1. Introduction
Virtually all cases of cryptococcosis are caused by
Cryptococcus neoformans and the closely related species complex,
C. gattii [
1]. The opportunistic pathogen predominantly infects hosts with CD4
+ T cell dysfunction. Persons with AIDS are particularly vulnerable. It is estimated that in 2020, 112,000 HIV-infected persons died of cryptococcal meningitis, accounting for 19% of AIDS-related deaths [
2]. Initial exposure occurs following the inhalation of yeast cells or basidiospores from environmental sources such as trees and soil contaminated with pigeon guano [
1]. Two varieties of
C. neoformans, var.
grubii (serotype A) and var.
neoformans (serotype D), have been recognized with >95% of human cases caused by var.
grubii; though, some taxonomists posit that the two varieties are distinct species [
3,
4,
5]. Three genetically distinct subpopulations of
C. neoformans var.
grubii, designated VNI, VNII, and VNB have been identified [
6,
7]. The VNI and VNII lineages are found worldwide, while VNB is mostly found in sub-Saharan Africa, particularly Botswana [
5,
6,
7,
8].
Despite the need, there are currently no licensed fungal vaccines [
9]. In preclinical studies, several laboratories, including ours, have developed vaccines which protect mice in experimental models of cryptococcosis. We have studied both attenuated whole organism vaccines and subunit vaccines. For the former, an attenuated
C. neoformans var.
grubii strain KN99 was constructed in which three chitin deacetylase (Cda) genes (
Cda1,
Cda2, and
Cda3) were deleted. The resulting avirulent strain,
cda1Δ
2Δ
3Δ (heretofore referred to as “cda123”), lacks cell wall chitosan and is cleared from the lungs of severely immunodeficient mouse lines [
10,
11]. Pulmonary vaccination with cda123 protects several strains of mice against an otherwise lethal challenge with KN99 [
10,
11]. The subunit vaccines consist of cryptococcal proteins recombinantly expressed in
E. coli, adjuvanted with Cationic Adjuvant Formulation 01 (CAF01). We have shown that subcutaneous vaccination with four recombinant antigens adjuvanted with CAF01 stimulates robust, antigen-specific CD4+ T cell immune responses and protects mice against KN99 challenge [
12,
13,
14]. The four antigen vaccine, hereby referred to as “4-Ag”, contains
Cda1 and Cda2, carboxypeptidase 1 trimmed of its region of human homology (Cpd1Δ), and Barwin-like domain protein 4 (Blp4). CAF01 is a liposome-based Mincle agonist adjuvant composed of the cationic quaternary ammonium salt N,N’-dimethyl-N,N’-dioctadecylammonium and the glycolipid mycobacterial immunomodulator α,α′-trehalose 6,6′-dibehenate [
12,
15].
KN99 is a genetically defined virulent laboratory strain derived from a clinical strain, H99, that was originally isolated in the United States from a patient with lymphoma [
16,
17]. It is important to test vaccine candidates using diverse clinical strains isolated from regions of the world where disease is most prevalent. Clinical and environmental isolates of
C. neoformans differ in their virulence in mouse models [
18,
19,
20]. Additionally, there is evidence from human studies that fungal strain genotype informs clinical outcome [
21]. Therefore, in the present studies, we assembled a panel of six virulent
C. neoformans strains isolated from patients with cryptococcal meningitis in Vietnam, Uganda, and Botswana; analyzing two strains from each country. We then tested the ability of our two cryptococcal candidate vaccines to protect mice against pulmonary challenge with the six strains. We found that both vaccines significantly protected mice, as measured by prolonged survival and decreased fungal organ burdens. However, the degree of protection differed as a function of cryptococcal strain and vaccine. Moreover, lung leukocytes isolated from vaccinated and infected mice had robust antigen-specific interferon-gamma (IFNγ) responses following ex vivo stimulation.
2. Materials and Methods
C. neoformans strains. Strains used to infect mice are listed in
Table 1. Strains BK80 and BMD1338 were from Jeremy Day (University of Oxford) [
20,
22]. Strains UgCl302, UgCl395 [
19,
23], and the reference strain, KN99 [
24], were obtained from Kirsten Nielsen (Virginia Tech University). Strains PMH1063 and PMH1065 were from Jennifer Tenor and John Perfect (Duke University) and their collection of clinical strains from Princess Marina Hospital in Gaborone, Botswana [
25]. Strain BMD1338 was isolated from a patient without known immunodeficiency [
20]. The other clinical strains were isolated from persons coinfected with HIV. Stock cultures of the strains were stored at -80°C in 25% glycerol.
Mouse Vaccination and Infection Model. BALB/c mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were housed and bred in the animal facilities of University of Massachusetts Chan Medical School (UMCMS) in a specific pathogen-free environment. Animal husbandry and experimental procedures were conducted according to the protocols approved by Institutional Animal Care and Use Committee at UMCMS. Male and female mice were used in approximately equal numbers in all the experiments. At 6-10 weeks of age, mice were vaccinated with either live-attenuated cda123 or a combination of four recombinant proteins with Tris Buffer (10 mM, pH 7.0, supplemented with 2% glycerol) and CAF01 adjuvant (Statens Serum Institute, Copenhagen, Denmark). The live-attenuated cda123 was prepared and administered as a single orotracheal dose containing 2 x 10
7 in 50 μL PBS as described [
26]. The 4-Ag vaccine was prepared and administered as in previous studies [
12]. Briefly, His-tagged antigens were recombinantly expressed in
E. coli and purified on a nickel column. The 4-Ag vaccine consisted of two separate injections adjuvanted with CAF01. One injection contained a combination of Cda1 and Cda2, while the other contained a combination of Cpd1Δ and Blp4. The 4-Ag vaccine contained 5 μg of each protein. Injections were administered subcutaneously (100 μL each) and were repeated at two-week intervals for a total of three vaccinations. Unvaccinated mice served as controls.
To prepare the inoculum to infect the mice, C. neoformans strains were grown in yeast-peptone-dextrose (YPD) broth in a shaker for 18 hours at 30°C, washed in PBS, counted and resuspended in PBS containing 4 x 105 yeast cells/mL. The mice were then challenged orotracheally with 50 µL of the suspension (containing 2 x 104 yeast cells). Mice were monitored daily. Euthanasia with CO2 asphyxiation was performed for mice that reached humane endpoints and time-point organ collection experiments.
