Submitted:
30 September 2025
Posted:
01 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
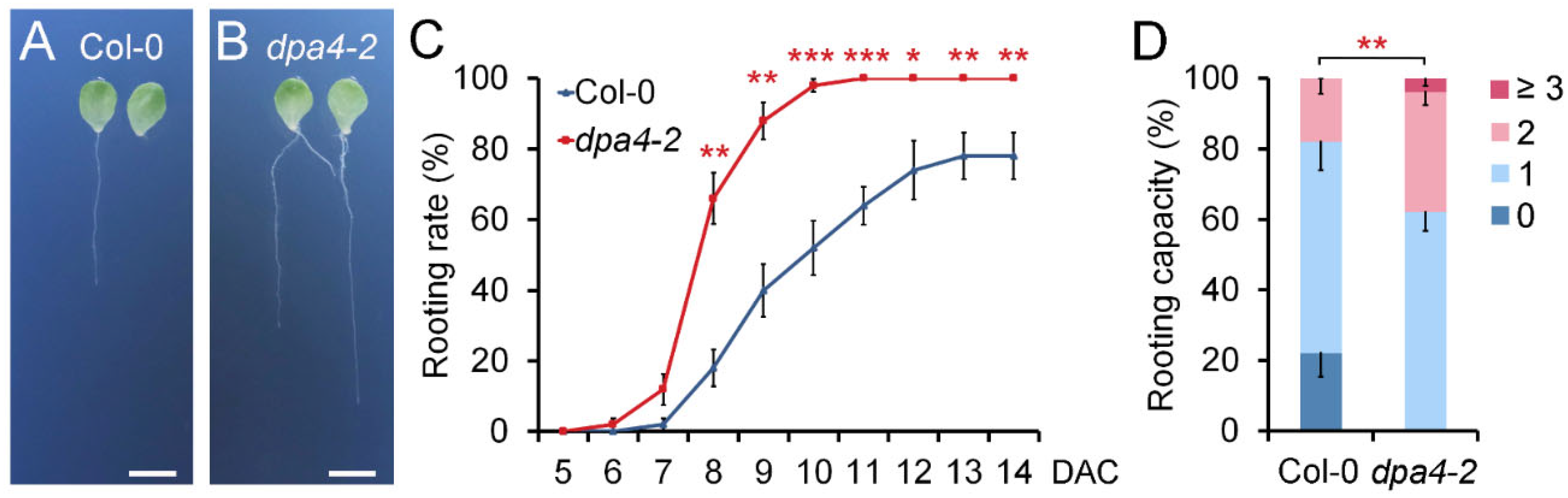
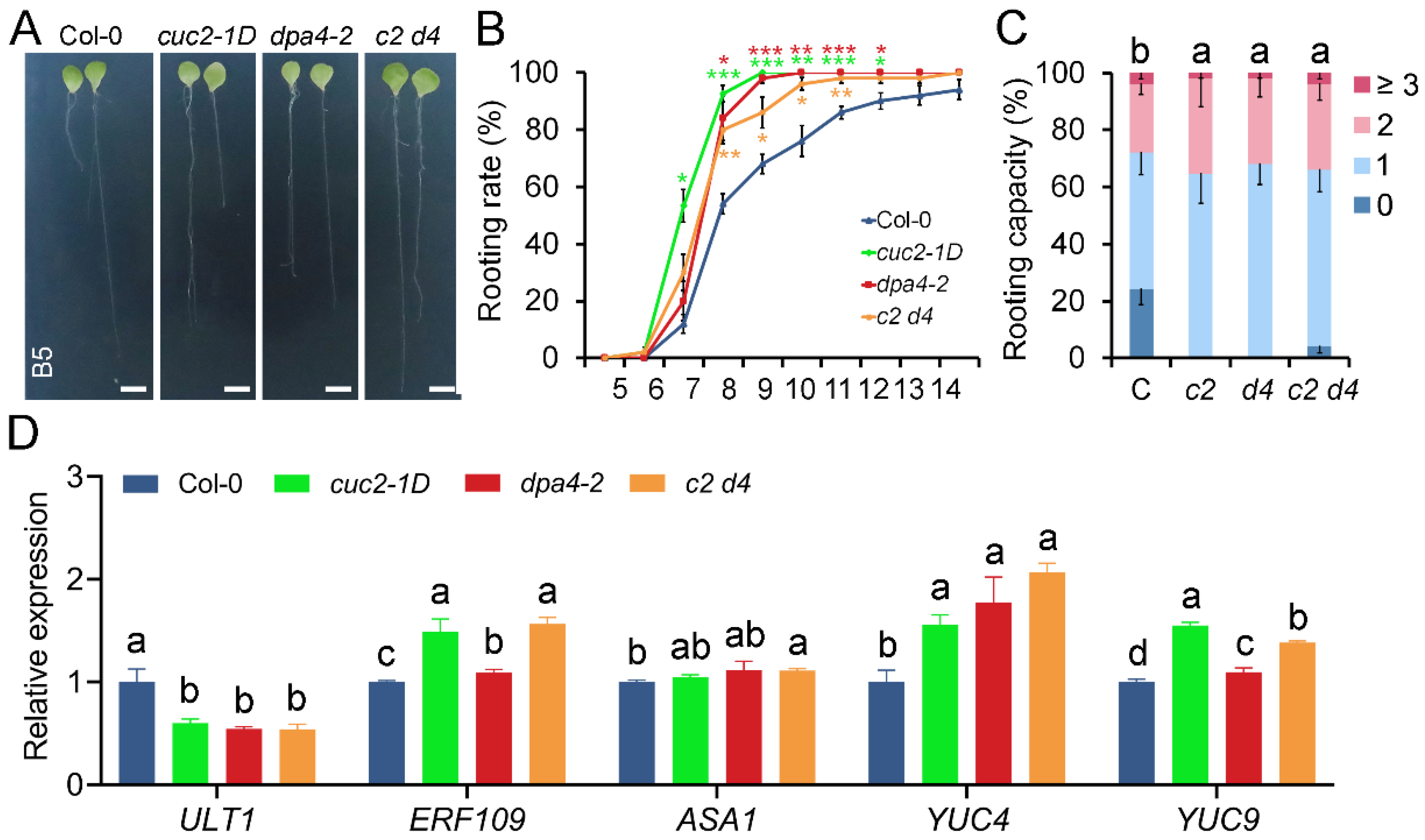
2.1. DPA4 Suppresses De Novo Root Regeneration (DNRR) from Leaf Explants
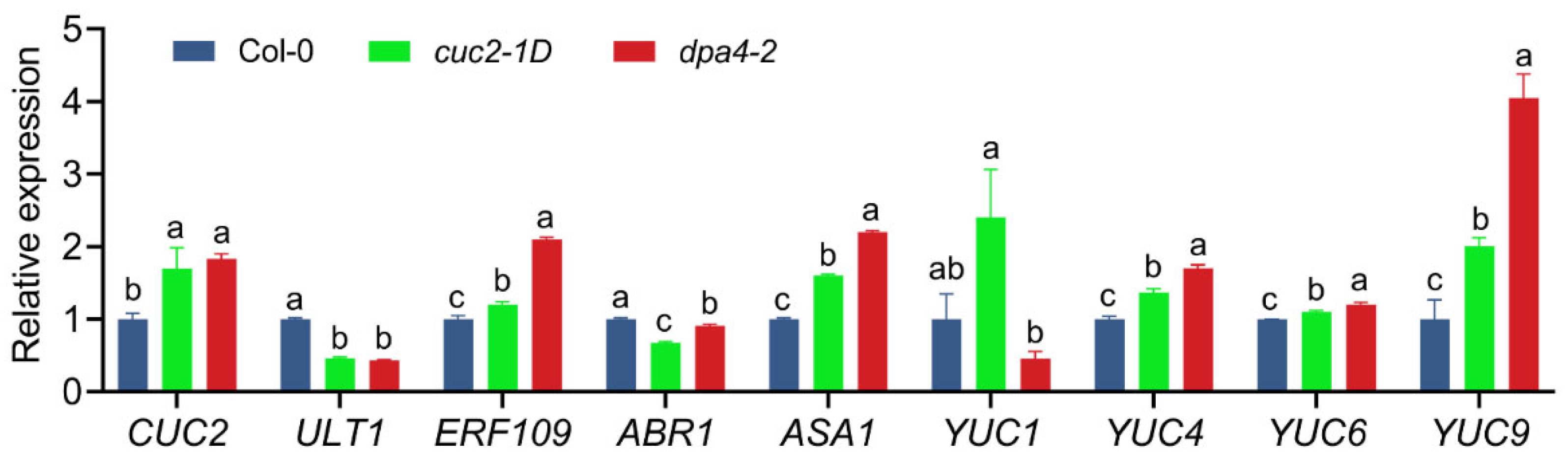
2.2. DPA4 Regulates Genes Known to be Involved in AR Formation and Auxin Biosynthesis
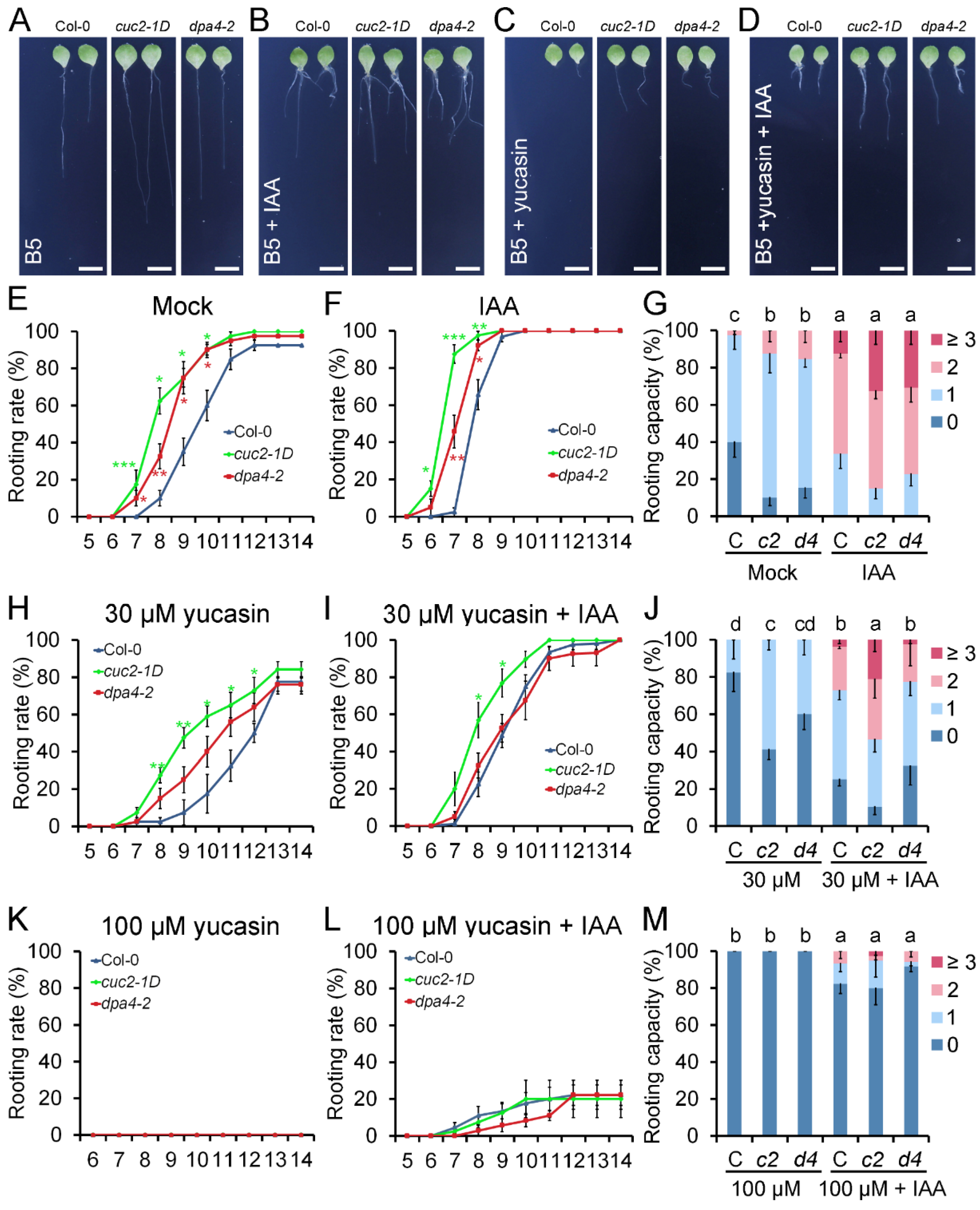
2.3. cuc2-1D Phenocopies the Increased AR Formation Phenotype of dpa4-2 Mutant Leaf Explants, While Both Mutants Display Similar Expression Changes in DNRR-Related Genes
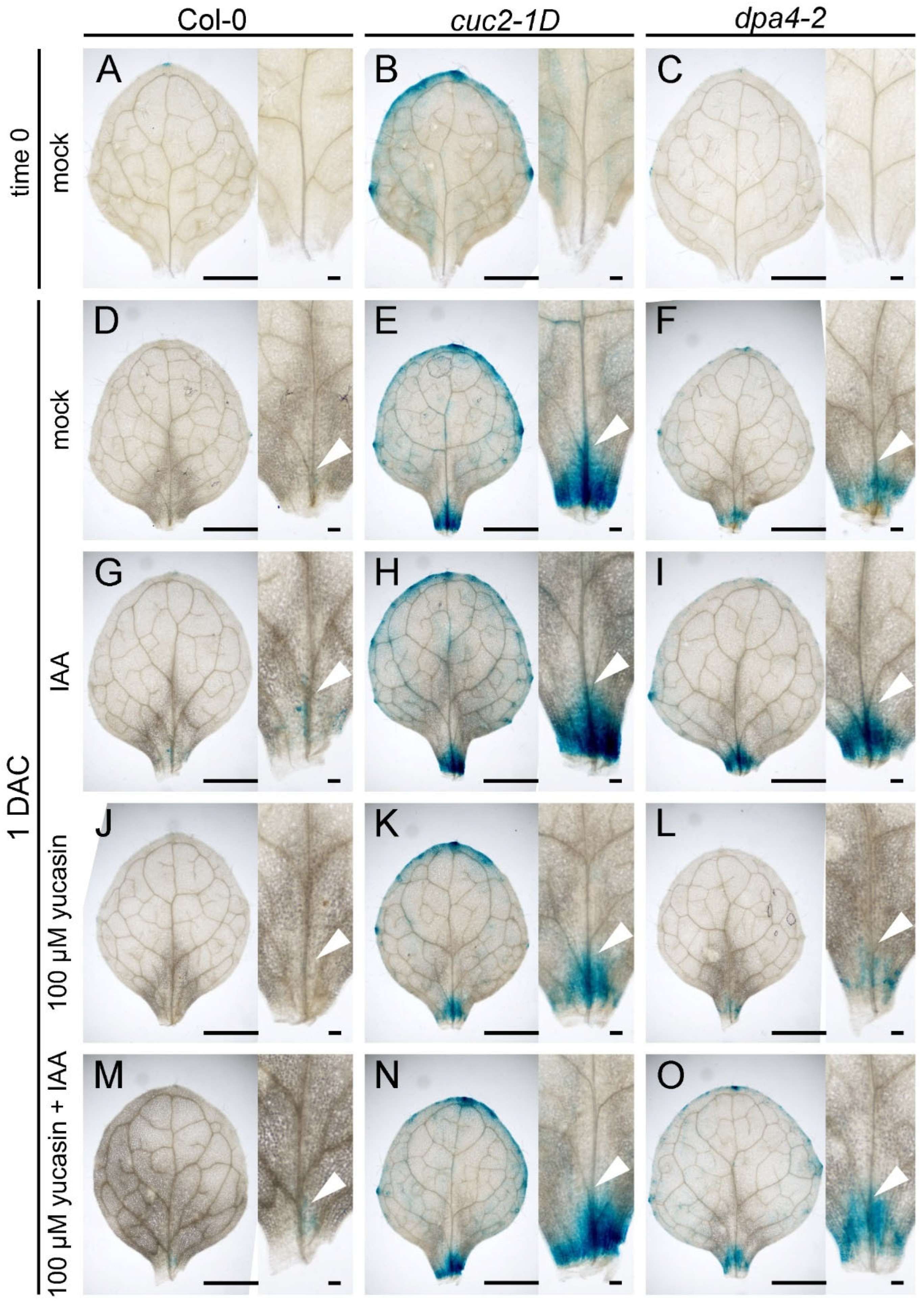
2.4. DPA4 and CUC2 Promote Endogenous Auxin Levels
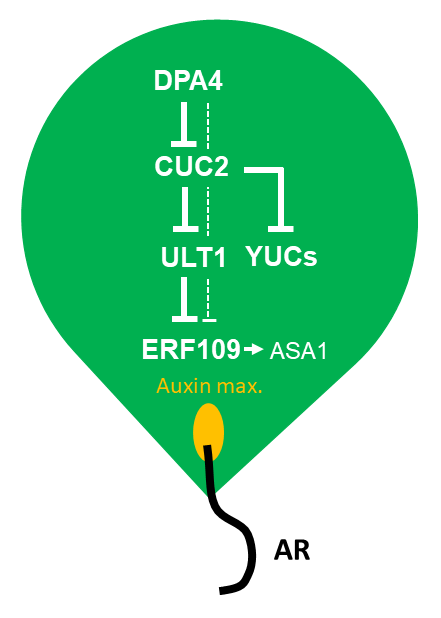
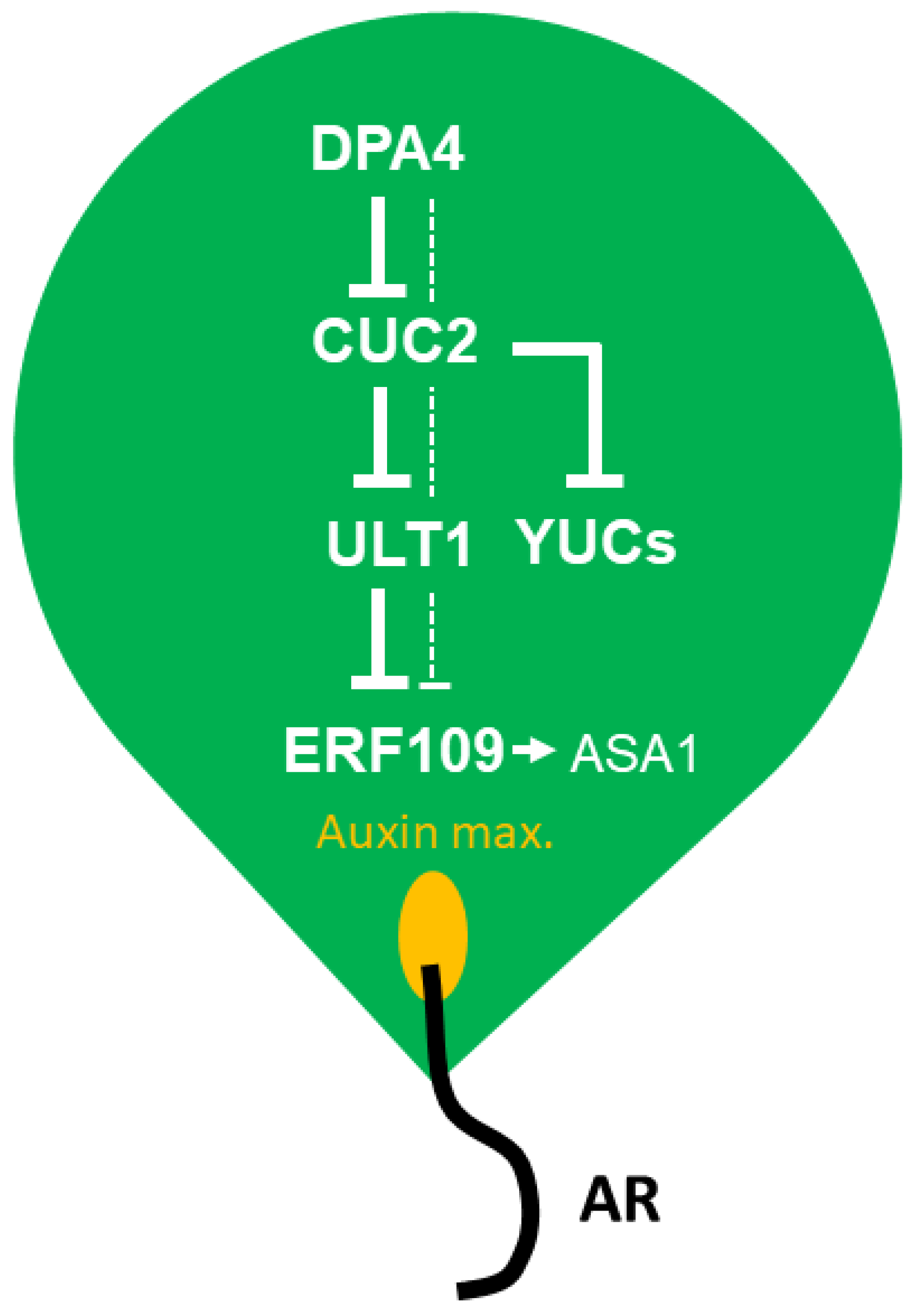
2.5. DPA4 and CUC2 Regulate AR Formation Likely Through a Common Genetic Pathway that Controls Auxin Biosynthesis
3. Discussion
4. Materials and Methods
4.1. DPA4 and CUC2 Regulate AR Formation Likely Through a Common Genetic Pathway that Controls Auxin Biosynthesis
4.2. De Novo Root Regeneration (DNRR) Assays with and Without Hormone Treatment
4.3. Quantitative RT-PCR and GUS Staining
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. iPSCs: A Comparison between Animals and Plants. Trends Plant Sci. 2018, 23, 660–666. [Google Scholar] [CrossRef]
