Submitted:
10 September 2025
Posted:
11 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Bacterial Cultivation
2.3. DNA Extraction
2.4. High Throughput WGS Shotgun Sequencing
2.5. WGS Metagenomic Analysis
2.6. WGS Assembly and Analysis
3. Results
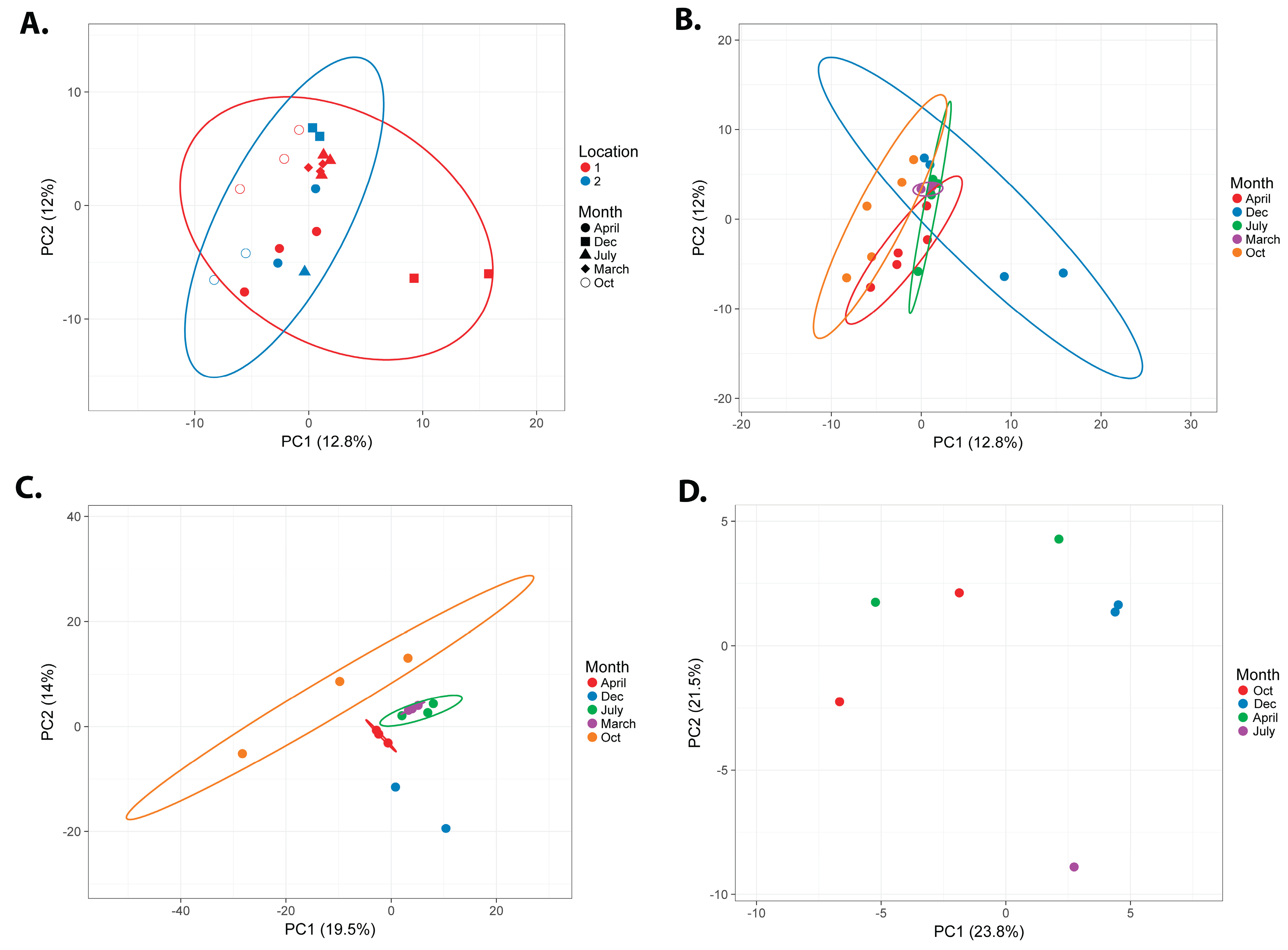
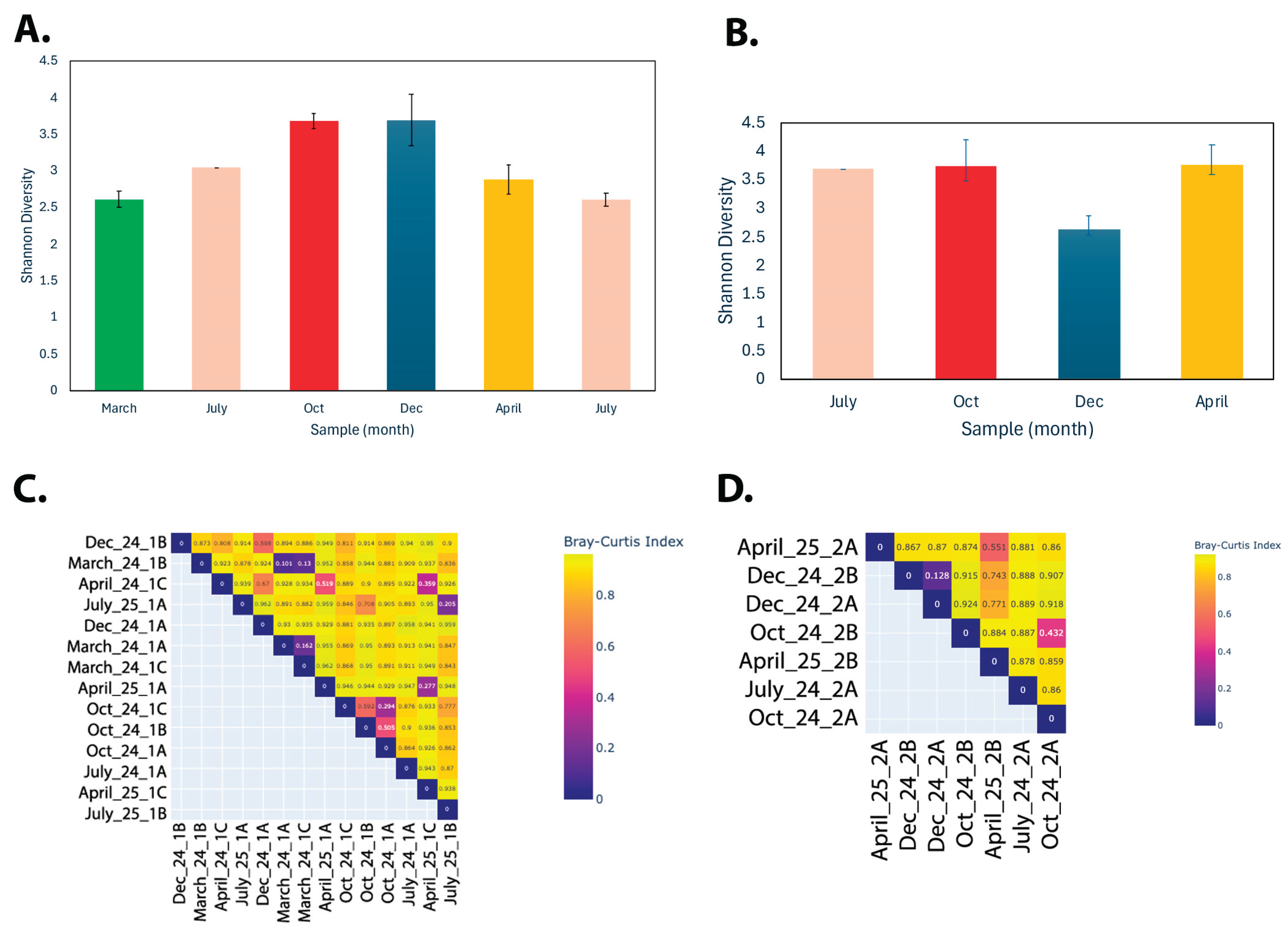
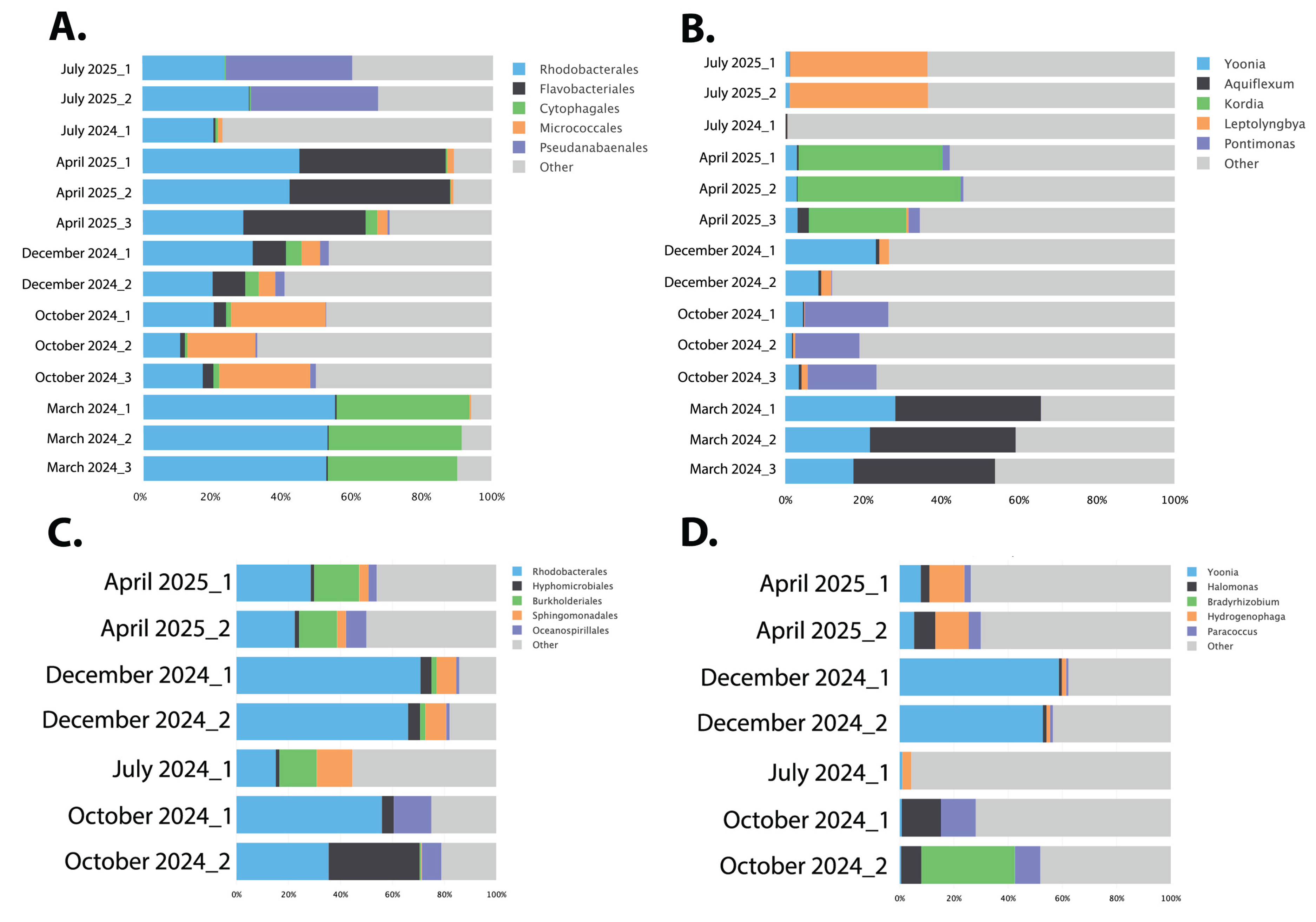
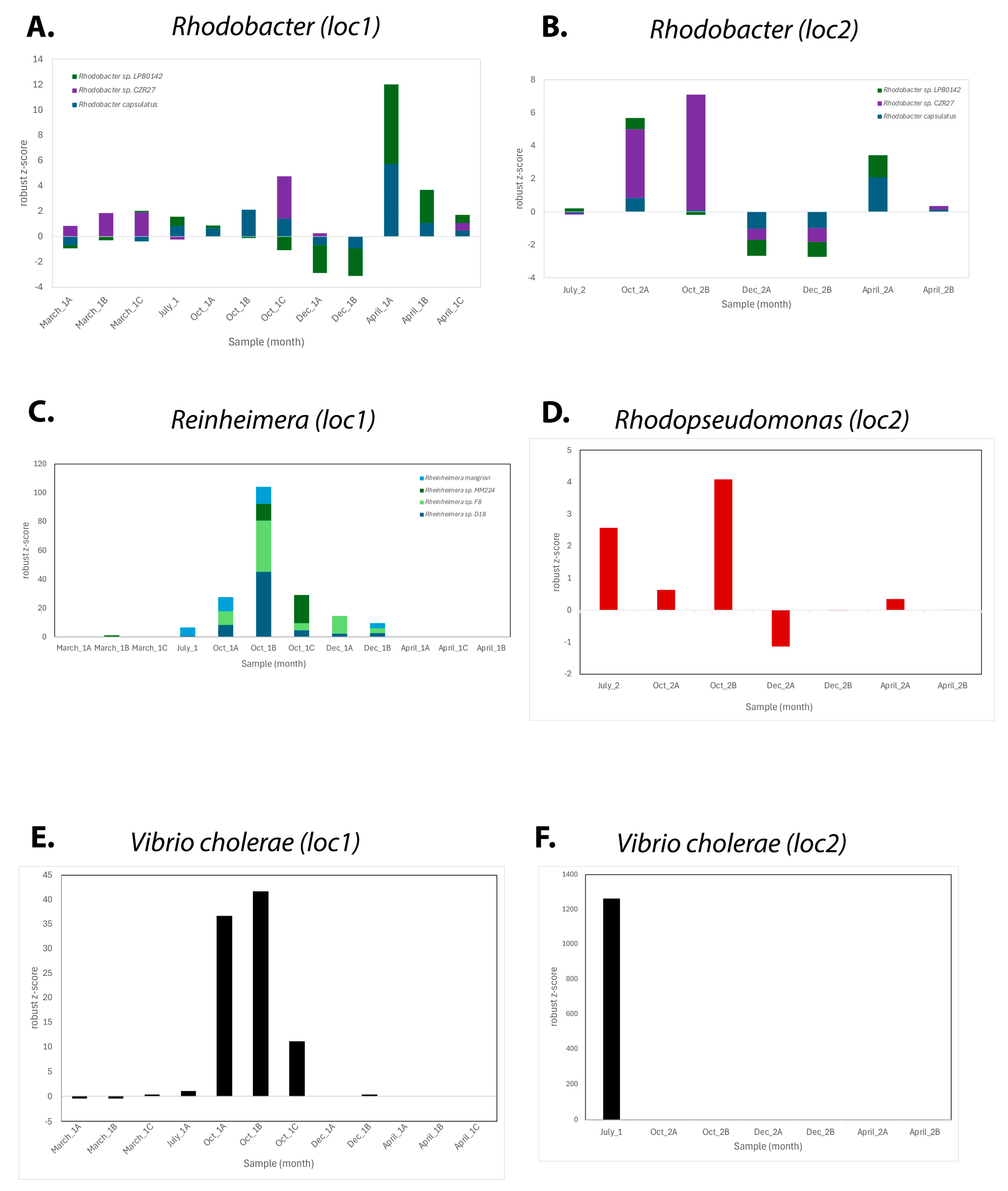
3.1. Seasonal Microbial Variations
3.1.1. Overall Comparison
3.1.2. Microbial Composition Analysis
3.1.3. Antibiotic Resistance Analysis
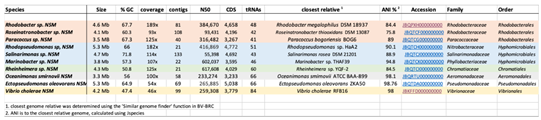
3.2. Isolated Species and Their Initial Characterization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ANI | Average nucleotide identity |
References
- Bhowmik, S. Ecological and economic importance of wetlands and their vulnerability. Research Anthology on Ecosystem Conservation and Preserving Biodiversity 2022, 11–27. [Google Scholar] [CrossRef]
- Xin, P.; Wilson, A.; Shen, C.; Ge, Z.; Moffett, K.B.; Santos, I.R.; et al. Surface water and groundwater interactions in salt marshes and their impact on plant ecology and coastal biogeochemistry. Reviews of Geophysics 2022, 60, e2021RG000740. [Google Scholar] [CrossRef]
- Bertness, M.; Silliman, B. Salt Marshes under siege. American Scientist 2004, 92, 54. [Google Scholar] [CrossRef]
- Campbell, A.D.; Fatoyinbo, L.; Goldberg, L.; et al. Global hotspots of salt marsh change and carbon emissions. Nature 2022, 612, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Joeckel, R.M.; Clement, B.A. Surface features of the Salt Basin of Lancaster County, Nebraska. CATENA 1999, 34, 243–275. [Google Scholar] [CrossRef]
- Johnsgard, P.A. The Nature of Nebraska: Ecology and Biodiversity; The University of Nebraska Press: Lincoln, NE, USA, 2001. [Google Scholar]
- Ungar, I.; Hogan, W.; McClelland, M. Plant communities of saline soils at Lincoln, Nebraska. Am. Midl. Nat. 1969, 82, 564–577. [Google Scholar] [CrossRef]
