Submitted:
24 September 2025
Posted:
25 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Enzyme Production by Solid-State Fermentation
2.2. Partial Purification of Xylanase
2.3. Gel Electrophoresis and Zymogram
2.4. Substrate Preference
2.5. Biochemical Characterization of Xylanase Aggregate
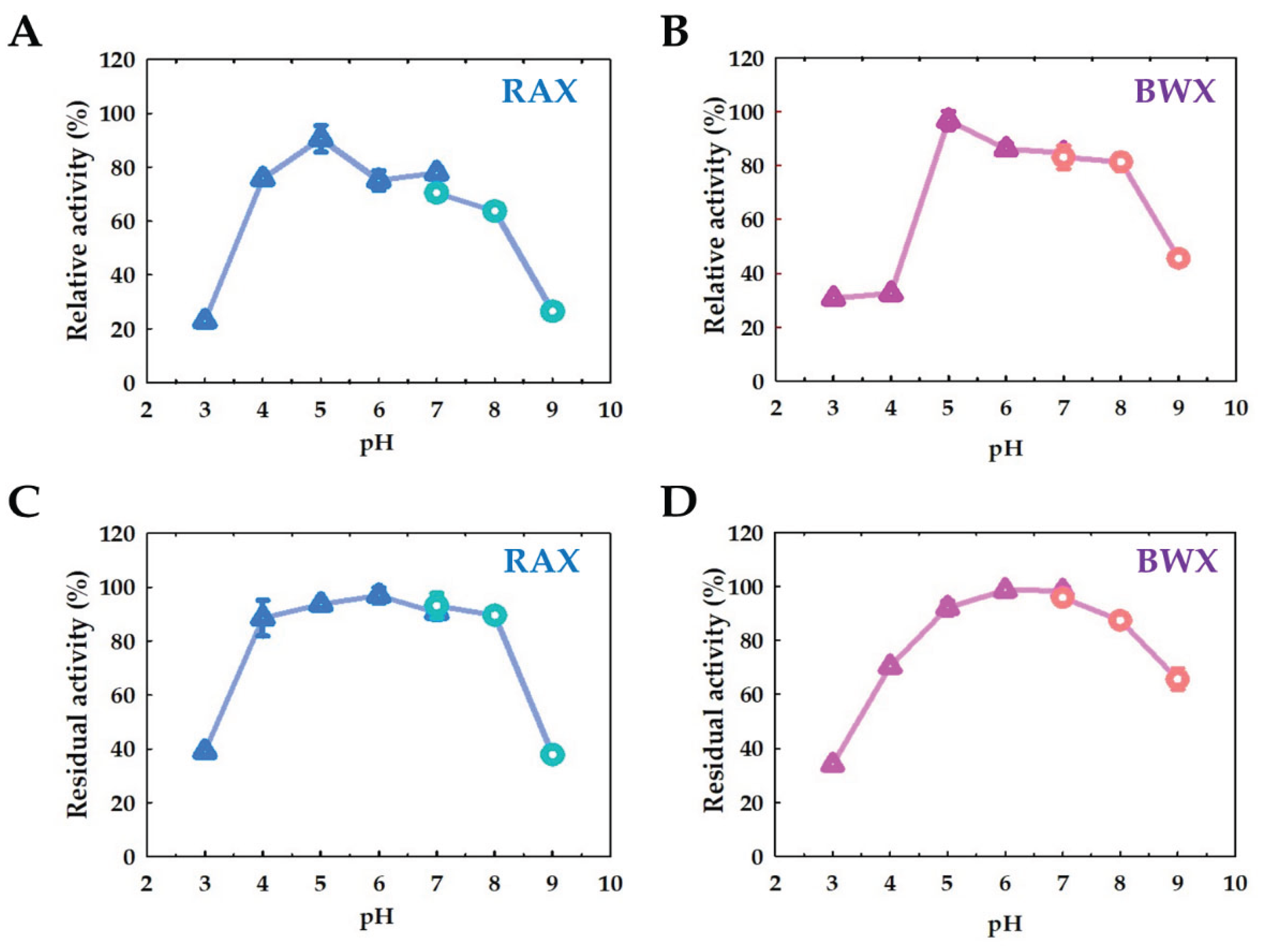
2.5.1. Effect of pH and Stability
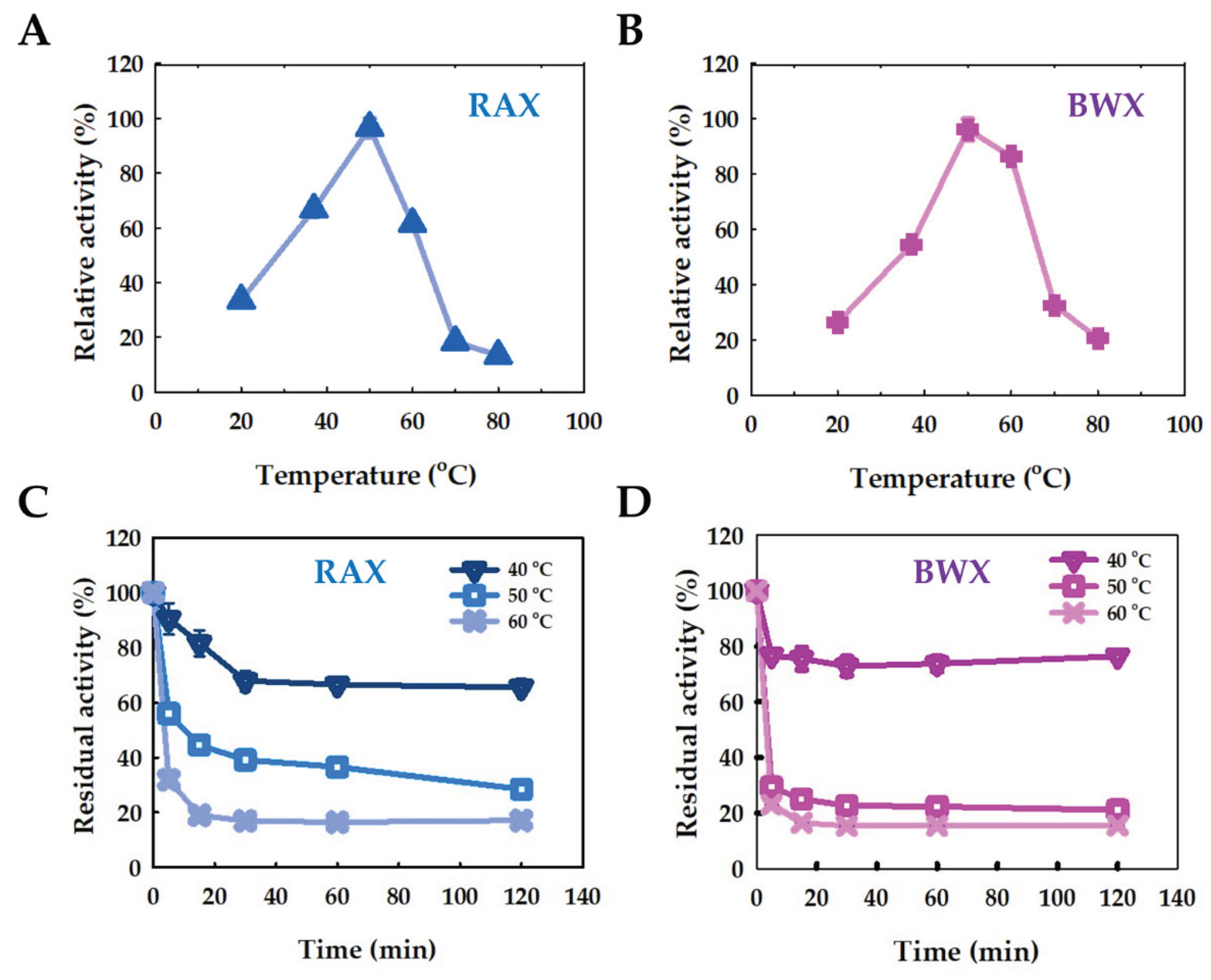
2.5.2. Effect of Temperature and Stability
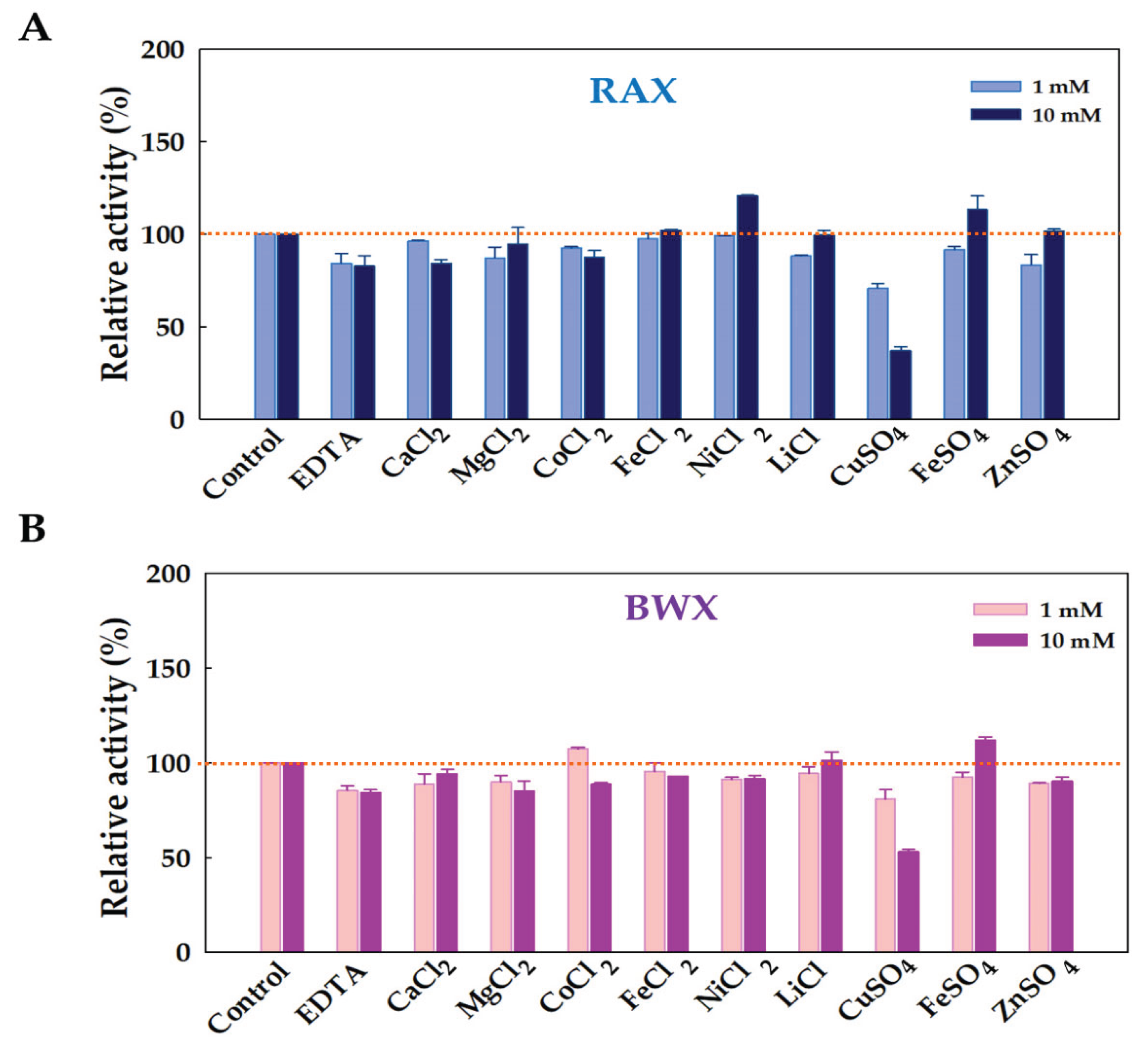
2.5.3. Effect of Metal Ion and Chemical Reagent
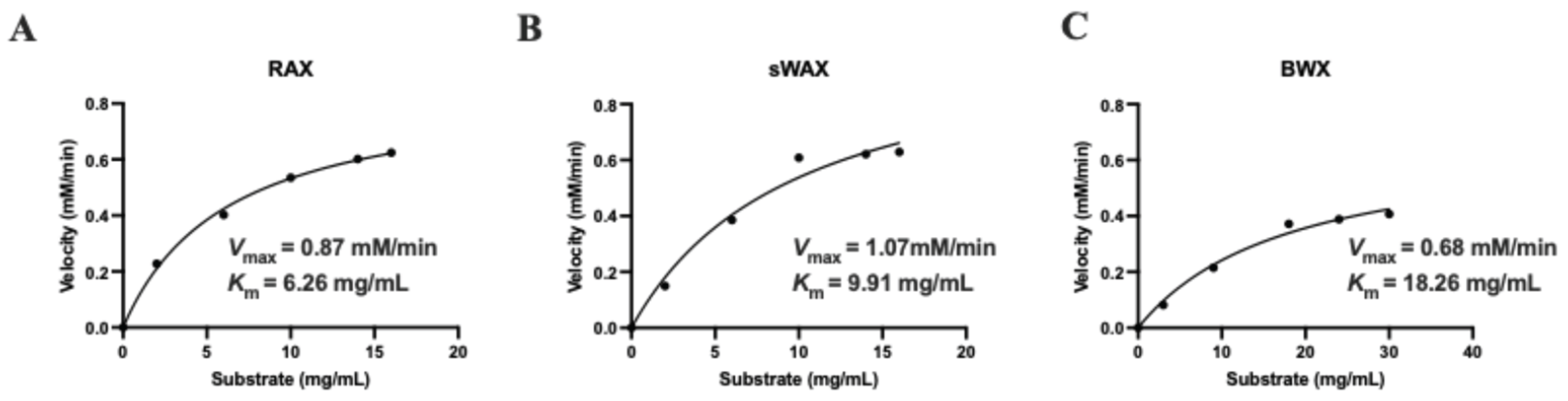
2.6. Kinetic Parameters of Xylanase Aggregate
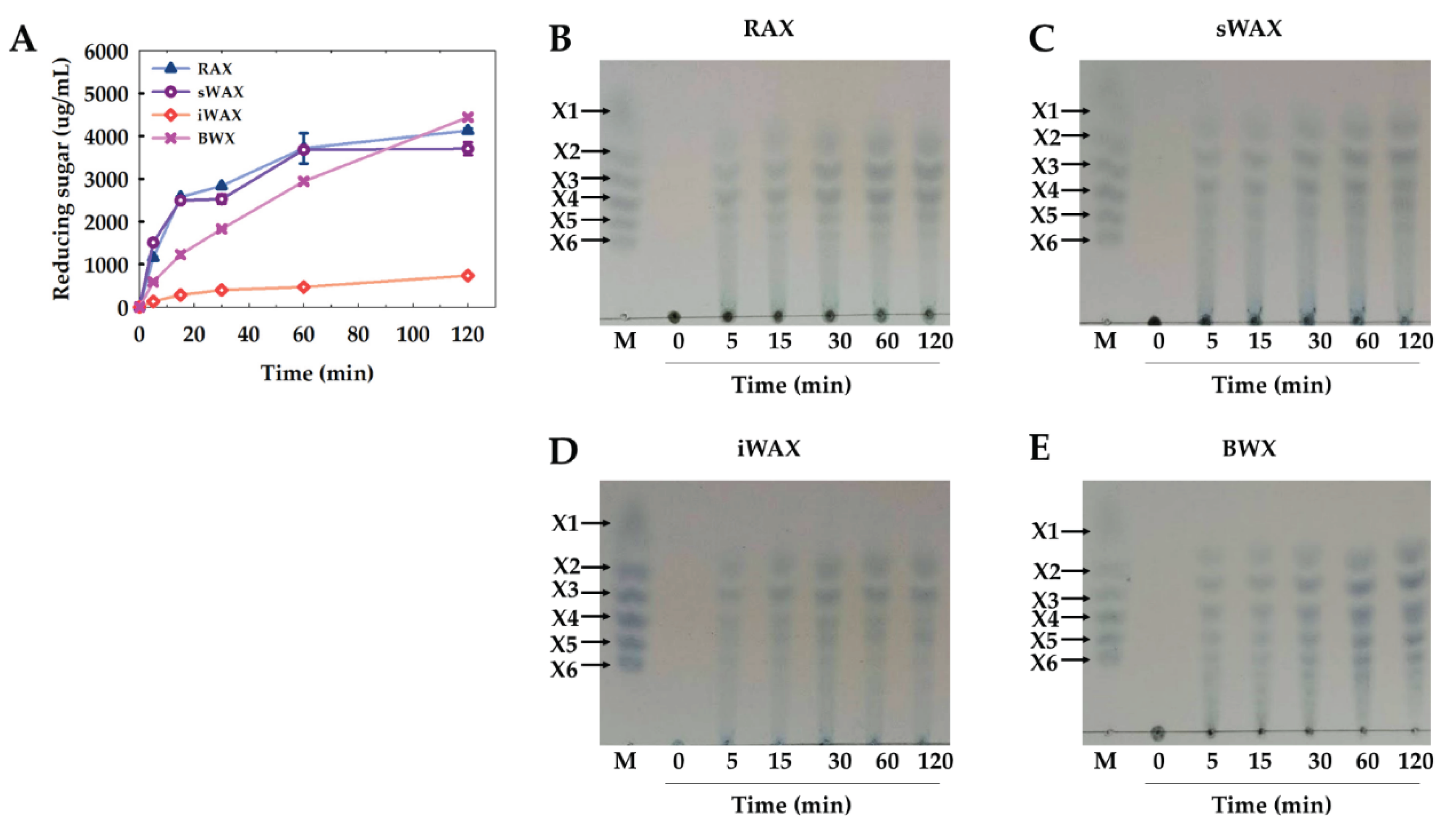
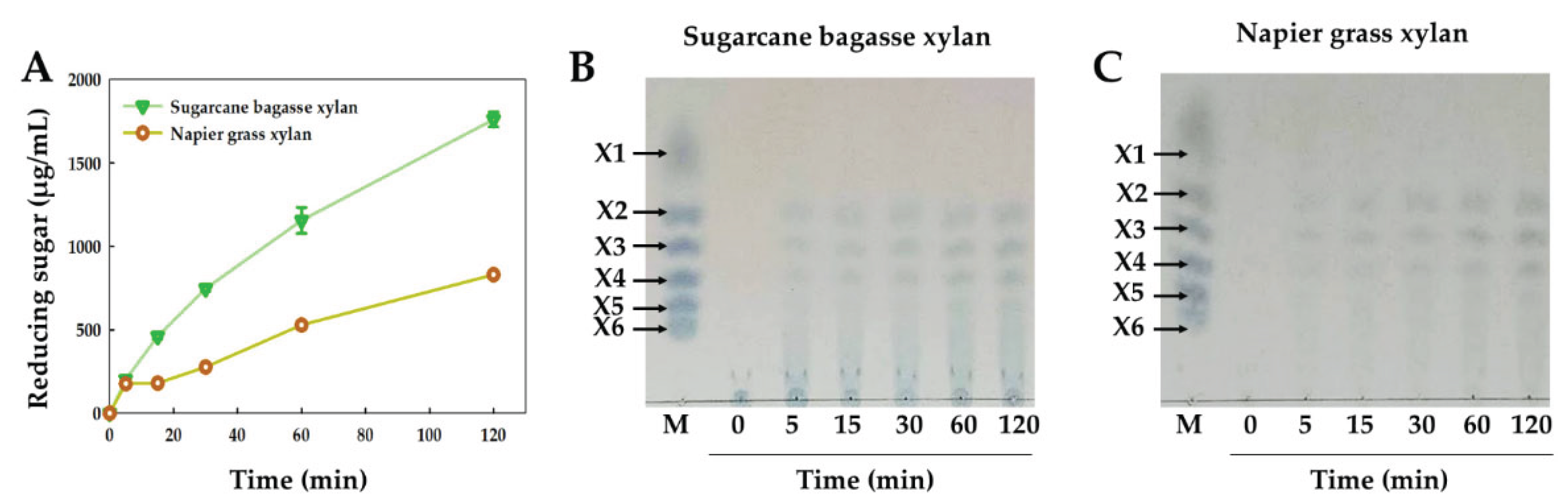
2.7. Time Course of Enzymatic Hydrolysis and Analysis of Hydrolysis Products
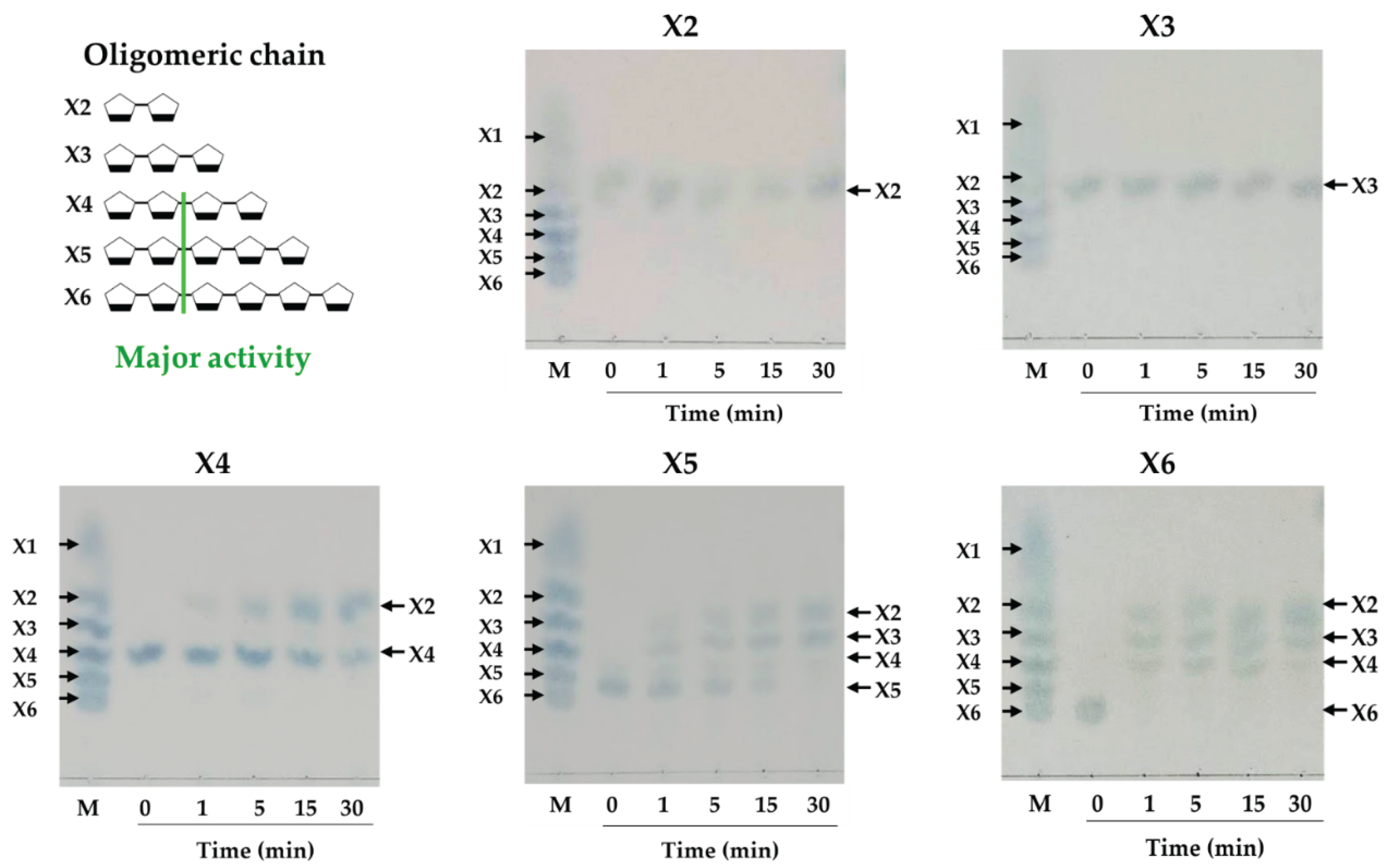
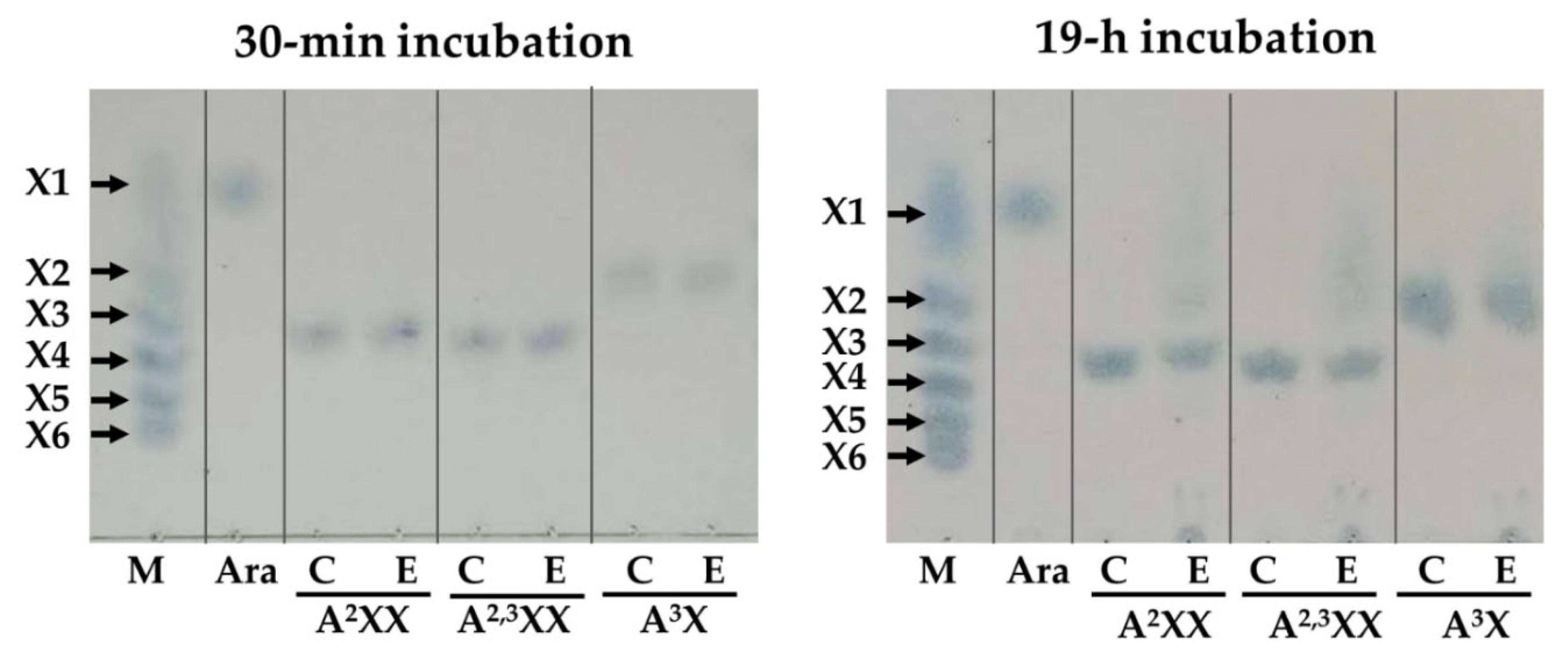
2.8. Modes of Action
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Enzyme Production by Solid-State Fermentation
4.3. Determination of Protein
4.4. Determination of Enzymatic Activities and Substrate Specificities
4.5. Xylanase Purification
4.5.1. Ammonium Sulfate Precipitation
4.5.2. Ion-Exchange Chromatography
4.5.3. Gel Filtration Chromatography
4.6. Biochemical Characterization of Partially Purified Xylanase