- Tazeb, A. Plant tissue culture technique as a novel tool in plant breeding: A review article. Environ Sci 2017, 17, 111–118. [Google Scholar]
- Liu, W.; Zhang, Y.; Fang, X.; Tran, S.; Zhai, N.; Yang, Z.; Guo, F.; Chen, L.; Yu, J.; Ison, M.S.; et al. Transcriptional landscapes of de novo root regeneration from detached Arabidopsis leaves revealed by time-lapse and single-cell RNA sequencing analyses. Plant Commun. 2022, 3, 100306. [Google Scholar] [CrossRef] [PubMed]
- Duclercq, J.; Sangwan-Norreel, B.; Catterou, M.; Sangwan, R.S. De novo shoot organogenesis: From art to science. Trends Plant Sci. 2011, 16, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Yang, Y.; Pan, G.; Shen, Y. New Insights Into Tissue Culture Plant-Regeneration Mechanisms. Front. Plant Sci. 2022, 13, 926752. [Google Scholar] [CrossRef]
- Doll, Y.; Ikeuchi, M. All roads lead to dome: Multicellular dynamics during de novo meristem establishment in shoot regeneration. Curr. Opin. Plant Biol. 2025, 85, 102733. [Google Scholar] [CrossRef]
- Šmeringai, J.; Schrumpfová, P.P.; Pernisová, M. Cytokinins - regulators of de novo shoot organogenesis. Front. Plant Sci. 2023, 14, 1239133. [Google Scholar] [CrossRef]
- Chen, X.; Qu, Y.; Sheng, L.; Liu, J.; Huang, H.; Xu, L. A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 2014, 5, 208. [Google Scholar] [CrossRef] [PubMed]