- Panella, M. Nebraska’s At-Risk Species Wildlife; Nebraska Game & Parks Commission: Lincoln, NE, USA, 2012. [Google Scholar]
- Brosius, T.R.; Higley, L.G. Behavioral niche partitioning in a sympatric tiger beetle assemblage and implications for the endangered Salt Creek tiger beetle. PeerJ 2013, 1, e169. [Google Scholar] [CrossRef]
- Gibbens, S. The remnants of a vast prehistoric sea lie hidden in Nebraska’s endangered marshes. In National Geographic; National Geographic Society: Washington, DC, USA, 2020. [Google Scholar]
- Gao, J.; Liu, M.; Shi, S.; Liu, Y.; Duan, Y.; Lv, X.; Bohu, T.; Li, Y.; Hu, Y.; Wang, N.; Wang, Q.; Zhuang, G.; & Zhuang, X. Disentangling responses of the subsurface microbiome to wetland status and implications for indicating ecosystem functions. Microorganisms 2021, 9, 211. [Google Scholar] [CrossRef]
- Athen, S.R.; Dubey, S.; & Kyndt, J.A. The Eastern Nebraska Salt Marsh Microbiome Is Well Adapted to an Alkaline and Extreme Saline Environment. Life 2021, 11, 446. [Google Scholar] [CrossRef]
- Núñez, A.; García, A.M.; Moreno, D.A.; Guantes, R. Seasonal changes dominate long-term variability of the urban air microbiome across space and time. Environ Int. 2021, 150, 106423. [Google Scholar] [CrossRef]
- Fiore, N.A.; Dunigan, D.D.; Shaffer, J.J.; Roberts, R.; Antony-Babu, S.; Plantz, B.A.; Nickerson, K.W.; Benson, A.K.; Weber, K.A. Microbial Community of Saline, Alkaline Lakes in the Nebraska Sandhills Based on 16S rRNA Gene Amplicon Sequence Data. Microbiol Resour Announc 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Pfennig, N. The phototrophic bacteria and their role in the sulfur cycle. Plant Soil 1975, 43, 1–16. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; Gerdes, S.; Henry, C.S.; Kenyon, R.W.; Machi, D.; Mao, C.; Nordberg, E.K.; Olsen, G.J.; Murphy-Olson, D.E.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Warren, A.; Xia, F.; Yoo, H.; Stevens, R.L. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; & Steinegger, M. Metagenome analysis using the Kraken Software Suite. Nature Protocols 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol 2019, 20, 257. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006, 9, 683–93. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecological Monographs 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Clausen, P.T.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC bioinformatics 2018, 19, 307. [Google Scholar] [CrossRef]
- Alcock, B.P. , et al., CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic acids research 2020, 48, D517–D525. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2014, 25, 1043–1055. [Google Scholar] [CrossRef]
- Aziz, R.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.; Formsma, K.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D. , et al., Mash: fast genome and metagenome distance estimation using MinHash. Genome biology 2016, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrobial agents and chemotherapy 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Lapthorne, S.; McWade, R.; Scanlon, N.; Ní Bhaoill, S.; Page, A.; O'Donnell, C.; Dornikova, G.; Hannan, M.; Lynch, B.; Lynch, M.; Brady, D. Rising clindamycin resistance in group A Streptococcus in an Irish healthcare institution. Access Microbiol. 2024, 6, 000772.v4. [Google Scholar] [CrossRef]
- Heß, S.; Gallert, C. Resistance behaviour of inducible clindamycin-resistant staphylococci from clinical samples and aquatic environments. J Med Microbiol. 2014, 63(Pt 11) Pt 11, 1446–1453. [Google Scholar] [CrossRef]
- Samreen, I.A.; Hesham, A.M.; Hussein, H.A. Environmental antimicrobial resistance and its drivers: a potential threat to public health. J. Global Antimicrobial Resistance 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Assefa, M. Inducible Clindamycin-Resistant Staphylococcus aureus Strains in Africa: A Systematic Review. Int J Microbiol. 2022, 2022, 1835603. [Google Scholar] [CrossRef]