4.6.1. Electrophoresis
4.6.2. Zymogram
4.6.3. Effect of pH and Stability
4.6.4. Effect of Temperature and Stability
4.6.5. Effect of Metal Ion and Chemical Reagent
4.6.6. Enzyme Kinetic (Michaelis-Menten Parameter)
4.7. Hydrolysis Products
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Bioresources and Bioprocessing 2019, 6. [CrossRef]
- Biely, P.; Singh, S.; Puchart, V. Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol Adv 2016, 34, 1260–1274. [CrossRef]
- Rennie, E.A.; Scheller, H.V. Xylan biosynthesis. Curr Opin Biotechnol 2014, 26, 100–107. [CrossRef]
- Marcotuli, I.; Hsieh, Y.S.; Lahnstein, J.; Yap, K.; Burton, R.A.; Blanco, A.; Fincher, G.B.; Gadaleta, A. Structural Variation and Content of Arabinoxylans in Endosperm and Bran of Durum Wheat (Triticum turgidum L.). J Agric Food Chem 2016, 64, 2883–2892. [CrossRef]
- Ye, Z.H.; Zhong, R. Outstanding questions on xylan biosynthesis. Plant Sci 2022, 325, 111476. [CrossRef]
- de O. Buanafina, M.M. Feruloylation in Grasses: Current and Future Perspectives. Molecular Plant 2, 861–872. [CrossRef]
- Zhang, B.; Zhong, Y.; Dong, D.; Zheng, Z.; Hu, J. Gut microbial utilization of xylan and its implication in gut homeostasis and metabolic response. Carbohydr Polym 2022, 286, 119271. [CrossRef]
