Submitted:
29 September 2025
Posted:
30 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study 1.. Description of Genetic Variability of PRRSV-2 Detected in Japan During 2020–2023
2.1.1. Study 1.1: Broad Regional Analysis (2020-2023)
2.1.2. Study 1.2: Regional Analysis of the Kanto Region (2020-2023)
2.1.2.1. Sample Collection
2.1.2.2. Sample Analysis
2.1.3. Statistical Analysis
2.2. Study 2. Assessment of the Relationship Between MLV Use and the Distribution of PRRSV-2 Clusters of Prevalent Strains
2.2.1. Sample Collection and Sequencing
2.2.2. Statistical Analysis
3. Results
3.1. Study 1: Description of Genetic Variability of PRRSV-2 Detected in Japan During 2020–2023 and Comparison of Sequence Homology with Previous Reports
3.1.1. Broad Regional Phylogenetic Analysis and Cluster Identification (Study 1.1)
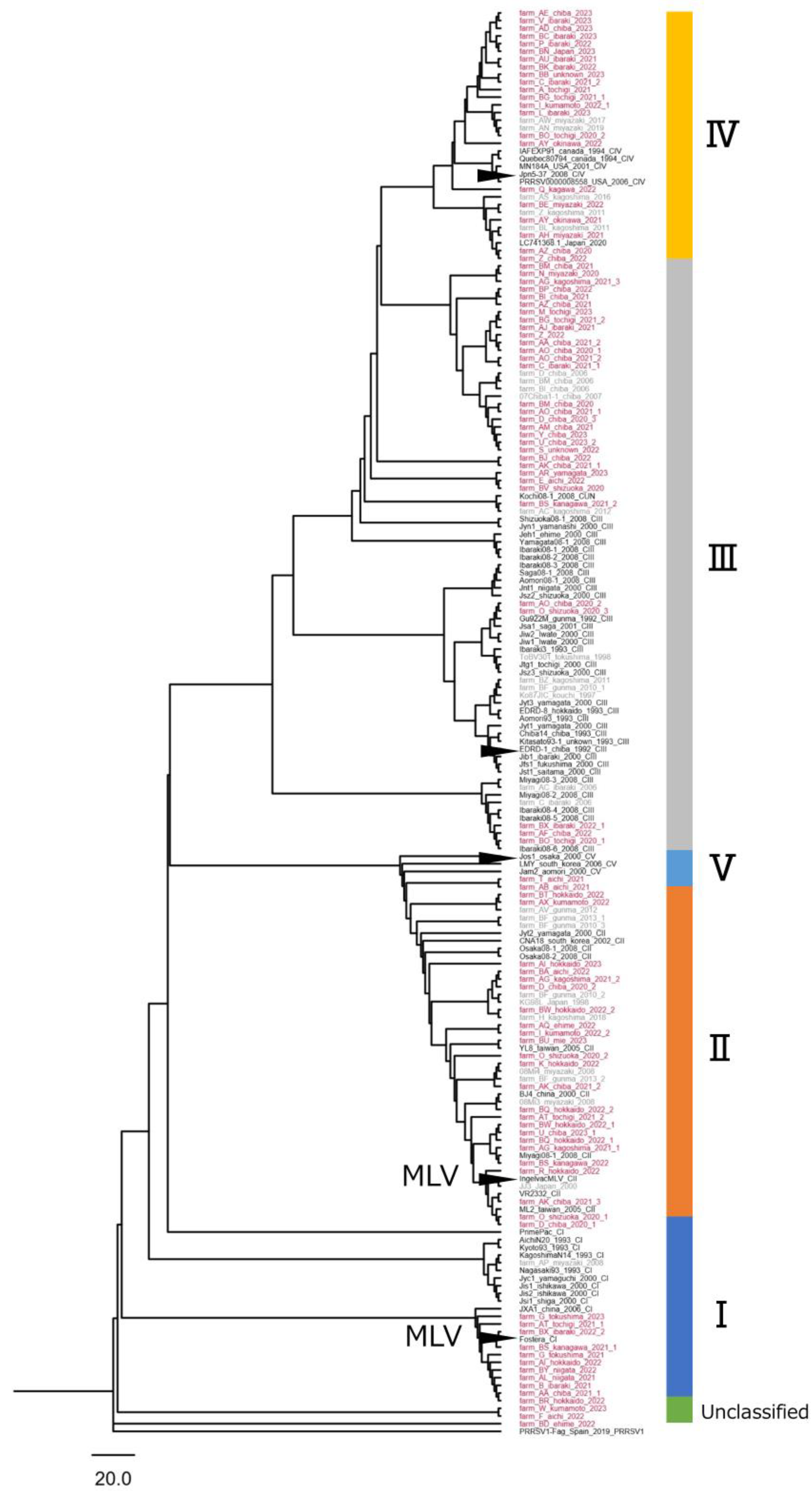
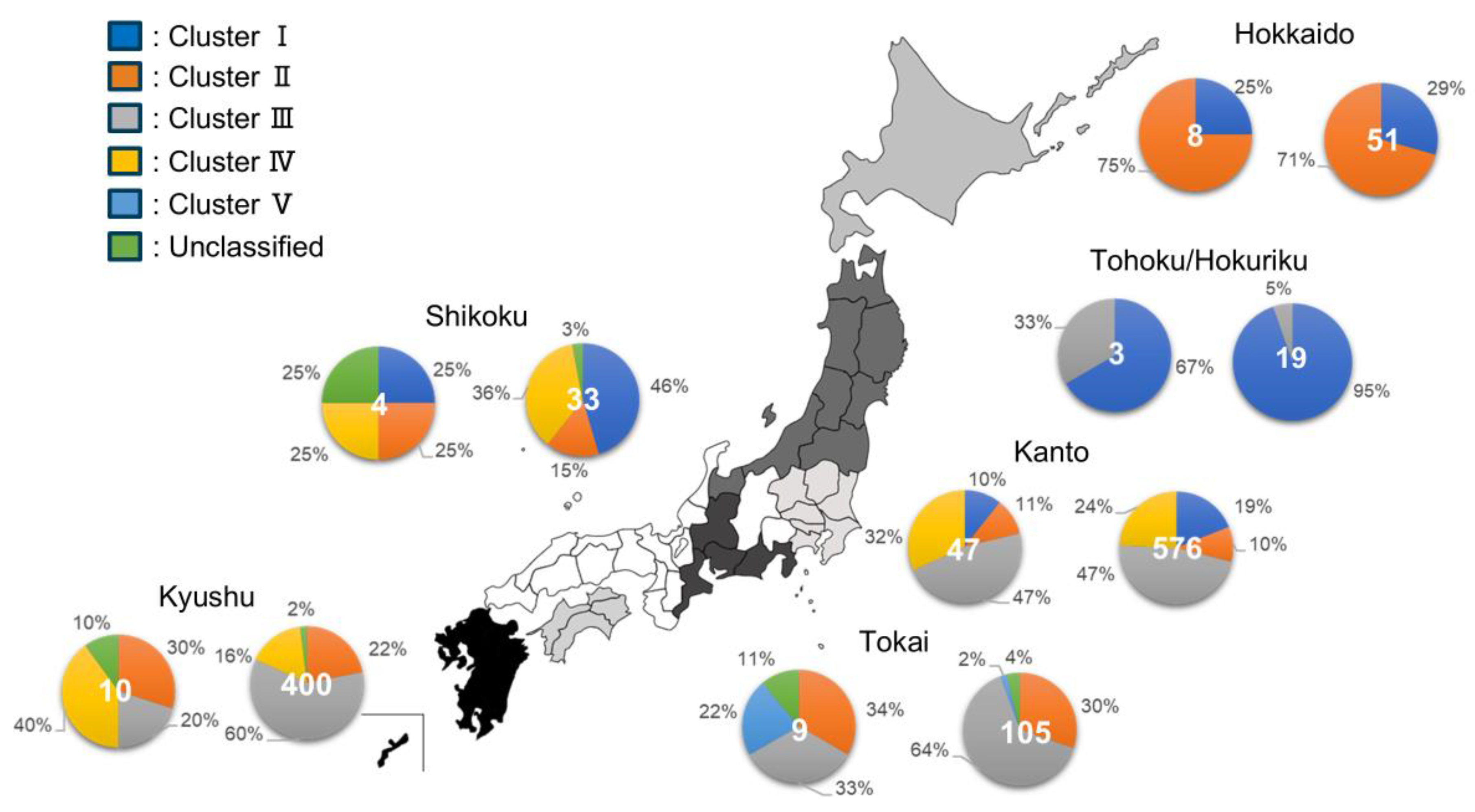
3.1.2. Descriptive Epizootiology of Distribution of Clusters Across Regions in Japan (Study 1.1)
3.1.3. Sequence Homology to the Reference Strains (Study 1.1)
3.2 Study 1.2: Regional Analysis of PRRSV-2 in the Kanto Region (2020-2023)
3.2.1. PRRSV-2 Detection Rates Across Different Production Stages (Study 1.2)
3.2.2. Distribution of PRRSV-2 Clusters Across Production Stages (Study 1.2)
3.3. Study 2: Assessment of the Relationship Between MLV Use and Distribution of PRRSV-2 Clusters of Prevalent Strains in the Kanto Region
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GP | glycoprotein |
| MLV | modified live vaccine |
| NIBS | Nippon Institute of Biological Science |
| ORF | open reading frame |
| PRRS | porcine reproductive and respiratory syndrome |
| PRRSV | porcine reproductive and respiratory syndrome virus |
| RFLP | restriction fragment length polymorphism |
| RT-PCR | qualitative reverse transcription polymerase chain reaction |
| RT-qPCR | quantitative reverse transcription polymerase chain reaction |
References
- Fiers, J.; Cay, A.B.; Maes, D.; Tignon, M. A Comprehensive Review on Porcine Reproductive and Respiratory Syndrome Virus with Emphasis on Immunity. Vaccines 2024, 12, 942. [Google Scholar] [CrossRef]
- Ruedas-Torres, I.; Rodríguez-Gómez, I.M.; Sánchez-Carvajal, J.M.; Larenas-Muñoz, F.; Pallarés, F.J.; Carrasco, L.; Gómez-Laguna, J. The jigsaw of PRRSV virulence. Vet. Microbiol. 2021, 260, 109168. [Google Scholar] [CrossRef]