Assessment of Organ Fungal Burden and Antigen-stimulated IFNγ Production. Mice were euthanized, exsanguinated by cardiac puncture, and spleens, lungs, and brains were harvested. Splenocytes and lung cells were prepared as described [
12,
27]. Briefly, spleens were gently macerated manually through a 70 µm strainer. Lung cells were prepared using the MACS Lung Dissociation Kit for mice, according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The remaining lung cells were passed through a 70 µm cell strainer. 200 µL portions of the lung and spleen samples were set aside to assess fungal burden. Leukocytes were then enriched on a 67% and 40% density gradient (Percoll, Cytiva) by centrifugation at 800xg for 20 minutes without braking, followed by the collection of cells from the interphase. Single cell lung leukocytes were washed and resuspended in complete medium (RPMI 1640 supplemented with 10% FBS, 1% GlutaMAX, 1% HEPES, and 1% Penicillin–Streptomycin, all purchased from ThermoFisher Scientific). Cell concentration and viability were then determined by Trypan Blue stain and a TC20 cell counter (Bio-Rad, Hercules, CA). Brain samples were collected and homogenized in 2 mL of PBS supplemented with 2% Penicillin–Streptomycin. Fungal burdens in spleens, lungs, and brains were determined by counting CFUs following dilution and spread plates on Sabouraud dextrose agar. The lower limit of detection was 20 CFUs for lungs and 10 CFUs for brains and spleens. If no CFUs were detected, a value for the lower limit of detection was assigned.
Heat-killed
C. neoformans strains for use as ex vivo stimuli were prepared as described [
27,
28]. Each strain was cultured in a shaker for 18 hours in YPD at 30°C. A 20 μL aliquot was then taken and put into 4 mL of fresh broth and cultured for an additional 48 hours. Cells were counted and diluted with PBS to 2.6 x 10
7 cells/mL, which corresponds to a dry weight of 1 mg/mL. The strains were then heat-killed at 70°C for 30 minutes. Fungal death was confirmed by plating samples on Sabouraud dextrose agar and demonstrating the absence of colony forming units (CFU). Aliquots were stored at -80°C until used for assays.
Antigen-stimulated IFNγ production by lung leukocytes was determined as in our previous studies [
12,