- Juturu, V.; Wu, J.C. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv 2012, 30, 1219–1227. [CrossRef]
- Heinen, P.R.; Bauermeister, A.; Ribeiro, L.F.; Messias, J.M.; Almeida, P.Z.; Moraes, L.A.B.; Vargas-Rechia, C.G.; de Oliveira, A.H.C.; Ward, R.J.; Filho, E.X.F.; et al. GH11 xylanase from Aspergillus tamarii Kita: Purification by one-step chromatography and xylooligosaccharides hydrolysis monitored in real-time by mass spectrometry. Int J Biol Macromol 2018, 108, 291–299. [CrossRef]
- Chadha, B.S.; Kaur, B.; Basotra, N.; Tsang, A.; Pandey, A. Thermostable xylanases from thermophilic fungi and bacteria: Current perspective. Bioresour Technol 2019, 277, 195–203. [CrossRef]
- Li, X.; Dilokpimol, A.; Kabel, M.A.; de Vries, R.P. Fungal xylanolytic enzymes: Diversity and applications. Bioresour Technol 2022, 344, 126290. [CrossRef]
- Gibson, D.M.; King, B.C.; Hayes, M.L.; Bergstrom, G.C. Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr Opin Microbiol 2011, 14, 264–270. [CrossRef]
- Cruz-Davila, J.; Perez, J.V.; Castillo, D.S.D.; Diez, N. Fusarium graminearum as a producer of xylanases with low cellulases when grown on wheat bran. Biotechnol Rep (Amst) 2022, 35, e00738. [CrossRef]