- Yamane, I.; Kure, K.; Ishikawa, H.; Tagagi, M.; Miyazaki, A.; Suzuki, T.; Shibahara, T.; Kubo, M.; Kobayashi, H.; Kokuho, T.; et al. Evaluation of the economical losses due to the outbreaks of porcine reproductive and respiratory syndrome. (A questionnaire-based epidemiological survey and estimation of the total economical losses in Japan). Proc. Jpn. Pig Vet. Soc. 2009, 55, 33–37. (In Japanese) [Google Scholar]
- Holtkamp, D.; Kliebenstein, J.; Neumann, E.; Zimmerman, J.; Rotto, H.; Yoder, T.; Wang, C.; Yeske, P.; Mowrer, C.; Haley, C. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on U.S. pork producers. J. Swine Health Prod. 2013, 21, 72–84. [Google Scholar] [CrossRef]
- Chae, C. Commercial PRRS Modified-Live Virus Vaccines. Vaccines 2021, 9, 185. [Google Scholar] [CrossRef]
- Luo, Q.; Zheng, Y.; Zhang, H.; Yang, Z.; Sha, H.; Kong, W.; Zhao, M.; Wang, N. Research Progress on Glycoprotein 5 of Porcine Reproductive and Respiratory Syndrome Virus. Animals: an Open Access Journal from MDPI 2023, 13, 813. [Google Scholar] [CrossRef] [PubMed]
- Kim, W. I. , Kim, J. J., Cha, S. H., Wu, W. H., Cooper, V., Evans, R., Choi, E. J., & Yoon, K. J. Significance of genetic variation of PRRSV ORF5 in virus neutralization and molecular determinants corresponding to cross neutralization among PRRS viruses. Vet. Microbiol. 2013, 162, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Bálint, Á.; Molnár, T.; Kecskeméti, S.; Kulcsar, G.; Soós, T.; Szabo, P.; Kaszab, E.; Fornyos, K.; Zádori, Z.; Bányai, K.; et al. Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary. Vaccines 2021, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Yim-Im, W.; Anderson, T.K.; Paploski, I.A.D.; VanderWaal, K.; Gauger, P.; Krueger, K.; Shi, M.; Main, R.; Zhang, J. Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes. Microbiol. Spectr. 2023, 11, e0291623. [Google Scholar] [CrossRef]
- Jian, Y.; Lu, C.; Shi, Y.; Kong, X.; Song, J.; Wang, J. Genetic evolution analysis of PRRSV ORF5 gene in five provinces of Northern China in 2024. BMC Vet. Res. 2025, 21, 4679. [Google Scholar] [CrossRef]
- Zeller, M.A.; Chang, J.; Trevisan, G.; Main, R.G.; Gauger, P.C.; Zhang, J. Rapid PRRSV-2 ORF5-based lineage classification using Nextclade. Front. Vet. Sci. 2024, 11, 1419340. [Google Scholar] [CrossRef]