- Garg, T.; Yadav, M.; Mushahary, K.K.K.; Kumar, A.; Pal, V.; Singh, H.; Jain, M.; Yadav, S.R. Spatially activated conserved auxin-transcription factor regulatory module controls de novo root organogenesis in rice. Planta 2023, 258, 52. [Google Scholar] [CrossRef]
- Ye, B.-B.; Shang, G.-D.; Pan, Y.; Xu, Z.-G.; Zhou, C.-M.; Mao, Y.-B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.-W. AP2/ERF Transcription Factors Integrate Age and Wound Signals for Root Regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.-X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef]
- Yan, J.; Song, Y.; Li, M.; Hu, T.; Hsu, Y.-F.; Zheng, M. IRR1 contributes to de novo root regeneration from Arabidopsis thaliana leaf explants. Physiol. Plant. 2023, 175, e14047. [Google Scholar] [CrossRef]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Sousa Barros, J.A.; Gude, I.; Perianez-Rodriguez, J.; Micol, J.L.; Del Pozo, J.C.; Moreno-Risueno, M.A.; Pérez-Pérez, J.M. Regulation of Hormonal Control, Cell Reprogramming, and Patterning during De Novo Root Organogenesis. Plant Physiol. 2018, 176, 1709–1727. [Google Scholar] [CrossRef]