- Moonjely, S.; Ebert, M.; Paton-Glassbrook, D.; Noel, Z.A.; Roze, L.; Shay, R.; Watkins, T.; Trail, F. Update on the state of research to manage Fusarium head blight. Fungal Genet Biol 2023, 169, 103829. [CrossRef]
- Tini, F.; Beccari, G.; Benfield, A.H.; Gardiner, D.M.; Covarelli, L. Role of the XylA gene, encoding a cell wall degrading enzyme, during common wheat, durum wheat and barley colonization by Fusarium graminearum. Fungal Genet Biol 2020, 136, 103318. [CrossRef]
- Bertonha, L.C.; Leal Neto, M.; Garcia, J.A.A.; Vieira, T.F.; Castoldi, R.; Bracht, A.; Peralta, R.M. Screening of Fusarium sp. for xylan and cellulose hydrolyzing enzymes and perspectives for the saccharification of delignified sugarcane bagasse. Biocatalysis and Agricultural Biotechnology 2018, 16, 385–389. [CrossRef]
- Dong, X.; Meinhardt, S.W.; Schwarz, P.B. Isolation and characterization of two endoxylanases from Fusarium graminearum. J Agric Food Chem 2012, 60, 2538–2545. [CrossRef]
- Heinen, P.R.; Caroline, H.; Rosane, M.P.; Adelar, B.R.d.C.G.S.; Jose, L.d.C.S.; Maria, d.L.T.M.P.; Marina, K.K. Xylanase from Fusarium heterosporum: Properties and influence of thiol compounds on xylanase activity. African Journal of Biotechnology 2014, 13, 1047–1055. [CrossRef]
- Saha, B.C. Production, purification and properties of xylanase from a newly isolated Fusarium proliferatum. Process Biochemistry 2002.