- Chandra, S.; Cezar, G.; Rupasinghe, K.; Magalhães, E.; Silva, G.; Almeida, M.; Crim, B.; Burrough, E.; Gauger, P.; Madson, D.; et al. Harnessing sequencing data for porcine reproductive and respiratory syndrome virus (PRRSV): tracking genetic evolution dynamics and emerging sequences in US swine industry. Front. Vet. Sci. 2025, 12, 1571020. [Google Scholar] [CrossRef] [PubMed]
- Wesley, R.D.; Mengeling, W.L.; Lager, K.M.; Clouser, D.F.; Landgraf, J.G.; Frey, M.L. Differentiation of a Porcine Reproductive and Respiratory Syndrome Virus Vaccine Strain from North American Field Strains by Restriction Fragment Length Polymorphism Analysis of ORF 5. J. Vet. Diagn. Invest. 1998, 10, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, G.; Sharma, A.; Gauger, P.; Harmon, K.M.; Zhang, J.; Main, R.; Zeller, M.; Linhares, L.C.M.; Linhares, D.C.L. PRRSV2 genetic diversity defined by RFLP patterns in the United States from 2007 to 2019. J. Vet. Diagn. Invest. 2021, 33, 920–931. [Google Scholar] [CrossRef]
- Shi, M.; Lam, T.T.Y.; Hon, C.C.; Hui, R.K.H.; Faaberg, K.S.; Wennblom, T.; Murtaugh, M.P.; Stadejek, T.; Leung, F.C.C. Molecular epidemiology of PRRSV: A phylogenetic perspective. Virus Res. 2010, 154, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Paploski, I.A.D.; Corzo, C.; Rovira, A.; Murtaugh, M.P.; Sanhueza, J.M.; Vilalta, C.; Schroeder, D.C.; VanderWaal, K. Temporal Dynamics of Co-circulating Lineages of Porcine Reproductive and Respiratory Syndrome Virus. Front. Microbiol. 2019, 10, 2486. [Google Scholar] [CrossRef]
- Paploski, I.A.D.; Pamornchainavakul, N.; Makau, D.N.; Rovira, A.; Corzo, C.A.; Schroeder, D.C.; Cheeran, M.C.J.; Doeschl-Wilson, A.; Kao, R.R.; Lycett, S.; et al. Phylogenetic Structure and Sequential Dominance of Sub-Lineages of PRRSV Type-2 Lineage 1 in the United States. Vaccines 2021, 9, 608. [Google Scholar] [CrossRef]
- Yoshii, M.; Kaku, Y.; Murakami, Y.; Shimizu, M.; Kato, K.; Ikeda, H. Genetic variation and geographic distribution of porcine reproductive and respiratory syndrome virus in Japan. Arch. Virol. 2005, 150, 2313–2324. [Google Scholar] [CrossRef]
- Iseki, H.; Takagi, M.; Miyazaki, A.; Katsuda, K.; Mikami, O.; Tsunemitsu, H. Genetic analysis of ORF5 in porcine reproductive and respiratory syndrome virus in Japan. Microbiol. Immunol. 2011, 55, 211–216. [Google Scholar] [CrossRef]