- Jing, T.; Ardiansyah, R.; Xu, Q.; Xing, Q.; Müller-Xing, R. Reprogramming of Cell Fate During Root Regeneration by Transcriptional and Epigenetic Networks. Front. Plant Sci. 2020, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Xu, L. De novo root regeneration from leaf explants: Wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 2018, 41, 39–45. [Google Scholar] [CrossRef]
- Lee, K.; Yoon, H.; Park, O.-S.; Seo, P.J. ENHANCER OF SHOOT REGENERATION1 promotes de novo root organogenesis after wounding in Arabidopsis leaf explants. Plant Cell 2024, 36, 2359–2374. [Google Scholar] [CrossRef]
- Cai, X.-T.; Xu, P.; Zhao, P.-X.; Liu, R.; Yu, L.-H.; Xiang, C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Di, D.-W. Precise Regulation of the TAA1/TAR-YUCCA Auxin Biosynthesis Pathway in Plants. Int. J. Mol. Sci. 2023, 24, 8514. [Google Scholar] [CrossRef]
- Sun, B.; Chen, L.; Liu, J.; Zhang, X.; Yang, Z.; Liu, W.; Xu, L. TAA family contributes to auxin production during de novo regeneration of adventitious roots from Arabidopsis leaf explants. Science Bulletin 2016, 61, 1728–1731. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription Factors WOX11/12 Directly Activate WOX5/7 to Promote Root Primordia Initiation and Organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Müller-Xing, R.; Xing, Q. The plant stem-cell niche and pluripotency: 15 years of an epigenetic perspective. Front. Plant Sci. 2022, 13, 1047. [Google Scholar] [CrossRef]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. Plant stem cells and de novo organogenesis. New Phytol. 