27]. Cells were cultured in round-bottom 96-well plates and stimulated with the indicated antigens in a final volume per well of 200 µL RPMI complete medium supplemented with 0.5 µg/mL amphotericin B (ThermoFisher). Final concentrations of recombinant proteins and heat-killed strains were 5 µg/mL and 50 µg/mL respectively. Lung leukocytes were cultured for 18 hours at 4 x 10
5 cells/well. The cultures were maintained at 37°C within a controlled humidified atmosphere with 5% CO
2. Supernatants were then collected for IFNγ analysis via ELISA (Mouse DuoSet ELISA Kits, R&D Systems, Bio-Techne, Minneapolis, MN).
Graphs and Statistics GraphPad Prism, version 10.1.1 (GraphPad Software Inc., Dotmatics, San Diego, CA, USA) was used to create survival curves, graphs and perform statistical analysis. Kaplan-Meier survival curves were analyzed using the Mantel-Cox log-rank test while the organ fungal burdens were analyzed with the Kruskal-Wallis test with multiple comparisons. For all statistical analyses, a P value of <0.05 following corrections for multiple comparisons was considered statistically significant.
3. Results
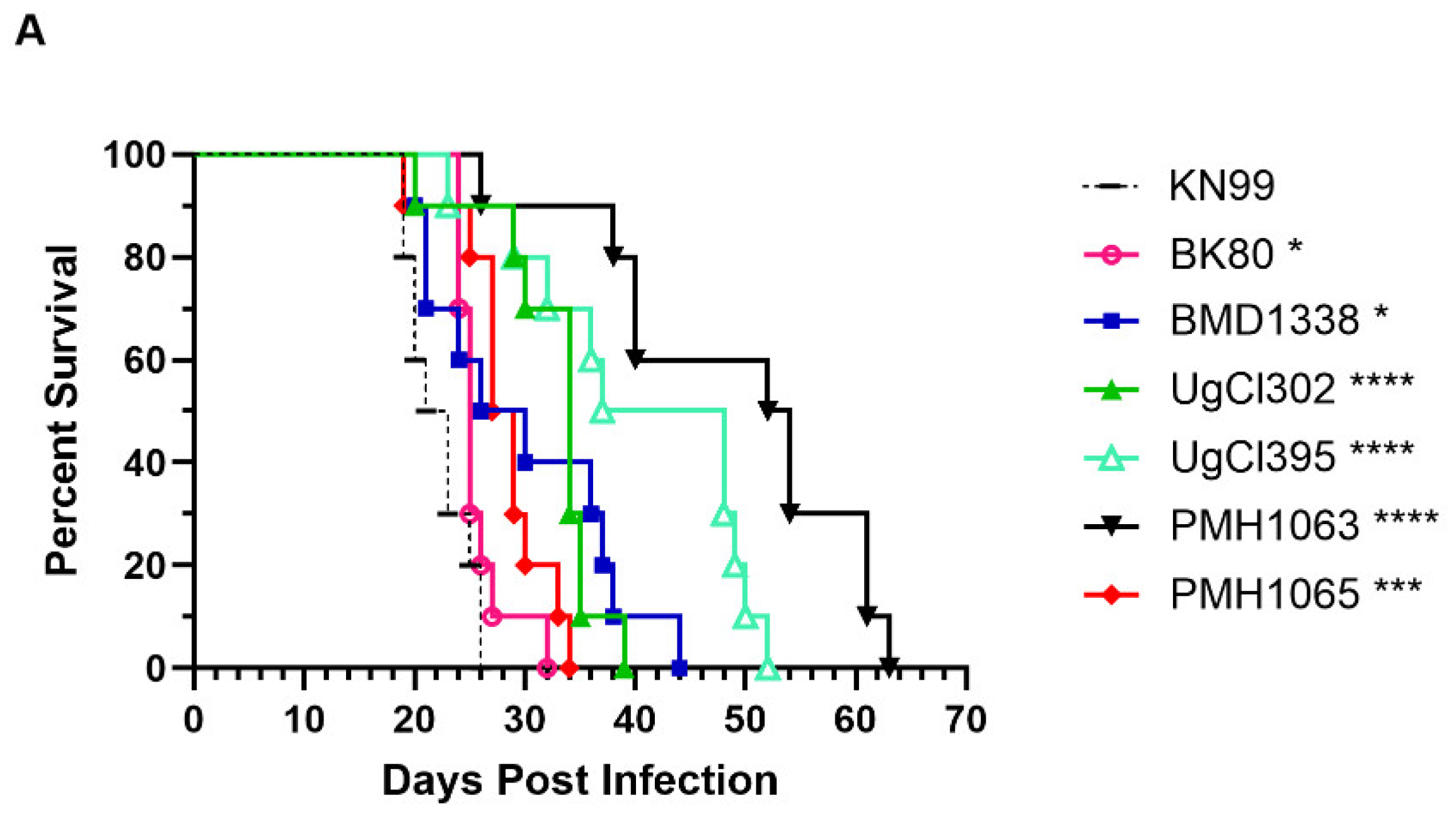
Two strains each from Vietnam, Uganda, and Botswana were chosen for study along with the reference strain KN99 (
Table 1). Initial experiments were designed to demonstrate the virulence of the isolates in unvaccinated BALB/c mice following orotracheal challenge with 2 x 10
4 yeast cells. While all mice succumbed by day 63 post-infection, there was variability in median survival with the mice infected with clinical strains surviving significantly longer than those infected with KN99 (
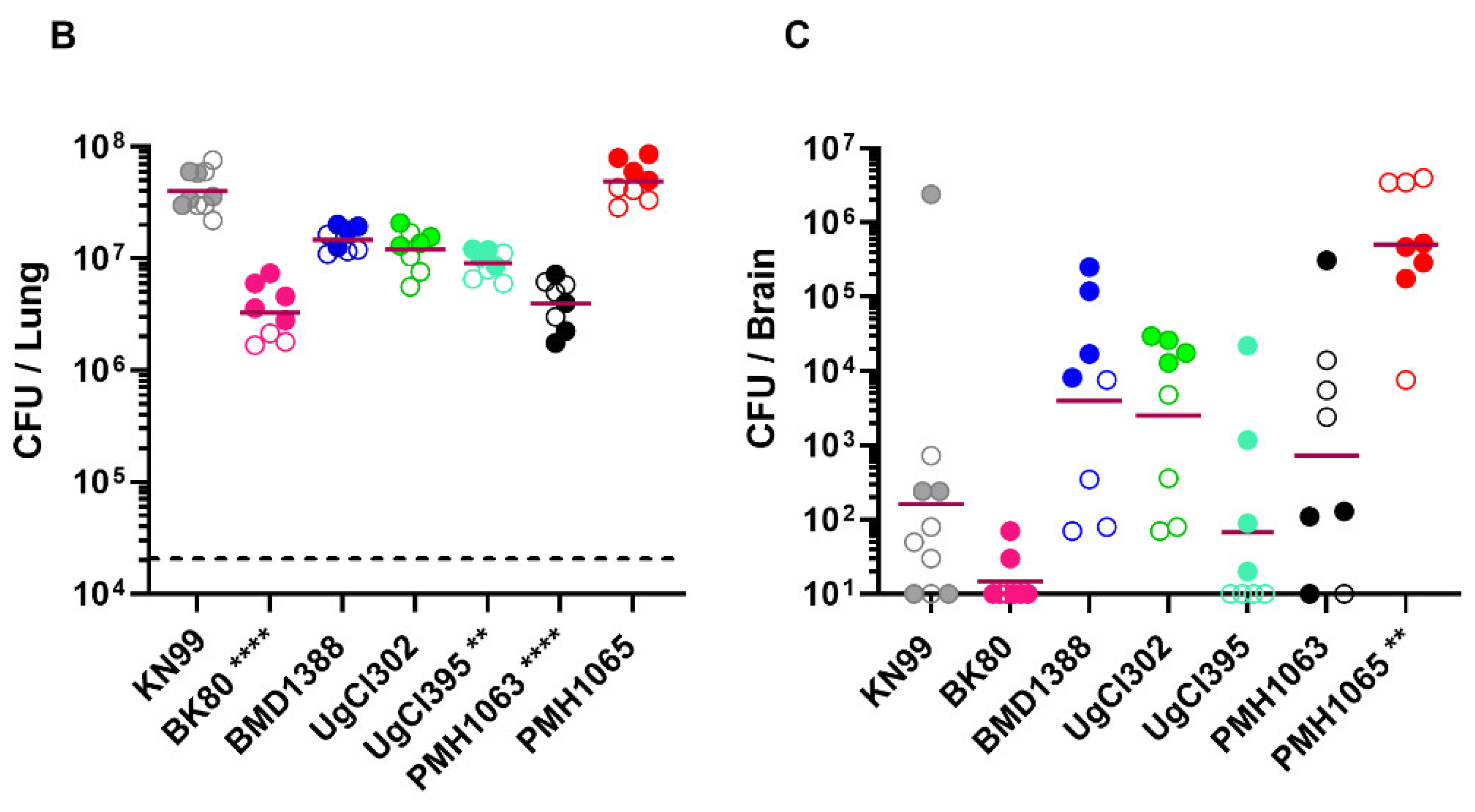
Figure 1). Based on times to 100% mortality, strains BK80, PMH1065 were the most virulent, followed by UgCl302, BMD1338, UgCl395 and PMH1063. Next, we examined lung and brain CFUs at 14 days post infection. For each of the strains, the mice had lung CFU at least 100 times higher than the inoculum, with mice infected with BK80, UgCl395, and PMH1063 having significantly fewer lung CFUs than KN99-infected mice. PMH1065-infected mice had significantly higher CFU in the brain than KN99-infected mice. Brain CFUs from mice infected with BMD1388, UgCl302, and PMH1063 trended higher but this difference was not statistically significant.