- Kyutoku, F.; Yokoyama, T.; Sugiura, K. Genetic Diversity and Epidemic Types of Porcine Reproductive and Respiratory Syndrome (PRRS) Virus in Japan from 2018 to 2020. Epidemiologia 2022, 3, 285–296. [Google Scholar] [CrossRef]
- Charerntantanakul, W. Porcine reproductive and respiratory syndrome virus vaccines: Immunogenicity, efficacy and safety aspects. World J. Virol. 2012, 1, 23. [Google Scholar] [CrossRef]
- Cho, J.G.; Dee, S.A. Porcine reproductive and respiratory syndrome virus. Theriogenology 2006, 66, 655–662. [Google Scholar] [CrossRef]
- Li, J.; Miller, L.C.; Sang, Y. Current Status of Vaccines for Porcine Reproductive and Respiratory Syndrome: Interferon Response, Immunological Overview, and Future Prospects. Vaccines 2024, 12, 606. [Google Scholar] [CrossRef]
- Montaner-Tarbes, S.; del Portillo, H.A.; Montoya, M.; Fraile, L. Key gaps in the knowledge of the porcine respiratory reproductive syndrome virus (PRRSV). Front. Vet. Sci. 2019, 6, 38. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Ye, M.; Li, X.; Li, S.; Xie, N.; Wei, Y.; Wang, J.; Zhu, J. High genetic diversity of Chinese porcine reproductive and respiratory syndrome viruses from 2016 to 2019. Res. Vet. Sci. 2020, 131, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2024. https://www.R-project.org/.
- Posit Team. RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA, 2024. https://www.posit.co/.
- Shen, Y.F.; Arruda, A.G.; Koscielny, M.P.; Cheng, T.Y. Contrasting PRRSV temporal lineage patterns at the individual farm, production system, and regional levels in Ohio and neighboring states from 2017 to 2021. Prev. Vet. Med. 2024, 226, 106186. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, Y.; Taira, O.; Omori, T.; Tsutsumi, N.; Sugiura, K. First detection of NADC34-like porcine reproductive and respiratory syndrome virus strains in Japan. J. Vet. Med. Sci. 2025, 87, 110–114. [Google Scholar] [CrossRef]
- Kikuti, M.; Sanhueza, J.; Vilalta, C.; Paploski, I.A.D.; VanderWaal, K.; Corzo, C.A. Porcine reproductive and respiratory syndrome virus 2 (PRRSV-2) genetic diversity and occurrence of wild type and vaccine-like strains in the United States swine industry. PLoS One 2021, 16, e0259531. [Google Scholar] [CrossRef]