- Arunasri, K.; Venkata Ramana, V.; Spröer, C.; Sasikala, Ch.; Ramana, Ch. V. Rhodobacter megalophilus sp. nov., a phototroph from the Indian Himalayas possessing a wide temperature range for growth. Int J Syst Evol Microbiol 2008, 58, 1792–1796. [Google Scholar] [CrossRef]
- Koh, R.-H.; Song, H.-G. Effects of application of Rhodopseudomonas sp. on seed germination and growth of tomato under axenic conditions. J Microbiol Biotechnol. 2007, 17, 1805–1810. [Google Scholar] [PubMed]
- Xu, J.; Feng, Y.; Wang, Y.; Lin, X. Effect of Rhizobacterium Rhodopseudomonas palustris Inoculation on Stevia rebaudiana Plant Growth and Soil Microbial Community. Pedosphere 2018, 28, 793–803. [Google Scholar] [CrossRef]
- Hsu, S.-H.; Shen, M-W.; Chen, J-C.; et al. The Photosynthetic Bacterium Rhodopseudomonas palustris Strain PS3 Exerts Plant Growth-Promoting Effects by Stimulating Nitrogen Uptake and Elevating Auxin Levels in Expanding Leaves. Front Plant Sci. 2021, 12, 573634. [Google Scholar] [CrossRef] [PubMed]
- Batool, K.; Tuz Zahra, F.; Rehman, Y. Arsenic-Redox Transformation and Plant Growth Promotion by Purple Nonsulfur Bacteria Rhodopseudomonas palustris CS2 and Rhodopseudomonas faecalis SS5. Biomed Res Int 2017, 6250327. [Google Scholar] [CrossRef]
- Kantha, T.; Kantachote, D.; Klongdee, N. Potential of biofertilizers from selected Rhodopseudomonas palustris strains to assist rice (Oryza sativa L. subsp. indica) growth under salt stress and to reduce greenhouse gas emissions. Ann Microbiol. 2015, 65, 2109–2118. [Google Scholar] [CrossRef]
- Kantachote, D.; Nunkaew, T.; Kantha, T.; Chaiprapat, S. Biofertilizers from Rhodopseudomonas palustris strains to enhance rice yields and reduce methane emissions. Applied Soil Ecology 2016, 100, 154–161. [Google Scholar] [CrossRef]