2018, 218, 1334–1339. [Google Scholar] [CrossRef]
- Jing, T.; Xing, Q.; Shi, Y.; Liu, X.; Müller-Xing, R. Depletion of Gibberellin Signaling Up-Regulates LBD16 Transcription and Promotes Adventitious Root Formation in Arabidopsis Leaf Explants. Int. J. Mol. Sci. 2024, 25. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.-W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Larue, C.T.; Wen, J.; Walker, J.C. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 2009, 58, 450–463. [Google Scholar] [CrossRef]
- Nicolas, A.; Maugarny-Calès, A.; Adroher, B.; Chelysheva, L.; Li, Y.; Burguet, J.; Bågman, A.-M.; Smit, M.E.; Brady, S.M.; Li, Y.; et al. De novo stem cell establishment in meristems requires repression of organ boundary cell fate. Plant Cell 2022, 34, 4738–4759. [Google Scholar] [CrossRef] [PubMed]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3424–3429. [Google Scholar] [CrossRef]
- Li, X.; Zheng, Y.; Xing, Q.; Ardiansyah, R.; Zhou, H.; Ali, S.; Jing, T.; Tian, J.; Song, X.S.; Li, Y.; et al. Ectopic expression of the transcription factor CUC2 restricts growth by cell cycle inhibition in Arabidopsis leaves. Plant Signal. Behav. 2020, 15, 1706024. [Google Scholar] [CrossRef]