- Klinge, K.L.; Vaughn, E.M.; Roof, M.B.; Bautista, E.M.; Murtaugh, M.P. Age-dependent resistance to Porcine reproductive and respiratory syndrome virus replication in swine. Virol. J. 2009, 6, 177. [Google Scholar] [CrossRef]
- Plaza-Soriano, Á.; Martínez-Lobo, F.J.; Garza-Moreno, L.; Castillo-Pérez, J.; Caballero, E.; Castro, J.M.; Simarro, I.; Prieto, C. Determination of the frequency of individuals with broadly cross-reactive neutralizing antibodies against PRRSV in the sow population under field conditions. Porcine Health Manag. 2024, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Lebret, A.; Berton, P.; Normand, V.; Messager, I.; Robert, N.; Bouchet, F.; Brissonnier, M.; Boulbria, G. PRRSV detection by qPCR in processing fluids and serum samples collected in a positive stable breeding herd following mass vaccination of sows with a modified live vaccine. Porcine Health Manag. 2021, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Matamoros, A.; Camprodon, A.; Maldonado, J.; Pedrazuela, R.; Miranda, J. Safety and long-lasting immunity of the combined administration of a modified-live virus vaccine against porcine reproductive and respiratory syndrome virus 1 and an inactivated vaccine against porcine parvovirus and Erysipelothrix rhusiopathiae in breeding pigs. Porcine Health Manag. 2019, 5, 11. [Google Scholar] [CrossRef] [PubMed]
| Phylogenetic cluster |
Year of isolation |
Prefecture | Name of isolate | Accession no. |
|---|---|---|---|---|
| I | 1993 | Aichi | Aichi N20 | AB175715 |
| I | 1993 | Kyoto | Kyoto 93 | AB175724 |
| I | 1993 | Nagasaki | Nagasaki 93 | AB175725 |
| I | 1993 | Kagoshima | Kagoshima N14 | AB175723 |
| I | 2000 | Ishikawa | Jis1 | AB175694 |
| I | 2000 | Ishikawa | Jis2 | AB175695 |
| I | 2000 | Shiga | Jsi1 | AB175701 |
| I | 2000 | Yamaguchi | Jyc1 | AB175709 |
| II | 2000 | Aomori | Jam2 | AB175690 |
| II | 2000 | Yamagata | Jyt2 | AB175713 |
| II | 2007–2008** | Miyagi | Miyagi08-1 | AB546104 |
| II | 2007–2008** | Osaka | Osaka08-1 | AB546120 |
| II | 2007–2008** | Osaka | Osaka08-2 | AB546121 |
| III | 1992 | Chiba | EDRD-1* | D45852 |
| III | 1992 | Gunma | Gu922M | AB175721 |
| III | 1993 | Hokkaido | EDRD-8 | AB175720 |
| III | 1993 | Aomori | Aomori93 | AB175716 |
| III | 1993 | Ibaraki | Ibaraki3 | AB175722 |
| III | 1993 | Chiba | Chiba 14 | AB175717 |