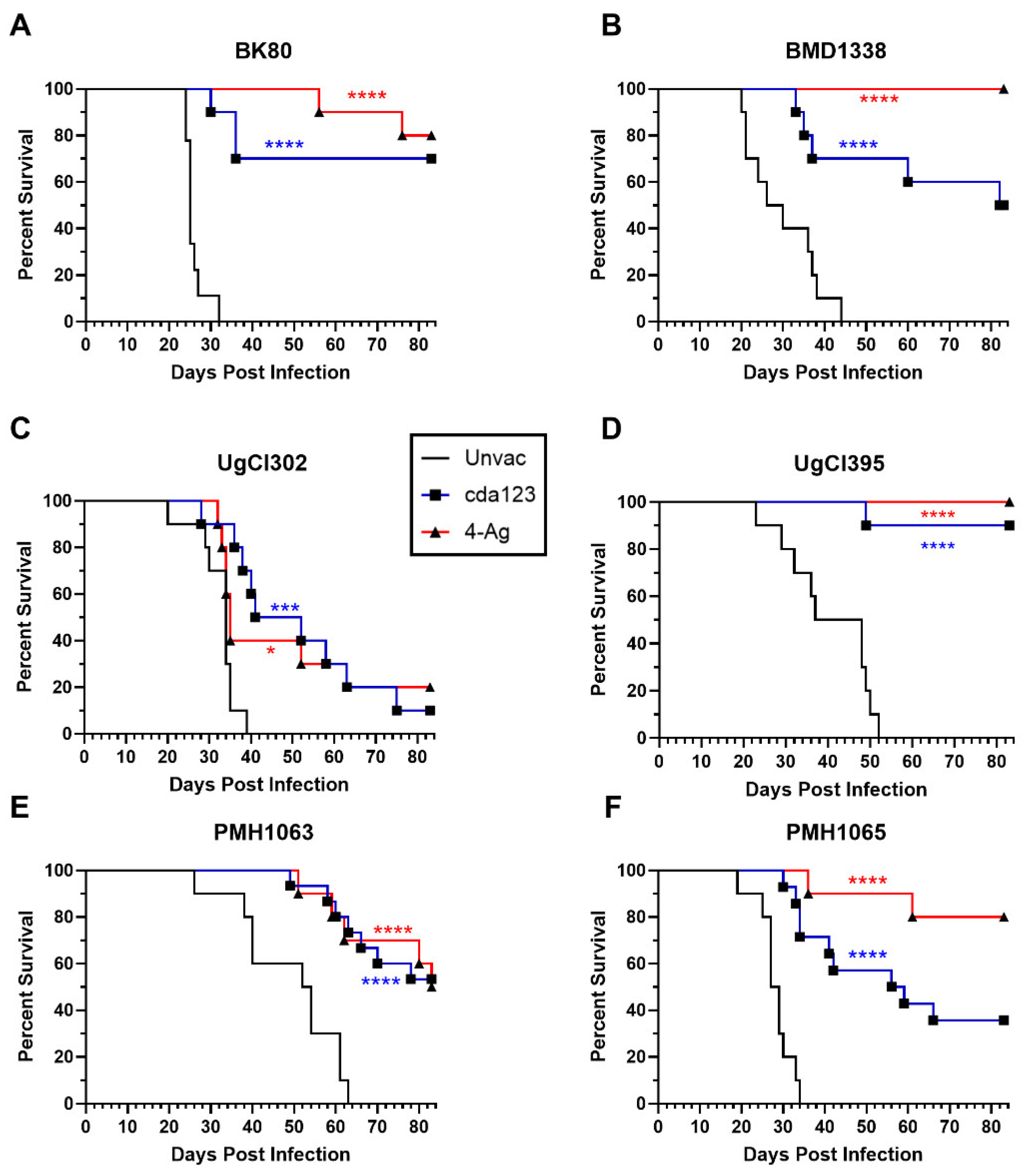
We next determined whether our two candidate cryptococcal vaccines, the live-attenuated cda123 vaccine and the 4-Ag CAF01-adjuvanted vaccine, could protect BALB/c mice challenged with the clinical cryptococcal strains, as we reported previously using the reference strain KN99 [
12]. Regardless of the infecting clinical strain, vaccination significantly improved survival for all groups (
Figure 2). However, the degree of protection varied as a function of vaccine and clinical strain. For example, mice vaccinated with the 4-Ag vaccine had 100% survival following infection with BMD1338 and UgCl395. Conversely, for strain UgCl302, which is closely related to UgCl395 [
19], only 20% of the mice were alive when the experiment was terminated on day 83 post-infection. For mice that received the cda123 vaccine, survival at day 83 post infection ranged from 10% for strain UgCl302 to 90% for strain UgCl395.
At the time of euthanasia, lung and brain CFUs were determined for the mice that survived the infection, (Supplemental
Figure S1). The best control of infection, as determined by organ CFUs, was seen for mice that received the 4-Ag vaccine and were infected with BK80. For that group, CFUs in the lung were below the infecting inoculum and all but one mouse had undetectable brain CFUs. Other groups had higher CFUs, although there was a lot of intragroup variation.
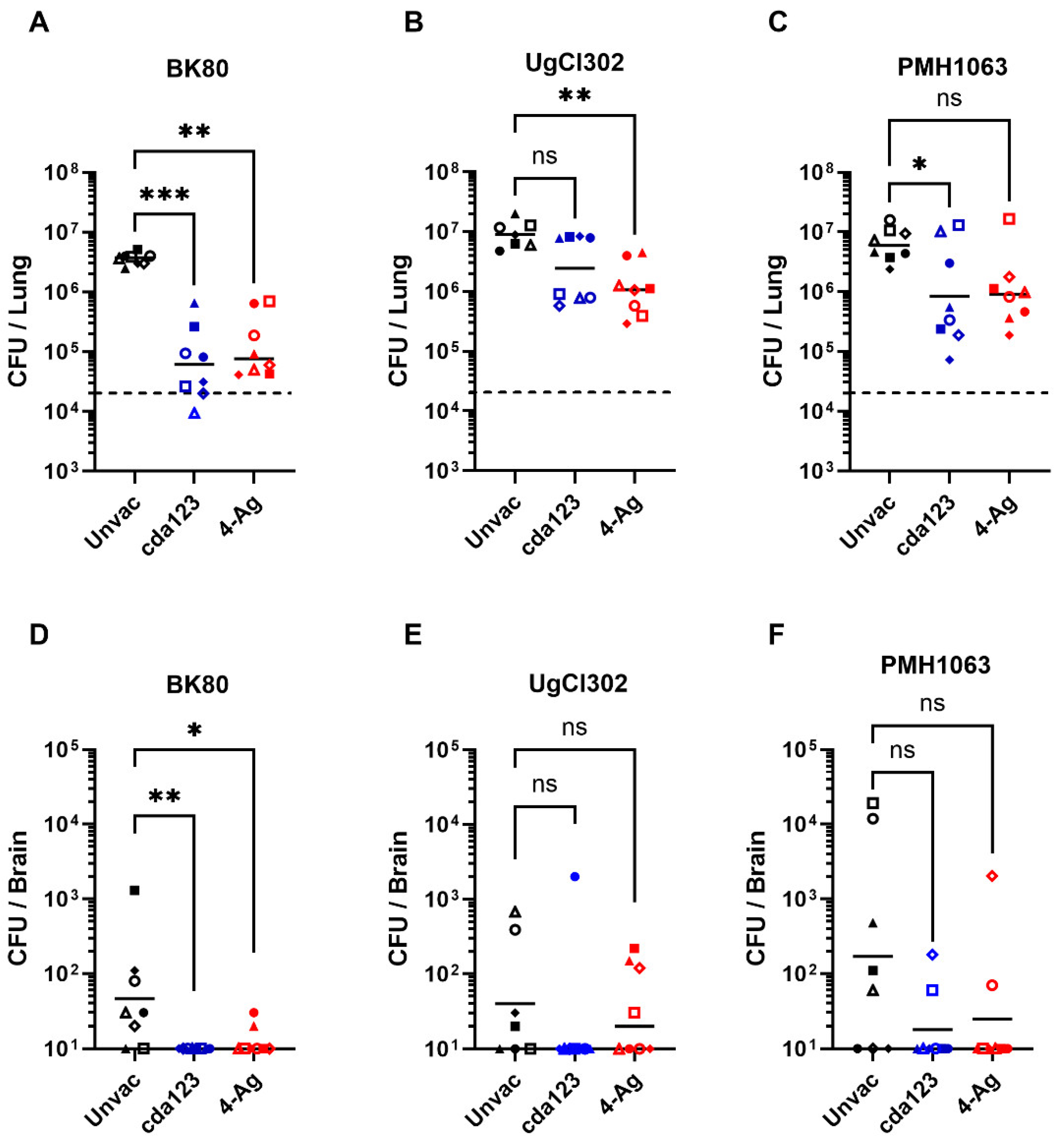
Having demonstrated that vaccination improved survival following challenge with the clinical strains, the next set of experiments explored how well the vaccines reduced fungal burden. CFUs in the lungs, spleens, and brains were determined in unvaccinated and vaccinated mice 14 days following infection (
Figure 3). For these experiments, we chose one strain each from Vietnam (BK80), Uganda (UgCl302), and Botswana (PMH1063). Fungal burdens in lungs and brains were decreased in vaccinated mice compared to unvaccinated mice, regardless of infecting strain. However, statistical significance was achieved for only some comparisons. Of the three fungal strains tested, the most robust vaccine-mediated reductions in organ fungal burdens occurred in mice infected with strain BK80. These vaccinated mice also had the closest lung CFUs to the initial inoculum. Survival following vaccination was also greatest in mice that received BK80 compared with UgCl302 and PMH1063 (
Figure 2). Not shown on
Figure 3, CFUs were undetectable in the spleens of all unvaccinated and vaccinated mice.