- Daimon, Y.; Takabe, K.; Tasaka, M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003, 44, 113–121. [Google Scholar] [CrossRef]
- Gordon, S.P.; Heisler, M.G.; Reddy, G.V.; Ohno, C.; Das, P.; Meyerowitz, E.M. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 2007, 134, 3539–3548. [Google Scholar] [CrossRef]
- Kareem, A.; Durgaprasad, K.; Sugimoto, K.; Du, Y.; Pulianmackal, A.J.; Trivedi, Z.B.; Abhayadev, P.V.; Pinon, V.; Meyerowitz, E.M.; Scheres, B.; et al. PLETHORA Genes Control Regeneration by a Two-Step Mechanism. Current Biology 2015, 25, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Verstraeten, I.; Werbrouck, S.; Geelen, D. CUC2 as an early marker for regeneration competence in Arabidopsis root explants. J. Plant Physiol. 2011, 168, 1598–1601. [Google Scholar] [CrossRef]
- Engelhorn, J.; Reimer, J.J.; Leuz, I.; Göbel, U.; Huettel, B.; Farrona, S.; Turck, F. Development-related PcG target in the apex 4 controls leaf margin architecture in Arabidopsis thaliana. Development 2012, 139, 2566–2575. [Google Scholar] [CrossRef]