- He, Y.; Zhou, X.; Li, J.; Li, H.; Li, Y.; Nie, Y. In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4. Biomolecules 2021, 11. [CrossRef]
- Bundidamorn, D.; Salaiphet, L.; Vichitsoonthonkul, T.; Ratanakhanokchai, K.; Phitsuwan, P. Screening of fungi isolated from damaged plant materials for the production of lignocellulolytic enzymes with decolorizing ability. Asia-Pacific Journal of Science and Technology 2021, 26, APST–26. [CrossRef]
- Guo, Z.; Yu, Z.; Li, Q.; Tang, L.; Guo, T.; Huang, S.; Mo, J.; Hsiang, T.; Luo, S. Fusarium species associated with leaf spots of mango in China. Microb. Pathog. 2021, 150, 104736. [CrossRef]
- Paccanaro, M.C.; Sella, L.; Castiglioni, C.; Giacomello, F.; Martinez-Rocha, A.L.; D’Ovidio, R.; Schafer, W.; Favaron, F. Synergistic Effect of Different Plant Cell Wall-Degrading Enzymes Is Important for Virulence of Fusarium graminearum. Mol Plant Microbe Interact 2017, 30, 886–895. [CrossRef]
- Tundo, S.; Moscetti, I.; Faoro, F.; Lafond, M.; Giardina, T.; Favaron, F.; Sella, L.; D’Ovidio, R. Fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci 2015, 240, 161–169. [CrossRef]
- Martinez-Pacheco, M.M.; Flores-Garcia, A.; Zamudio-Jaramillo, M.A.; Chavez-Parga, M.C.; Alvarez-Navarrete, M. Optimization of production of xylanases with low cellulases in Fusarium solani by means of a solid state fermentation using statistical experimental design. Rev Argent Microbiol 2020, 52, 328–338. [CrossRef]
- Huang, Y.; Busk, P.K.; Lange, L. Cellulose and hemicellulose-degrading enzymes in Fusarium commune transcriptome and functional characterization of three identified xylanases. Enzyme Microb Technol 2015, 73-74, 9–19. [CrossRef]
- Faria, S.P.; de Melo, G.R.; Cintra, L.C.; Ramos, L.P.; Amorim Jesuino, R.S.; Ulhoa, C.J.; de Faria, F.P. Production of cellulases and xylanases by Humicola grisea var. thermoidea and application in sugarcane bagasse arabinoxylan hydrolysis. Industrial Crops and Products 2020, 158. [CrossRef]
- Ezeilo, U.R.; Wahab, R.A.; Mahat, N.A. Optimization studies on cellulase and xylanase production by Rhizopus oryzae UC2 using raw oil palm frond leaves as substrate under solid state fermentation. Renewable Energy 2020, 156, 1301–1312. [CrossRef]
- Leschonski, K.P.; Kaasgaard, S.G.; Spodsberg, N.; Krogh, K.B.R.M.; Kabel, M.A. Two Subgroups within the GH43_36 α-l-Arabinofuranosidase Subfamily Hydrolyze Arabinosyl from Either Mono-or Disubstituted Xylosyl Units in Wheat Arabinoxylan. International Journal of Molecular Sciences 2022, 23. [CrossRef]
- Li, T.; Wu, Q.; Wang, Y.; John, A.; Qu, H.; Gong, L.; Duan, X.; Zhu, H.; Yun, Z.; Jiang, Y. Application of Proteomics for the Investigation of the Effect of Initial pH on Pathogenic Mechanisms of Fusarium proliferatum on Banana Fruit. Front Microbiol 2017, 8, 2327. [CrossRef]
- Neves Junior, A.; Mansoldo, F.R.P.; Godoy, M.G.; Firpo, R.M.; Cedrola, S.M.L.; Vermelho, A.B. Production of an endo-polygalacturonase from Fusarium proliferatum isolated from agro-industrial waste. Biocatalysis and Agricultural Biotechnology 2021, 38. [CrossRef]
- Perincherry, L.; Urbaniak, M.; Pawlowicz, I.; Kotowska, K.; Waskiewicz, A.; Stepien, L. Dynamics of Fusarium Mycotoxins and Lytic Enzymes during Pea Plants’ Infection. Int J Mol Sci 2021, 22. [CrossRef]
- Perincherry, L.; Ajmi, C.; Oueslati, S.; Waskiewicz, A.; Stepien, L. Induction of Fusarium lytic Enzymes by Extracts from Resistant and Susceptible Cultivars of Pea (Pisum sativum L.). Pathogens 2020, 9. [CrossRef]
- Chang, K.F.; Hwang, S.F.; Conner, R.L.; Ahmed, H.U.; Zhou, Q.; Turnbull, G.D.; Strelkov, S.E.; McLaren, D.L.; Gossen, B.D. First report of Fusarium proliferatum causing root rot in soybean (Glycine max L.) in Canada. Crop Protection 2015, 67, 52–58. [CrossRef]
- Faraz, A.; Haq, I.U.; Ijaz, S.; Mubeen, F.; Habib, A.; Qadri, R.W.K.; Khan, N.A. Morphgenomics based identification of Fusarium proliferatum causing Syagrus romanzoffiana wilt and exploitation of antifungal potential of Trichoderma species against this pathogen. Journal of Plant Pathology 2020, 102, 1097–1105. [CrossRef]
- Lei, S.; Wang, L.; Liu, L.; Hou, Y.; Xu, Y.; Liang, M.; Gao, J.; Li, Q.; Huang, S. Infection and Colonization of Pathogenic Fungus Fusarium proliferatum in Rice Spikelet Rot Disease. Rice Science 2019, 26, 60–68. [CrossRef]
- Das, S.P.; Ravindran, R.; Deka, D.; Jawed, M.; Das, D.; Goyal, A. Bioethanol production from leafy biomass of mango (Mangifera indica) involving naturally isolated and recombinant enzymes. Prep. Biochem. Biotechnol. 2013, 43, 717–734. [CrossRef]
- Sultan, A.; Frisvad, J.C.; Andersen, B.; Svensson, B.; Finnie, C. Investigation of the indigenous fungal community populating barley grains: Secretomes and xylanolytic potential. J Proteomics 2017, 169, 153–164. [CrossRef]
- Saha, B.C. Purification and properties of an extracellular β-xylosidase from a newly isolated Fusarium proliferatum. Bioresource Technology 2002, 90, 33–38. [CrossRef]
- Shin, H.-D.; Chen, R.R. Production and characterization of a type B feruloyl esterase from Fusarium proliferatum NRRL 26517. Enzyme and Microbial Technology 2006, 38, 478–485. [CrossRef]
- Chen, C.; Zhao, X.; Wang, X.; Wang, B.; Li, H.; Feng, J.; Wu, A. Mutagenesis of UDP-xylose epimerase and xylan arabinosyl-transferase decreases arabinose content and improves saccharification of rice straw. Plant Biotechnol J 2021, 19, 863–865. [CrossRef]
- Yoshida, S.; Kusakabe, I.; Matsuo, N.; Shimizu, K.; Yasui, T.; Murakami, K. Structure of Rice-straw Arabinoglucuronoxylan and Specificity ofStreptomycesXylanase toward the Xylan. Agric. Biol. Chem. 2014, 54, 449–457. [CrossRef]
- Mardetko, N.; Trontel, A.; Novak, M.; Pavlecic, M.; Ljubas, B.D.; Grubisic, M.; Tominac, V.P.; Ludwig, R.; Santek, B. Screening of Lignocellulolytic Enzyme Activities in Fungal Species and Sequential Solid-State and Submerged Cultivation for the Production of Enzyme Cocktails. Polymers (Basel) 2021, 13. [CrossRef]
- Prasoulas, G.; Gentikis, A.; Konti, A.; Kalantzi, S.; Kekos, D.; Mamma, D. Bioethanol Production from Food Waste Applying the Multienzyme System Produced On-Site by Fusarium oxysporum F3 and Mixed Microbial Cultures. Fermentation 2020, 6. [CrossRef]
- Najjarzadeh, N.; Matsakas, L.; Rova, U.; Christakopoulos, P. Effect of Oligosaccharide Degree of Polymerization on the Induction of Xylan-Degrading Enzymes by Fusarium oxysporum f. sp. Lycopersici. Molecules 2020, 25. [CrossRef]
- Amadi, O.C.; Egong, E.J.; Nwagu, T.N.; Okpala, G.; Onwosi, C.O.; Chukwu, G.C.; Okolo, B.N.; Agu, R.C.; Moneke, A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon 2020, 6, e04566. [CrossRef]
- Ezeilo, U.R.; Lee, C.T.; Huyop, F.; Zakaria, II; Wahab, R.A. Raw oil palm frond leaves as cost-effective substrate for cellulase and xylanase productions by Trichoderma asperellum UC1 under solid-state fermentation. J Environ Manage 2019, 243, 206–217. [CrossRef]
- Marques, N.P.; de Cassia Pereira, J.; Gomes, E.; da Silva, R.; Araújo, A.R.; Ferreira, H.; Rodrigues, A.; Dussán, K.J.; Bocchini, D.A. Cellulases and xylanases production by endophytic fungi by solid state fermentation using lignocellulosic substrates and enzymatic saccharification of pretreated sugarcane bagasse. Industrial Crops and Products 2018, 122, 66–75. [CrossRef]
- de Amo, G.S.; Bezerra-Bussoli, C.; da Silva, R.R.; Kishi, L.T.; Ferreira, H.; Mariutti, R.B.; Arni, R.K.; Gomes, E.; Bonilla-Rodriguez, G.O. Heterologous expression, purification and biochemical characterization of a new xylanase from Myceliophthora heterothallica F.2.1.4. Int J Biol Macromol 2019, 131, 798–805. [CrossRef]
- Singh, G.; Samuchiwal, S.; Hariprasad, P.; Sharma, S. Melioration of Paddy Straw to produce cellulase-free xylanase and bioactives under Solid State Fermentation and deciphering its impact by Life Cycle Assessment. Bioresour Technol 2022, 360, 127493. [CrossRef]
- Yang, X.; Shi, P.; Huang, H.; Luo, H.; Wang, Y.; Zhang, W.; Yao, B. Two xylose-tolerant GH43 bifunctional beta-xylosidase/alpha-arabinosidases and one GH11 xylanase from Humicola insolens and their synergy in the degradation of xylan. Food Chem 2014, 148, 381–387. [CrossRef]
- Ohtsuki, T.; Suyanto; Yazaki, S.; Ui, S.; Mimura, A.; Suyanto. Production of large multienzyme complex by aerobic thermophilic fungus Chaetomium sp. nov. MS-017 grown on palm oil mill fibre. Lett Appl Microbiol 2005, 40, 111–116. [CrossRef]
- Phitsuwan, P.; Ratanakhanokchai, K. The recovery and bioproperties of a xylanolytic multi-enzyme complex from Tepidimicrobium xylanilyticum BT14. Journal of Molecular Catalysis B: Enzymatic 2015, 120, 28–37. [CrossRef]
- Phitsuwan, P.; Morag, E.; Tachaapaikoon, C.; Pason, P.; Kosugi, A.; Ratanakhanokchai, K. Behavior and supportive evidence of a large xylanase-containing multienzyme complex of tepidimicrobium xylanilyticum bt14. BioResources 2012, 7, 5934–5949.