- Hobbs, A.; Ochoa-Rojas, D.; Humphrey, C.E.; Kyndt, J.A.; Moore, T.C. Soil microbiome perturbation impedes growth of Bouteloua curtipendula and increases relative abundance of soil microbial pathogens. PLOS One bioRxiv 2025. [Google Scholar] [CrossRef]
- Oda, Y.; Larimer, F.W.; Chain, P.S.; Malfatti, S.; Shin, M.V.; Vergez, L.M.; Hauser, L.; Land, M.L.; Braatsch, S.; Beatty, J.T.; Pelletier, D.A.; Schaefer, A.L.; Harwood, C.S. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci U S A. 2008, 105, 18543–8. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Meyer, T.E.; Kyndt, J. Genomic and genetic sequence information of strains assigned to the genus Rhodopseudomonas reveal the great heterogeneity of the group and identify strain Rhodopseudomonas palustris DSM 123T as the authentic type strain of this species. Int J Syst Evol Microbiol. 2020, 70, 3932–3938. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Kyndt, J.A.; Meyer, T.E. Genomic Comparison, Phylogeny and Taxonomic Reevaluation of the Ectothiorhodospiraceae and Description of Halorhodospiraceae fam. nov. and Halochlorospira gen. nov. Microorganisms 2022, 10, 295. [Google Scholar] [CrossRef]
- Mao, C.; Abraham, D.; Wattam, A.R.; Wilson, M.J.; Shukla, M.; Yoo, H.S.; Sobral, B.W. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 2015, 31, 252–258. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Kyndt, J.A. Vibrio cholerae genome isolated from the Nebraska salt marshes contains several antibiotic resistance markers. Microbiol Resour Announc. [CrossRef]
- Stal, L.J.; Gemerden, H.; Krumbein, W.E. Structure and development of a benthic marine microbial mat. FEMS Microbiol. Lett. 1985, 31, 111–125. [Google Scholar] [CrossRef]
- Bolhuis, H.; Stal, L.J. Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. ISME J. 2011, 5, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Zhe, Z.; Yuxiang, Z.; Federico, M.; Maripat, X.; Huan, L.; Junquan, X.; Baolan, H.; Min, W. The microbial community structure and nitrogen cycle of high-altitude pristine saline lakes on the Qinghai-Tibetan plateau. Frontiers in Microbiology, 2024, 15, DOI–10.3389. [Google Scholar] [CrossRef]