| III | 1993 | Tokyo | Kitasato 93-1 | AB023782 |
| III | 2000 | Iwate | Jiw1 | AB175696 |
| III | 2000 | Iwate | Jiw2 | AB175697 |
| III | 2000 | Yamagata | Jyt1 | AB175712 |
| III | 2000 | Yamagata | Jyt3 | AB175714 |
| III | 2000 | Niigata | Jnt1 | AB175698 |
| III | 2000 | Ibaraki | Jib1 | AB175693 |
| III | 2000 | Tochigi | Jtg1 | AB175708 |
| III | 2000 | Saitama | Jst1 | AB175702 |
| III | 2000 | Yamanashi | Jyn1 | AB175710 |
| III | 2000 | Shizuoka | Jsz2 | AB175704 |
| III | 2000 | Ehime | Jeh1 | AB175691 |
| III | 2001 | Saga | Jsa1 | AB175700 |
| III | 2007–2008** | Aomori | Aomori08-1 | AB546102 |
| III | 2007–2008** | Miyagi | Miyagi08-2 | AB546105 |
| III | 2007–2008** | Miyagi | Miyagi08-3 | AB546106 |
| III | 2007–2008** | Yamagata | Yamagata08-1 | AB546108 |
| III | 2007–2008** | Ibaraki | Ibaraki08-1 | AB546109 |
| III | 2007–2008** | Ibaraki | Ibaraki08-2 | AB546110 |
| III | 2007–2008** | Ibaraki | Ibaraki08-3 | AB546111 |
| III | 2007–2008** | Ibaraki | Ibaraki08-4 | AB546112 |
| III | 2007–2008** | Ibaraki | Ibaraki08-5 | AB546113 |
| III | 2007–2008** | Ibaraki | Ibaraki08-6 | AB546114 |
| III | 2007–2008** | Shizuoka | Shizuoka08-1 | AB546118 |
| III | 2007–2008** | Kochi | Kochi08-1 | AB546123 |
| IV | 2008 | Jpn5-37* | - | AB546125 |
| IV | 2020 | 6145-1L | - | LC741368 |
| III | 2007–2008** | Saga | Saga08-1 | AB546124 |
| V | 2000 | Osaka | Jos1* | AB175699 |
| * Reference strains used for homology analysis in this study | ||||
| **The exact obtained year is not provided as the annotation | ||||
| Phylogenetic cluster |
Year of isolation | Country/ Region |
Name of isolate | Accession no. |
|---|---|---|---|---|
| I | 2006 | China | JXA1 | EF112445 |
| I | 2019 | USA | Fostera® PRRS* | Fostera® PRRS* |
| II | 1992 | USA | VR2332 | EF536003 |
| II | 1997 | USA | Ingelvac® PRRS MLV* | AF020048 |
| II | 2000 | China | BJ-4 | AF331831 |
| II | 2002 | South Korea | CNA18 | DQ473472 |
| II | 2005 | Taiwan | ML2 | EU273672 |
| II | 2006 | Taiwan | YL8 | EU273700 |
| IV | 1994 | Canada | IAF-EXP91 | L40898 |
| IV | 1994 | Canada | Quebec 807/94 | Z82995 |
| IV | 2001 | USA | MN184A | DQ176019 |
| IV | 2006 | USA | PRRSV0000008558 | EU758599 |
| V | 2006 | South Korea | LMY | DQ473474 |
| PRRSV-1 | 2019 | Spain | PRRSV1-Fag | MZ318699 |