- van Dyk, J.S.; Sakka, M.; Sakka, K.; Pletschke, B.I. The cellulolytic and hemi-cellulolytic system of Bacillus licheniformis SVD1 and the evidence for production of a large multi-enzyme complex. Enzyme and Microbial Technology 2009, 45, 372–378. [CrossRef]
- Jiang, Z.Q.; Deng, W.; Li, X.T.; Ai, Z.L.; Li, L.T.; Kusakabe, I. Characterization of a novel, ultra-large xylanolytic complex (xylanosome) from Streptomyces olivaceoviridis E-86. Enzyme and Microbial Technology 2005, 36, 923–929. [CrossRef]
- Dou, T.Y.; Chen, J.; Liu, C. Isolation and subunit compositions of the xylanosome complexes produced by Cellulosimicrobium species. Enzyme Microb Technol 2020, 133, 109445. [CrossRef]
- Jiang, Z.; Dang, W.; Yan, Q.; Zhai, Q.; Li, L.; Kusakabe, I. Subunit composition of a large xylanolytic complex (xylanosome) from Streptomyces olivaceoviridis E-86. J Biotechnol 2006, 126, 304–312. [CrossRef]
- Duarte, M.; Alves, V.D.; Correia, M.; Caseiro, C.; Ferreira, L.M.A.; Romao, M.J.; Carvalho, A.L.; Najmudin, S.; Bayer, E.A.; Fontes, C.; et al. Structure-function studies can improve binding affinity of cohesin-dockerin interactions for multi-protein assemblies. Int J Biol Macromol 2022. [CrossRef]
- Wang, W.; Yu, Y.; Dou, T.Y.; Wang, J.Y.; Sun, C. Species of family Promicromonosporaceae and family Cellulomonadeceae that produce cellulosome-like multiprotein complexes. Biotechnol Lett 2018, 40, 335–341. [CrossRef]
- Dou, T.Y.; Luan, H.W.; Ge, G.B.; Dong, M.M.; Zou, H.F.; He, Y.Q.; Cui, P.; Wang, J.Y.; Hao, D.C.; Yang, S.L.; et al. Functional and structural properties of a novel cellulosome-like multienzyme complex: efficient glycoside hydrolysis of water-insoluble 7-xylosyl-10-deacetylpaclitaxel. Sci Rep 2015, 5, 13768. [CrossRef]
- Hamann, P.R.V.; de, M.B.S.L.; Gomes, T.C.; Noronha, E.F. Assembling mini-xylanosomes with Clostridium thermocellum XynA, and their properties in lignocellulose deconstruction. Enzyme Microb Technol 2021, 150, 109887. [CrossRef]
- Sunna, A.; Antranikian, G. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 1997, 17, 39–67.
- Silva, L.A.O.; Terrasan, C.R.F.; Carmona, E.C. Purification and characterization of xylanases from Trichoderma inhamatum. Electronic Journal of Biotechnology 2015, 18, 307–313. [CrossRef]
- Sharma, S.; Verma, P.; Agrawal, K. Harnessing the potential of fungal xylanases: An insight into its application and technological advancements. Industrial Crops and Products 2024, 222. [CrossRef]
- Polizeli, M.L.; Rizzatti, A.C.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol 2005, 67, 577–591. [CrossRef]
- Schmid, M.; Prinz, T.K.; Stabler, A.; Sangerlaub, S. Effect of Sodium Sulfite, Sodium Dodecyl Sulfate, and Urea on the Molecular Interactions and Properties of Whey Protein Isolate-Based Films. Front Chem 2017, 4, 49. [CrossRef]
- Garcia Medeiros, R.; Hanada, R.; Filho, E.X.F. Production of xylan-degrading enzymes from Amazon forest fungal species. International Biodeterioration & Biodegradation 2003, 52, 97–100. [CrossRef]
- Pauchet, Y.; Ruprecht, C.; Pfrengle, F. Analyzing the Substrate Specificity of a Class of Long-Horned-Beetle-Derived Xylanases by Using Synthetic Arabinoxylan Oligo- and Polysaccharides. ChemBioChem 2020, 21, 1517–1525. [CrossRef]
- Aronsson, A.; Guler, F.; Petoukhov, M.V.; Crennell, S.J.; Svergun, D.I.; Linares-Pasten, J.A.; Nordberg Karlsson, E. Structural insights of RmXyn10A - A prebiotic-producing GH10 xylanase with a non-conserved aglycone binding region. Biochim Biophys Acta Proteins Proteom 2018, 1866, 292–306. [CrossRef]
- Labourel, A.; Crouch, L.I.; Bras, J.L.; Jackson, A.; Rogowski, A.; Gray, J.; Yadav, M.P.; Henrissat, B.; Fontes, C.M.; Gilbert, H.J.; et al. The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans. J Biol Chem 2016, 291, 22149–22159. [CrossRef]
- Vardakou, M.; Flint, J.; Christakopoulos, P.; Lewis, R.J.; Gilbert, H.J.; Murray, J.W. A family 10 Thermoascus aurantiacus xylanase utilizes arabinose decorations of xylan as significant substrate specificity determinants. J Mol Biol 2005, 352, 1060–1067. [CrossRef]
- Liu, B.; Stevens-Green, R.; Johal, D.; Buchanan, R.; Geddes-McAlister, J. Fungal pathogens of cereal crops: Proteomic insights into fungal pathogenesis, host defense, and resistance. J. Plant Physiol. 2022, 269, 153593. [CrossRef]
- Sella, L.; Gazzetti, K.; Faoro, F.; Odorizzi, S.; D’Ovidio, R.; Schafer, W.; Favaron, F. A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol Biochem 2013, 64, 1–10. [CrossRef]
- Moscetti, I.; Tundo, S.; Janni, M.; Sella, L.; Gazzetti, K.; Tauzin, A.; Giardina, T.; Masci, S.; Favaron, F.; D’Ovidio, R. Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in durum wheat transgenic plants. Mol Plant Microbe Interact 2013, 26, 1464–1472. [CrossRef]
- Fülöp, L. The inhibition of xylanase enzymes by oligosaccharides produced during the degradation of biopolymers in biomass. Biomass and Bioenergy 2025, 195. [CrossRef]
- Rudjito, R.C.; Jimenez-Quero, A.; Munoz, M.; Kuil, T.; Olsson, L.; Stringer, M.A.; Krogh, K.; Eklof, J.; Vilaplana, F. Arabinoxylan source and xylanase specificity influence the production of oligosaccharides with prebiotic potential. Carbohydr Polym 2023, 320, 121233. [CrossRef]
- Khaire, K.C.; Sharma, K.; Thakur, A.; Moholkar, V.S.; Goyal, A. Extraction and characterization of xylan from sugarcane tops as a potential commercial substrate. J Biosci Bioeng 2021, 131, 647–654. [CrossRef]
- MILLER, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428,.