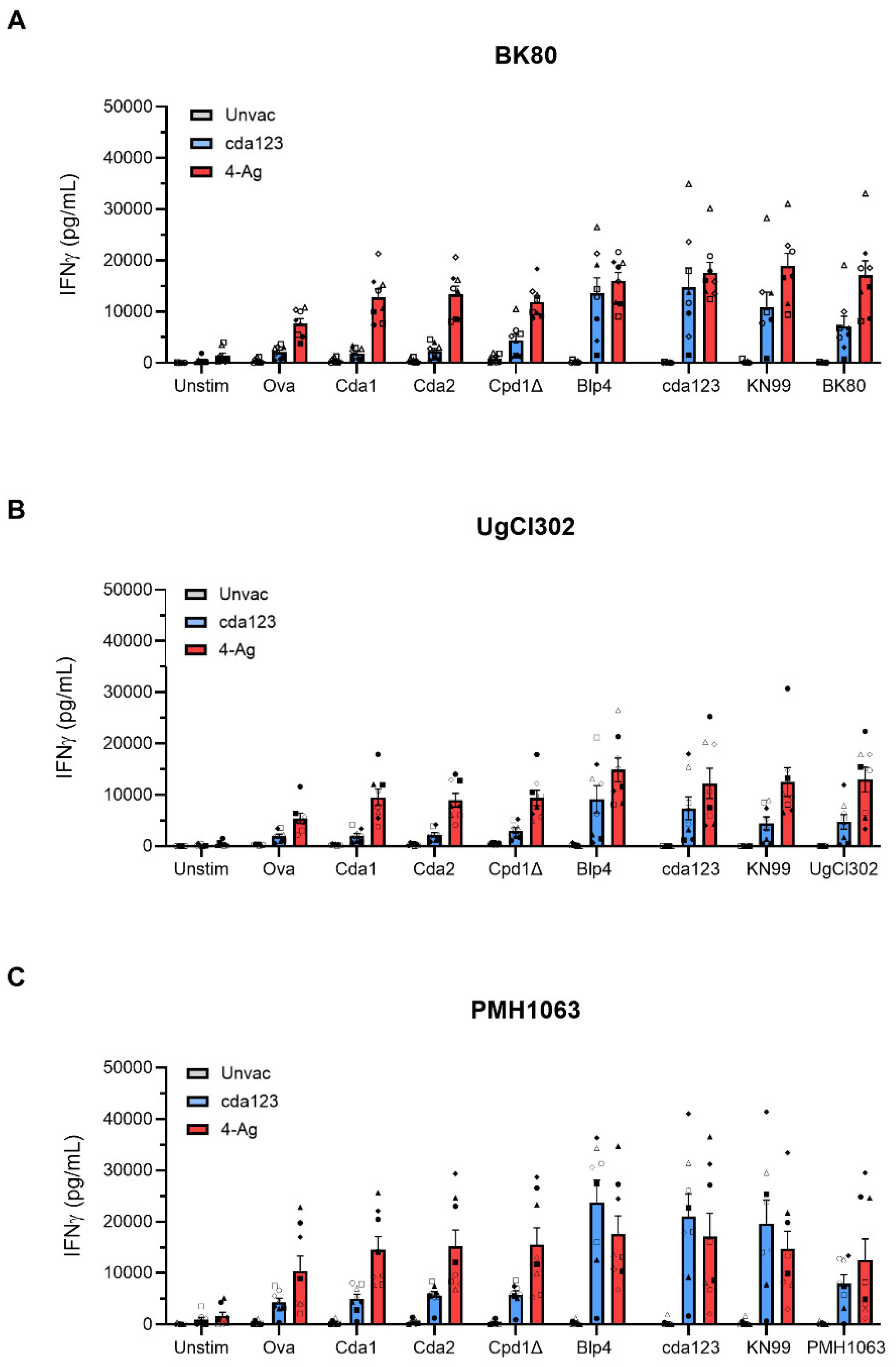
IFNγ is required for protection of cda123 and 4-Ag vaccinated mice against challenge with the KN99 strain [
12,
26]. Therefore, we measured antigen-stimulated IFNγ release by lung leukocytes (
Figure 4). Mice were left unvaccinated or vaccinated with cda123 or 4-Ag. The animals were then given a pulmonary challenge with BK80, UgCl302, or PMH1063. Lung leukocytes were harvested at 14 DPI and simulated with recombinant antigens or heat-killed
C. neoformans cells (including cda123, KN99, and the infecting strain). His-tagged recombinant ovalbumin (Ova), prepared in the same manner as the recombinant
E. coli proteins, was used as a control stimulus.
Regardless of the antigen tested, IFNγ release was generally low following stimulation of lung leukocytes from unvaccinated mice infected with BK80, UgCl302, or PMH1063 (
Figure 4), which has been previously seen with KN99 infection [
12]. In addition, irrespective of the infecting cryptococcal strain, some IFNγ release was seen in Ova-stimulated lung leukocytes from vaccinated mice, suggesting responses to the His-tag or contaminating
E. coli proteins in the preparation. For mice vaccinated with cda123 and infected with any of the three strains, levels of IFNγ following stimulation of lung leukocytes with recombinant Cda1, Cda2, and Cpd1Δ were similar to that seen following Ova stimulation. However, recombinant Blp4 and the heat-killed cryptococcal strains stimulated significant IFNγ release, suggesting antigen-specific responses to Blp4 developed. For the 4-Ag vaccinated and infected mice, significant IFNγ release was assayed following stimulation with each of the recombinant cryptococcal antigens and heat-killed strains. For each antigen and vaccination group, lung leukocyte IFNγ release was similar when comparing lung leukocytes from mice infected with BK80, UgCl302, and PMH1063.
