 634, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Nooruzzaman, M.; Covaleda, L. M.; Martin, N. H.; Koebel, K.; Ivanek, R.; Alcaine, S. D.; Diel, D. G. Thermal Inactivation Spectrum of Influenza a H5N1 Virus in Raw Milk. bioRxiv (Cold Spring Harbor Laboratory) 2024. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).