- Imhoff, J.F. Rhodobacter. In Bergey's Manual of Systematics of Archaea and Bacteria (eds M.E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, F.A. Rainey and W.B. Whitman). 2015. [Google Scholar] [CrossRef]
- Boldareva, E.N.; Moskalenko, A.A.; Makhneva, Z.K.; Tourova, T.P.; Kolganova, T.V.; Gorlenko, V.M. Rubribacterium polymorphum gen. nov., sp. nov., a novel alkaliphilic nonsulfur purple bacterium from an Eastern Siberian soda lake. Microbiology 2009, 78, 732–740. [Google Scholar] [CrossRef]
- Milford, A.D.; Achenbach, L.A.; Jung, D.O.; Madigan, M.T. Rhodobaca bogoriensis gen. nov. and sp. nov. alcaliphilic purple nonsulfur bacterium from African Rift valley soda lakes. Arch. Microbiol. 2000, 174, 18–27. [Google Scholar] [CrossRef]
- Boldareva, E.N.; Akimov, V.N.; Boychenko, V.A.; Stadnichuk, I.N.; Moskalenko, A.A.; Makhneva, Z.K.; Gorlenko, V.M. Rhodobaca barguzinensis sp. nov., a new alkaliphilic purple sulfur bacterium isolated from a soda lake of the Barguzin valley (Buryat Republic,Eastern Siberia). Microbiology 2008, 77, 206–218. [Google Scholar] [CrossRef]
- Jang, G.I.; Cho, Y.; Cho, B.C. Pontimonas salivibrio gen. nov., sp. nov., a new member of the family Microbacteriaceae isolated from a seawater reservoir of a solar saltern. Int J Syst Evol Microbiol. 2013, 63 Pt 6, 2124–2131. [Google Scholar] [CrossRef]
- Vreeland, R.H. 2015. "Halomonas". In Whitman WB (ed.). Bergey's Manual of Systematics of Archaea and Bacteria. pp. 1–19. [CrossRef]
- Peng, Z.; Liu, G.; Huang, K. Cold Adaptation Mechanisms of a Snow Alga Chlamydomonas nivalis During Temperature Fluctuations. Frontiers in Microbiology 2021, 11. [Google Scholar] [CrossRef]