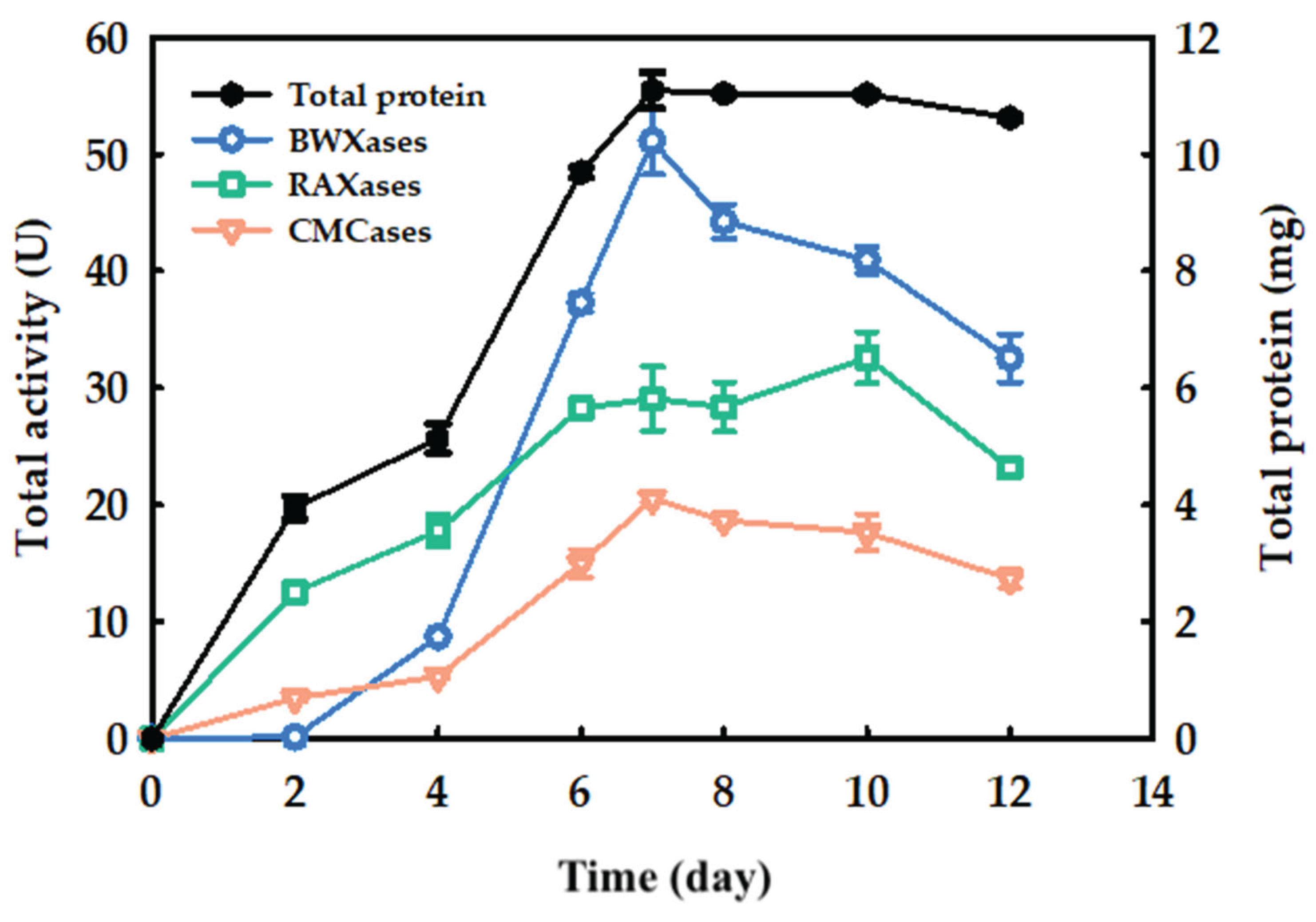
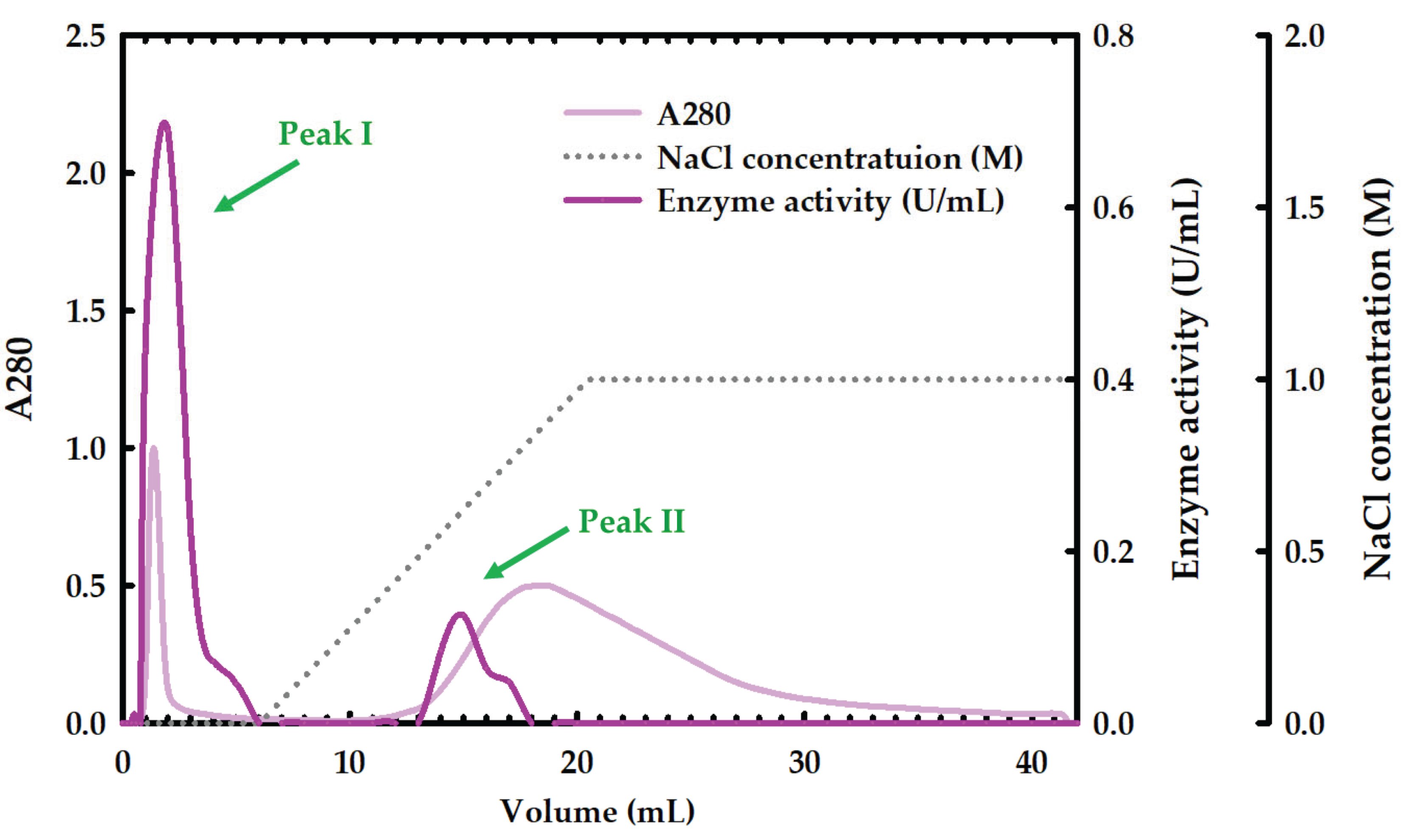
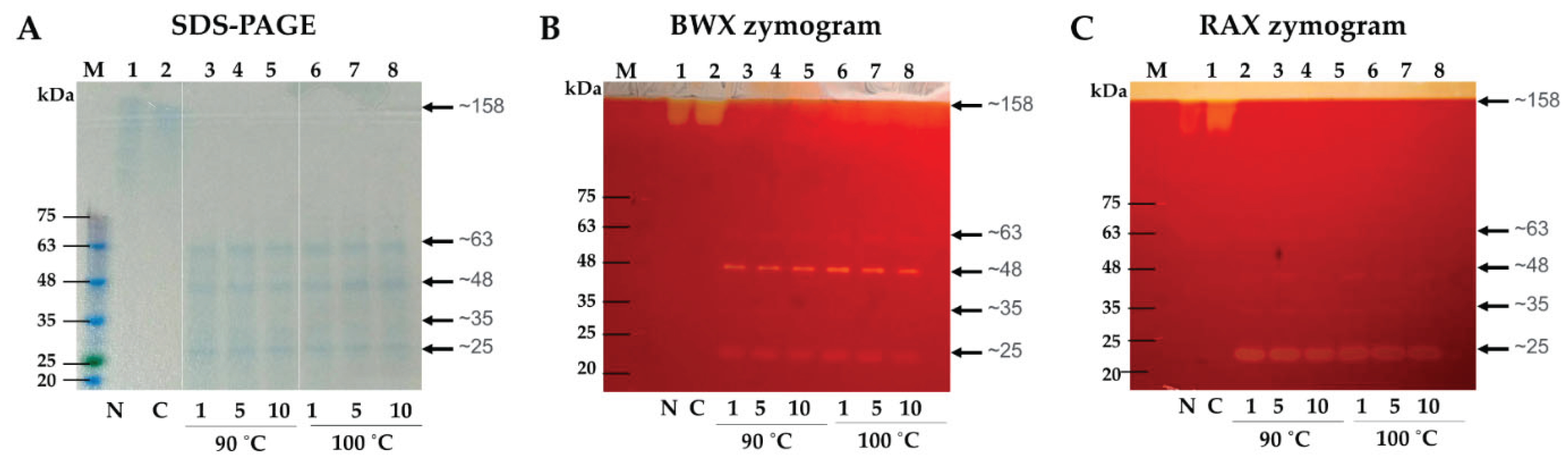
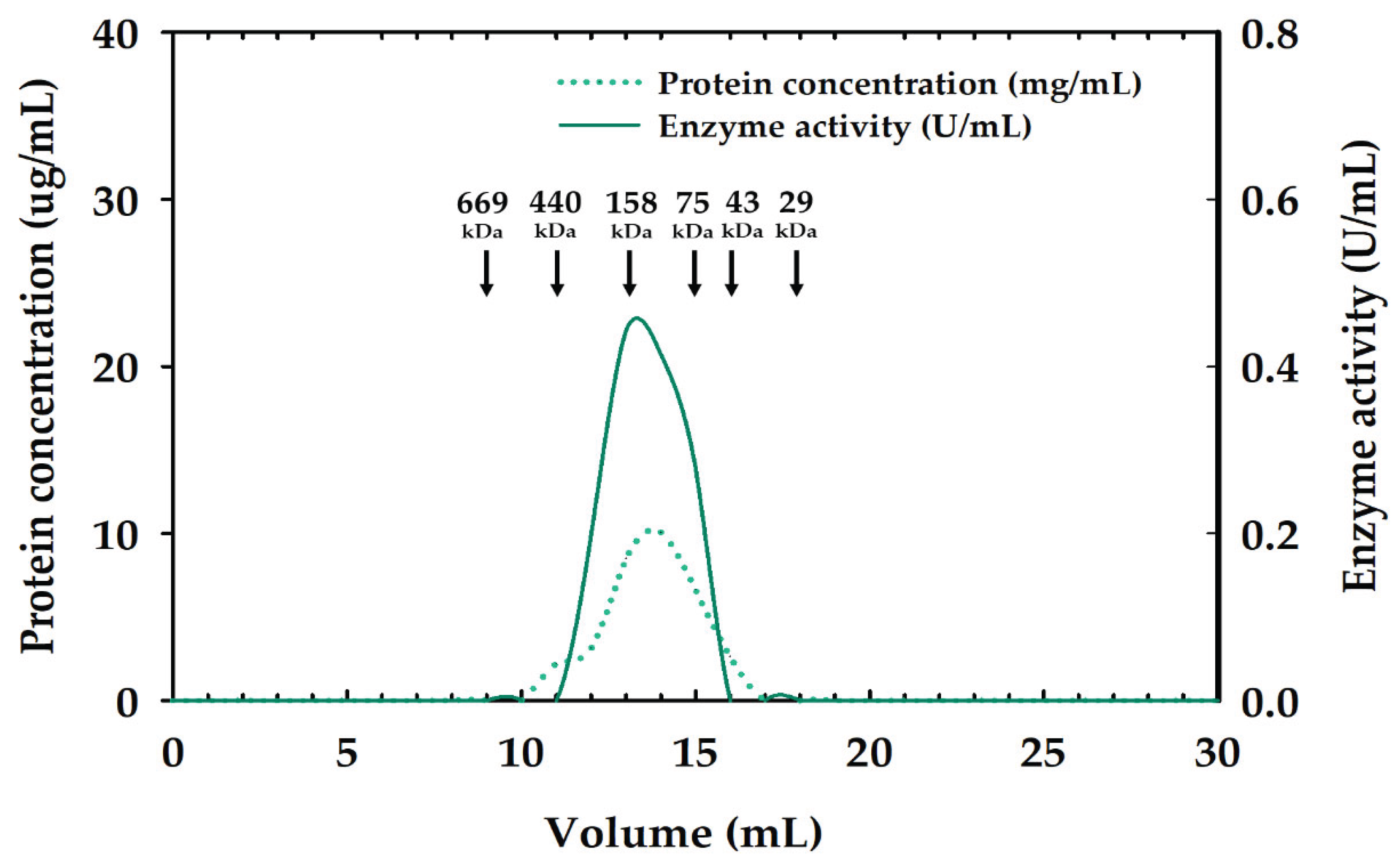
 and Tris-HCl bufffer (
and Tris-HCl bufffer ( ).
).
 and Tris-HCl bufffer (
and Tris-HCl bufffer ( ).
).
| Purification step | Volume (mL) | Protein concentration (mg/mL) | Enzyme activity (U/mL) | Total protein (mg) |
Total activity (U) |
Specific activity (U/mg) | Yield (%) | Purification (fold) | |
| Crude enzyme | 242 | 0.41±0.01 | 3.79±0.11 | 99.28±3.14 | 917.21±27.63 | 9.24±0.28 | 100 | 1 | |
| (NH₄)₂SO₄ precipitation | 45 | 0.74±0.01 | 9.15±0.40 | 33.46±0.44 | 411.84±18.15 | 12.37±0.55 | 44.90 | 1.33 | |
| DEAE sepharose | 1 | 0.16±0.02 | 25.04±3.03 | 0.16±0.02 | 25.04±3.03 | 156.47±18.94 | 2.72 | 17.19 | |
| Substrate | Total activity (U) | Specificity (U/mg) |