| * Reference strains used for homology analysis in this study | ||||
| ** Year of submission to GenBank | ||||
| Cluster | Reference strain |
No. of Obtained sequences |
Homology distribution (%) |
Mean (%) |
Median (%) |
25th percentile (%) |
75th percentile (%) |
|---|---|---|---|---|---|---|---|
| I | Fostera® PRRS | 158 | 95.2-100 | 98.9 | 99.2 | 98.3 | 99.6 |
| II | Ingelvac®PRRS MLV | 221 | 88.1-100 | 98.9 | 99.6 | 98.8 | 99.7 |
| III | EDRD-1 | 577 | 85.3-96.7 | 90.0 | 90.5 | 87.4 | 91.9 |
| IV | Jpn5-37 | 219 | 79.5-95.3 | 84.7 | 84.9 | 84.2 | 85.2 |
| V | Jos1 | 2 | 82.7-88.5 | 85.6 | 85.6 | 84.1 | 87.0 |
| Unclassified | 13 | ||||||
| Total | 1190 |
| Pig production stage | |||||||
| Gilt | Sow | Fetus | Piglet | Grower | Finisher | Unknown | |
| No. of positive samples / No. of samples |
35/61 | 70/249 | 9/10 | 49/116 | 601/927 | 85/262 | 27/45 |
| Positivity rate (%) |
57.4% | 28.1% | 90.0% | 42.2% | 64.8% | 32.4% | 60.0% |
| Pig production stage | ||||||||||||||
| Cluster | Gilt (%) (n/N) |
Sow (%) (n/N) |
Fetus (%) (n/N) |
Piglet (%) (n/N) |
Grower (%) (n/N) |
Finisher (%) (n/N) |
Unknown (%) (n/N) |
|||||||
| I | 0.0% | (0/7) | 0.0% | (0/23) | 33.3% | (1/3) | 3.6% | (1/28) | 17.9% | (82/459) | 50.0% | (17/34) | 36.4% | (8/22) |
| II | 0.0% | (0/7) | 0.0% | (0/23) | 0.0% | (0/3) | 10.7% | (3/28) | 11.5% | (53/459) | 2.9% | (1/34) | 4.6% | (1/22) |
| III | 28.6% | (2/7) | 17.4% | (4/23) | 0.0% | (0/3) | 35.7% | (10/28) | 52.3% | (240/459) | 11.8% | (4/34) | 40.9% | (9/22) |
| IV | 71.4% | (5/7) | 82.6% | (19/23) | 66.7% | (2/3) | 50.0% | (14/28) | 18.3% | (84/459) | 35.3% | (12/34) | 18.2% | (4/22) |
| V | 0.0% | (0/0) | 0.0% | (0/0) | 0.0% | (0/0) | 0.0% | (0/0) | 0.0% | (0/0) | 0.0% | (0/0) | 0.0% | (0/0) |
| *Values are expressed as percentages. N indicates the total number of sequences obtained for each production stage, and n denotes the number of sequences classified into each phylogenetic cluster within the corresponding stage. | ||||||||||||||
| Cluster | Sequences from farms using MLV | Sequences from farms not using MLV | Sequences from farms with unknown MLV status | Total |
| I | 109 | 0 | 0 | 109 |
| II | 58 | 0 | 0 | 58 |
| III | 213 | 55 | 1 | 269 |
| IV | 26 | 110 | 4 | 140 |
| Total | 406 | 165 | 5 | 576 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