- Morales-Sánchez, D.; Schulze, P.S.C.; Kiron, V.; Wijffels, R.H. Temperature-Dependent Lipid Accumulation in the Polar Marine Microalga Chlamydomonas malina RCC2488. Front Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Nelson, A.R.; Lopez, A.L.; Sack, D.A. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.A.; Vidal, R.M.; Velasco, J.; George, S.; Lucero, Y.; Gómez, L.A.; Carreño, L.J.; García-Betancourt, R.; O'Ryan, M. Vibrio cholerae, classification, pathogenesis, immune response, and trends in vaccine development. Front Med (Lausanne). 2023, 10, 1155751. [Google Scholar] [CrossRef] [PubMed]
- Grimes, D.J. Ecology of estuarine bacteria capable of causing human disease: A review. Estuaries 1991, 14, 345–360. [Google Scholar] [CrossRef]
- Brumfield, K.D.; Chen, A.J.; Gangwar, M.; Usmani, M.; Hasan, N.A.; Jutla, A.S.; Huq, A.; Colwell, R.R. Environmental Factors Influencing Occurrence of Vibrio parahaemolyticus and Vibrio vulnificus. Appl Environ Microbiol 2023, 89, e00307–23. [Google Scholar] [CrossRef]
- Shen, S.; Wu, W.; Grimes, D.J.; Saillant, E.A.; Griffitt, R.J. Community composition and antibiotic resistance of bacteria in bottlenose dolphins Tursiops truncatus – Potential impact of 2010 BP Oil Spill. Science of The Total Environment 2020, 732, 139125. [Google Scholar] [CrossRef]
- Pendergraft, M.A.; Grimes, D.J.; Giddings, S.N.; Feddersen, F.; Beall, C.M.; Lee, C.; Santander, M.V.; Prather, K.A. Airborne transmission pathway for coastal water pollution. PeerJ 11352021, 9, e11358. [Google Scholar] [CrossRef]
- Rouard, C.; Collet, L.; Njamkepo, E.; Jenkins, C.; Sacheli, R.; Benoit-Cattin, T.; Figoni, J.; Weill, F.X. Long-Distance Spread of a Highly Drug-Resistant Epidemic Cholera Strain. New England Journal of Medicine 2024, 391, 2271–2273. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, CF. Antibiotic resistance in the environment. Nat Rev Microbiol 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br J Biomed Sci 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Ifedinezi, O.V.; Nnaji, N.D.; Anumudu, C.K.; Ekwueme, C.T.; Uhegwu, C.C.; Ihenetu, F.C.; Obioha, P.; Simon, B.O.; Ezechukwu, P.S.; Onyeaka, H. Environmental Antimicrobial Resistance: Implications for Food Safety and Public Health. Antibiotics 2024, 13, 1087. [Google Scholar] [CrossRef] [PubMed]