4. Discussion
Most AIDS-related cryptococcosis cases occur in sub-Saharan Africa and Asia and are caused by C. neoformans var. grubii [
2,
5,
29]. By phylogenetic analysis, four lineages of C. neoformans var. grubii have been described, VNI, VNII, VNBI, and VNBII [
30]. The first two subpopulations have a worldwide distribution, whereas the VNBI and VNBII lineages are mainly confined to Southern Africa [
5,
30]. To assemble a panel of clinical isolates for our vaccine studies, we prioritized genotypically and phenotypically defined clinical isolates obtained from regions with high cryptococcal prevalence. An additional criterion was known virulence in mouse models of experimental cryptococcosis. The Uganda strains, UgCl302 and UgCl395, are in the VNI sequence-type 93 lineage, and have been shown to be highly virulent following intranasal infection of A/J mice [
19]. Sequence-type 93 isolates predominate in clinical samples from Uganda and Malawi but are also common in other parts of the world, such as Brazil [
22,
31]. The Vietnamese strains BMD1338 and BK80 are of the sequence-type 5 and sequence-type 4 lineages, respectively, which are frequently found in Southeast Asia [
22,
32]. The Botswana strains, PMH1063 and PMH1065 are of the VNBII and VNI lineages, respectively [
25]. They were isolated as part of a study of 34 patients with HIV-associated cryptococcal meningitis; 31 of the 34 fungal strains were C. neoformans while the remaining three were C. gattii. By RNAseq, genes encoding for two of our subunit vaccine antigens, Cda1 and Blp4, were among the top 50 expressed genes in fungi obtained from the cerebrospinal fluid [
25].
Many clinical strains isolated from persons with HIV-associated cryptococcosis are avirulent or hypovirulent in mouse models of disease [
19,
20,
21]. For our vaccination studies, we selected six clinical strains that were virulent in BALB/c mice, as measured by 100% mortality within 70 days and increased fungal burden in the lungs and brain at day 14. This strain collection could prove useful not only for vaccine studies but also for investigations into immune responses, virulence factors, and antifungal therapy. Future studies are needed with clinical strains representing other lineages within the C. neoformans species complex as well as the genetically diverse C. gattii species complex.
Compared with unvaccinated mice, mice that received the cda123 and the 4-Ag vaccines were significantly protected after challenge with the six clinical strains. However, the degree of protection afforded by vaccination differed among the strains. Interestingly, the disparity was most evident among the two Ugandan strains, UgCl302 and UgCl395, both of which are of the VNI sequence-type 93 lineage. Survival at the termination of the study was 90-100% in vaccinated mice challenged with UgCl395, but only 10-20% in vaccinated mice challenged with UgCl302. Closely related strains can differ markedly in terms of mouse virulence [
7,
18,
19,
20]. Using a closely related collection of C. neoformans sequence-type 93 isolates, mouse virulence could be mapped to single nucleotide polymorphisms in specific genes, some of which also correlated with IFNγ levels upon infection [
7]. Our findings extend this observation to include vaccine-mediated protection. Taken together, the studies suggest that caution must be applied before positing that a single strain is representative of a cryptococcal species, variety, or lineage [
33,
34,
35].
Lung leukocytes obtained from mice vaccinated and infected with any of the three clinical strains tested produced robust IFNγ when stimulated with vaccine antigens or heat-killed C. neoformans strains. This cytokine response is similar to that seen in vaccinated mice infected with C. neoformans strain KN99 [
12,
26,
27,
28]. In experimental models of cryptococcosis using C. neoformans strain KN99, IFNγ is required for protection mediated by the cda123 and 4-Ag vaccines [
26,
28]. A vaccine consisting of the H99 strain of C. neoformans recombinantly engineered to express murine IFNγ is highly protective in murine infection models [
36]. There is strong evidence for the importance of IFNγ in human infection as phase 2 clinical trials of adjuvant IFNγ therapy in HIV-infected patients with cryptococcal meningitis demonstrated salutary effects with regards to fungal clearance [
37,
38]. Moreover, peripheral blood mononuclear cells (PBMCs) obtained from subjects with a history of cryptococcosis have greater cryptococcal antigen-stimulated IFNγ production compared with PBMCs from healthy controls [
27]. Given the importance of IFNγ in clinical settings and preclinical models, our data demonstrating strong antigen-stimulated pulmonary IFNγ release in vaccinated and infected mice provide support for clinical development of our cryptococcal vaccines.
Our work demonstrates that two leading vaccine candidates protect against cryptococcal strains representative of those found in sub-Saharan Africa and Southeast Asia, regions of the world that account for the majority of cryptococcal meningitis cases found globally. The 4-Ag vaccine performed as well as and, in some cases, better than the cda123 vaccine, as measured by 83-day survival and 14-day fungal burden in the lungs and brain. However, caution needs to be exercised before concluding one vaccine is superior as different dosing regimens and vaccine schedules were used. The cda123 vaccine has the advantage of being easy to prepare and only requires a single dose but has the drawback in that it needs to be delivered to the lungs using a relatively high inoculum [
11,
26]. Moreover, live-attenuated vaccines must be used with caution in immunocompromised populations [
39]. The 4-Ag vaccine might be easier to advance clinically as it is a traditional subunit adjuvanted protein vaccine [
12]. Vaccine formulations using fewer antigens and chimeric antigens are being studied as having four antigens increases manufacturing expenses. Recently, protection of mice against challenge with C. neoformans strain H99 was demonstrated using an adjuvanted mRNA vaccine encoding for Cda1, one of the antigens in the 4-Ag vaccine [
40]. The promising vaccine studies in mice lend support to trialing vaccines in humans at risk for cryptococcosis.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Figure S1: Lung and brain fungal burdens of surviving mice following vaccination and infection.
Author Contributions
Conceptualization, S.M.L.; methodology, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., G.K.P., C.A.S., and S.M.L.; validation, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., C.A.S., and S.M.L.; formal analysis, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., C.A.S., and S.M.L.; investigation, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., J.L.T., and C.A.S.; resources, G.K.P., J.L.T., and J.R.P.; data curation, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., C.A.S., and S.M.L.; writing—original draft preparation, D.C. and S.M.L.; writing—review and editing, D.C., R.W., Z.H., L.V.N.O., M.M.H., N.R., G.K.P., J.L.T., J.R.P., C.A.S., and S.M.L.; supervision, J.R.P., C.A.S., S.M.L.; project administration, J.R.P., C.A.S., S.M.L.; funding acquisition, J.R.P., C.A.S., S.M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Institutes of Health grants R01AI125045, R01AI172154, R24AI192252, R25HL092610, RO1 AI 93257, RO1 AI189300, and contract 75N93019C00064.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Data will be shared upon request.