| Rye arabinoxylan (RAX) | 25.04±3.03 | 156.47±18.94 |
| Soluble wheat arabinoxylan (sWAX) | 17.65±0.44 | 110.28±2.73 |
| Insoluble wheat arabinoxylan (iWAX) | 5.01±0.41 | 31.31±2.59 |
| Beechwood xylan (BWX) | 10.15±0.27 | 63.42±1.69 |
| Carboxymethyl cellulose (CMC) | 6.59±0.22 | 41.86±1.41 |
| Microcrystalline cellulose (Avicel) | 0.00±0.00 | 0.00±0.00 |
| Compounds | Relative activity (%) of RAXase | Relative activity (%) of BWXase | ||
| 1% | 5% | 1% | 5% | |
| Tween-20 | 108.78±2.66 | 126.96±3.37 | 115.61±1.68 | 130.16±3.62 |
| Tween-80 | 96.34±4.73 | 117.09±6.13 | 117.73±2.22 | 118.86±1.40 |
| Triton X-100 | 83.46±3.83 | 77.00±8.58 | 97.45±1.91 | 94.83±5.49 |
| SDS | 73.45±3.17 | 53.41±0.19 | 71.56±2.33 | 61.48±1.77 |
| 2mM | 5 mM | 2mM | 5 mM | |
| β-mercaptoethanol | 80.23±0.31 | 80.65±1.72 | 107.09±4.10 | 106.78±1.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