- Shao, J.; Meng, J.; Wang, F.; Shou, B.; Chen, Y.; Xue, H.; Zhao, J.; Qi, Y.; An, L.; Yu, F.; et al. NGATHA-LIKEs Control Leaf Margin Development by Repressing CUP-SHAPED COTYLEDON2 Transcription. Plant Physiol. 2020, 184, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Peterson, K.; Jack, T. The plant B3 superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef]
- Tian, J.; Xing, Q.; Jing, T.; Fan, X.; Zhang, Q.; Müller-Xing, R. The epigenetic regulator ULTRAPETALA1 suppresses de novo root regeneration from Arabidopsis leaf explants. Plant Signal. Behav. 2022, 17, 2031784. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 2014, 78, 372–384. [Google Scholar] [CrossRef]
- Nishimura, T.; Hayashi, K.-I.; Suzuki, H.; Gyohda, A.; Takaoka, C.; Sakaguchi, Y.; Matsumoto, S.; Kasahara, H.; Sakai, T.; Kato, J.-I.; et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014, 77, 352–366. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Ray, A.; Mandal, D.; Nag Chaudhuri, R. ABI3 mediated repression of RAV1 gene expression promotes efficient dehydration stress response in Arabidopsis thaliana. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194582. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Tanaka, S.; Miyazaki, T.; Aida, M. Expression of the auxin biosynthetic genes YUCCA1 and YUCCA4 is dependent on the boundary regulators CUP-SHAPED COTYLEDON genes in the Arabidopsis thaliana embryo. Plant Biotechnol. (Tokyo) 2022, 39, 37–42. [Google Scholar] [CrossRef]