Acknowledgments
The authors are grateful to David Meya, Kirsten Nielsen, and Jeremy Day for generously providing cryptococcal strains used in this study, and to Jennifer Lodge and Rajendra Upadhya for constructive feedback.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| Cda |
chitin deacetylase |
| cda123 |
cda1Δ2Δ3Δ avirulent Cryptococcus strain |
| CAF01 |
Cationic Adjuvant Formulation 01 |
| 4-Ag |
Four antigen Vaccine |
| Cpd1Δ |
carboxypeptidase 1 trimmed of its region of human homology |
| Blp4 |
Barwin-like domain protein 4 |
| YPD |
yeast-peptone-dextrose |
| DPI |
Days post infection |
References
- Meya, D.B.; Williamson, P.R. Cryptococcal Disease in Diverse Hosts. N Engl J Med 2024, 390, 1597-1610. [CrossRef]
- Rajasingham, R.; Govender, N.P.; Jordan, A.; Loyse, A.; Shroufi, A.; Denning, D.W.; Meya, D.B.; Chiller, T.M.; Boulware, D.R. The global burden of HIV-associated cryptococcal infection in adults in 2020: a modelling analysis. The Lancet Infectious Diseases 2022, 22, 1748-1755.
- Hagen, F.; Khayhan, K.; Theelen, B.; Kolecka, A.; Polacheck, I.; Sionov, E.; Falk, R.; Parnmen, S.; Lumbsch, H.T.; Boekhout, T. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 2015, 78, 16-48. [CrossRef]
- Kwon-Chung, K.J.; Bennett, J.E.; Wickes, B.L.; Meyer, W.; Cuomo, C.A.; Wollenburg, K.R.; Bicanic, T.A.; Castaneda, E.; Chang, Y.C.; Chen, J.; et al. The Case for Adopting the “Species Complex” Nomenclature for the Etiologic Agents of Cryptococcosis. mSphere 2017, 2. [CrossRef]
- Desjardins, C.A.; Giamberardino, C.; Sykes, S.M.; Yu, C.H.; Tenor, J.L.; Chen, Y.; Yang, T.; Jones, A.M.; Sun, S.; Haverkamp, M.R.; et al. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res 2017, 27, 1207-1219. [CrossRef]
- Litvintseva, A.P.; Thakur, R.; Vilgalys, R.; Mitchell, T.G. Multilocus Sequence Typing Reveals Three Genetic Subpopulations of Cryptococcus neoformans var. grubii (Serotype A), Including a Unique Population in Botswana. Genetics 2006, 172, 2223-2238. [CrossRef]
- Jackson, K.M.; Kono, T.J.Y.; Betancourt, J.J.; Wang, Y.; Kabbale, K.D.; Ding, M.; Kezh, P.; Ha, G.; Yoder, J.M.; Fulton, S.R.; et al. Single nucleotide polymorphisms are associated with strain-specific virulence differences among clinical isolates of Cryptococcus neoformans. Nature Communications 2024, 15, 10491. [CrossRef]
- Kassaza, K.; Wasswa, F.; Nielsen, K.; Bazira, J. Cryptococcus neoformans Genotypic Diversity and Disease Outcome among HIV Patients in Africa. Journal of Fungi 2022, 8, 734.
- Oliveira, L.V.N.; Wang, R.; Specht, C.A.; Levitz, S.M. Vaccines for human fungal diseases: close but still a long way to go. NPJ Vaccines 2021, 6, 33. [CrossRef]
- Specht, C.A.; Wang, R.; Oliveira, L.V.N.; Hester, M.M.; Gomez, C.; Mou, Z.; Carlson, D.; Lee, C.K.; Hole, C.R.; Lam, W.C.; et al. Immunological correlates of protection mediated by a whole organism, Cryptococcus neoformans, vaccine deficient in chitosan. mBio 2024, 15, e0174624. [CrossRef]
- Upadhya, R.; Lam, W.C.; Maybruck, B.; Specht, C.A.; Levitz, S.M.; Lodge, J.K. Induction of Protective Immunity to Cryptococcal Infection in Mice by a Heat-Killed, Chitosan-Deficient Strain of Cryptococcus neoformans. mBio 2016, 7, e00547-00516. [CrossRef]
- Wang, R.; Oliveira, L.V.N.; Hester, M.M.; Carlson, D.; Christensen, D.; Specht, C.A.; Levitz, S.M. Protection against experimental cryptococcosis elicited by Cationic Adjuvant Formulation 01-adjuvanted subunit vaccines. PLOS Pathogens 2024, 20, e1012220. [CrossRef]
- Hester, M.M.; Lee, C.K.; Abraham, A.; Khoshkenar, P.; Ostroff, G.R.; Levitz, S.M.; Specht, C.A. Protection of mice against experimental cryptococcosis using glucan particle-based vaccines containing novel recombinant antigens. Vaccine 2020, 38, 620-626. [CrossRef]
- Specht, C.A.; Lee, C.K.; Huang, H.; Hester, M.M.; Liu, J.; Luckie, B.A.; Torres Santana, M.A.; Mirza, Z.; Khoshkenar, P.; Abraham, A.; et al. Vaccination with Recombinant Cryptococcus Proteins in Glucan Particles Protects Mice against Cryptococcosis in a Manner Dependent upon Mouse Strain and Cryptococcal Species. MBio 2017, 8, e01872-01817. [CrossRef]
- Pedersen, G.K.; Andersen, P.; Christensen, D. Immunocorrelates of CAF family adjuvants. Semin Immunol 2018, 39, 4-13. [CrossRef]
- Nielsen, K.; Cox, G.M.; Wang, P.; Toffaletti, D.L.; Perfect, J.R.; Heitman, J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infection and immunity 2003, 71, 4831-4841.