- Tyler, L.; Miller, M.J.; Fletcher, J.C. The Trithorax Group Factor ULTRAPETALA1 Regulates Developmental as Well as Biotic and Abiotic Stress Response Genes in Arabidopsis. G3 Genes|Genomes|Genetics 2019, 9, 4029–4043. [Google Scholar] [CrossRef]
- Wan, Q.; Yao, R.; Zhao, Y.; Xu, L. JA and ABA signaling pathways converge to protect plant regeneration in stress conditions. Cell Rep. 2025, 44, 115423. [Google Scholar] [CrossRef]
- Tu, T.; Zheng, S.; Ren, P.; Meng, X.; Zhao, J.; Chen, Q.; Li, C. Coordinated cytokinin signaling and auxin biosynthesis mediates arsenate-induced root growth inhibition. Plant Physiol. 2021, 185, 1166–1181. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Xing, Q.; Müller-Xing, R. A novel UV-B priming system reveals an UVR8-depedent memory, which provides resistance against UV-B stress in Arabidopsis leaves. Plant Signal. Behav. 2021, 16, 1879533. [Google Scholar] [CrossRef] [PubMed]
- Müller-Xing, R.; Ardiansyah, R.; Xing, Q.; Faivre, L.; Tian, J.; Wang, G.; Zheng, Y.; Wang, X.; Jing, T.; Leau, E. de; et al. Polycomb proteins control floral determinacy by H3K27me3-mediated repression of pluripotency genes in Arabidopsis thaliana. J. Exp. Bot. 2022, 73, 2385–2402. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