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J., 3rd; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS genetics 2014, 10, e1004261. [CrossRef]
- Litvintseva, A.P.; Mitchell, T.G. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infection and immunity 2009, 77, 3188-3195. [CrossRef]
- Mukaremera, L.; McDonald, T.R.; Nielsen, J.N.; Molenaar, C.J.; Akampurira, A.; Schutz, C.; Taseera, K.; Muzoora, C.; Meintjes, G.; Meya, D.B.; et al. The Mouse Inhalation Model of Cryptococcus neoformans Infection Recapitulates Strain Virulence in Humans and Shows that Closely Related Strains Can Possess Differential Virulence. Infect Immun 2019, 87. [CrossRef]
- Thanh, L.T.; Toffaletti, D.L.; Tenor, J.L.; Giamberardino, C.; Sempowski, G.D.; Asfaw, Y.; Phan, H.T.; Van Duong, A.; Trinh, N.M.; Thwaites, G.E.; et al. Assessing the virulence of Cryptococcus neoformans causing meningitis in HIV infected and uninfected patients in Vietnam. Med Mycol 2020, 58, 1149-1161. [CrossRef]
- Wiesner, D.L.; Moskalenko, O.; Corcoran, J.M.; McDonald, T.; Rolfes, M.A.; Meya, D.B.; Kajumbula, H.; Kambugu, A.; Bohjanen, P.R.; Knight, J.F.; et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 2012, 3. [CrossRef]
- Ashton, P.M.; Thanh, L.T.; Trieu, P.H.; Van Anh, D.; Trinh, N.M.; Beardsley, J.; Kibengo, F.; Chierakul, W.; Dance, D.A.B.; Rattanavong, S.; et al. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nature Communications 2019, 10, 2035. [CrossRef]
- Boulware, D.R.; von Hohenberg, M.; Rolfes, M.A.; Bahr, N.C.; Rhein, J.; Akampurira, A.; Williams, D.A.; Taseera, K.; Schutz, C.; McDonald, T.; et al. Human Immune Response Varies by the Degree of Relative Cryptococcal Antigen Shedding. Open Forum Infectious Diseases 2015, 3. [CrossRef]
- Nielsen, K.; Marra, R.E.; Hagen, F.; Boekhout, T.; Mitchell, T.G.; Cox, G.M.; Heitman, J. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 2005, 171, 975-983. [CrossRef]
- Yu, C.-H.; Sephton-Clark, P.; Tenor, J.L.; Toffaletti, D.L.; Giamberardino, C.; Haverkamp, M.; Cuomo, C.A.; Perfect, J.R. Gene Expression of Diverse Cryptococcus Isolates during Infection of the Human Central Nervous System. mBio 2021, 12, e02313-02321, doi:doi:10.1128/mBio.02313-21.
- Specht, C.A.; Wang, R.; Oliveira, L.V.N.; Hester, M.M.; Gomez, C.; Mou, Z.; Carlson, D.; Lee, C.K.; Hole, C.R.; Lam, W.C.; et al. Immunological correlates of protection mediated by a whole organism, Cryptococcus neoformans, vaccine deficient in chitosan. mBio 2024, e0174624. [CrossRef]
- Oliveira, L.V.N.; Hargarten, J.C.; Wang, R.; Carlson, D.; Park, Y.-D.; Specht, C.A.; Williamson, P.R.; Levitz, S.M. Peripheral blood CD4+ and CD8+ T cell responses to Cryptococcus candidate vaccine antigens in human subjects with and without cryptococcosis. Journal of Infection 2025, 91, 106521. [CrossRef]
- Wang, R.; Oliveira, L.V.N.; Lourenco, D.; Gomez, C.L.; Lee, C.K.; Hester, M.M.; Mou, Z.; Ostroff, G.R.; Specht, C.A.; Levitz, S.M. Immunological correlates of protection following vaccination with glucan particles containing Cryptococcus neoformans chitin deacetylases. NPJ Vaccines 2023, 8, 6. [CrossRef]
- Chen, Y.; Litvintseva, A.P.; Frazzitta, A.E.; Haverkamp, M.R.; Wang, L.; Fang, C.; Muthoga, C.; Mitchell, T.G.; Perfect, J.R. Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Molecular Ecology 2015, 24, 3559-3571. [CrossRef]
- Coelho, M.A.; David-Palma, M.; Aylward, J.; Pham, N.Q.; Visagie, C.M.; Fuchs, T.; Yilmaz, N.; Roets, F.; Sun, S.; Taylor, J.W.; et al. Decoding Cryptococcus: From African biodiversity to worldwide prevalence. PLOS Pathogens 2025, 21, e1012876. [CrossRef]
- Ferreira-Paim, K.; Andrade-Silva, L.; Fonseca, F.M.; Ferreira, T.B.; Mora, D.J.; Andrade-Silva, J.; Khan, A.; Dao, A.; Reis, E.C.; Almeida, M.T.G.; et al. MLST-Based Population Genetic Analysis in a Global Context Reveals Clonality amongst Cryptococcus neoformans var. grubii VNI Isolates from HIV Patients in Southeastern Brazil. PLOS Neglected Tropical Diseases 2017, 11, e0005223. [CrossRef]
- Day, J.N.; Qihui, S.; Thanh, L.T.; Trieu, P.H.; Van, A.D.; Thu, N.H.; Chau, T.T.H.; Lan, N.P.H.; Chau, N.V.V.; Ashton, P.M.; et al. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLOS Neglected Tropical Diseases 2017, 11, e0005628. [CrossRef]
- Van Dyke, M.C.C.; Chaturvedi, A.K.; Hardison, S.E.; Leopold Wager, C.M.; Castro-Lopez, N.; Hole, C.R.; Wozniak, K.L.; Wormley, F.L. Induction of Broad-Spectrum Protective Immunity against Disparate Cryptococcus Serotypes. Frontiers in Immunology 2017, 8, 1-16. [CrossRef]
- Yauch, L.E.; Lam, J.S.; Levitz, S.M. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathog 2006, 2, e120. [CrossRef]
- Kelly, R.M.; Chen, J.; Yauch, L.E.; Rottman, J.B.; Levitz, S.M. Opsonic requirements for dendritic cell-mediated responses to Cryptococcus neoformans. Infect Immun 2005, 73, 592-598.
- Wormley, F.L., Jr.; Perfect, J.R.; Steele, C.; Cox, G.M. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infection and immunity 2007, 75, 1453-1462. [CrossRef]
- Jarvis, J.N.; Meintjes, G.; Rebe, K.; Williams, G.N.; Bicanic, T.; Williams, A.; Schutz, C.; Bekker, L.G.; Wood, R.; Harrison, T.S. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS 2012, 26, 1105-1113. [CrossRef]
- Pappas, P.G.; Bustamante, B.; Ticona, E.; Hamill, R.J.; Johnson, P.C.; Reboli, A.; Aberg, J.; Hasbun, R.; Hsu, H.H. Recombinant interferon- gamma 1b as adjunctive therapy for AIDS-related acute cryptococcal meningitis. J Infect Dis 2004, 189, 2185-2191. [CrossRef]
- Levitz, S.M.; Golenbock, D.T. Beyond empiricism: informing vaccine development through innate immunity research. Cell 2012, 148, 1284-1292. [CrossRef]
- Li, Y.; Ambati, S.; Meagher, R.B.; Lin, X. Developing mRNA lipid nanoparticle vaccine effective for cryptococcosis in a murine model. npj Vaccines 2025, 10, 24. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).