- Saxena, D.; Gwalani, R.; Yadav, A.; Shah, R. Growing Concerns on Antimicrobial Resistance - Past, Present, and Future Trends. Indian J Community Med. 2025, 50, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.W.; Tan, T.M.; Poh, C.L. Genetic determinants of tetracycline resistance in Vibrio harveyi. Antimicrob Agents Chemother 2002, 46, 1038–45. [Google Scholar] [CrossRef]
- Faruque, A.S.; Alam, K.; Malek, M.A.; Khan, M.G.; Ahmed, S.; Saha, D.; Khan, W.A.; Nair, G.B.; Salam, M.A.; Luby, S.P.; Sack, D.A. Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J Health Popul Nutr. 2007, 25, 241–3. [Google Scholar]
- Kar, S.K.; Pal, B.B.; Khuntia, H.K.; Achary, K.G.; Khuntia, C.P. Emergence and spread of tetracycline resistant Vibrio cholerae O1 El Tor variant during 2010 cholera epidemic in the tribal areas of Odisha, India. Int J Infect Dis. 2015, 33, 45–49. [Google Scholar] [CrossRef]
- Ahmadi, M.H. Global status of tetracycline resistance among clinical isolates of Vibrio cholerae: a systematic review and meta-analysis. Antimicrob Resist Infect Control 2021, 10, 115. [Google Scholar] [CrossRef]
- Malmstrom, T. Saline Wetlands Conservation Partnership 2018 Progress Report; Lincoln Parks and Recreation Department: Lincoln, NE, USA, 2019. [Google Scholar]
- Lynum, C.A.; Bulseco, A.N.; Dunphy, C.M.; Osborne, S.M.; Vineis, J.H.; Bowen, J.L. Microbial community response to a passive salt marsh restoration. Estuaries Coasts 2020, 43, 1439–1455. [Google Scholar] [CrossRef]
- Iqbal, S.; Begum, F.; Nguchu, B.A.; Claver, U.P.; Shaw, P. The invisible architects: microbial communities and their transformative role in soil health and global climate changes. Environ Microbiome 2025, 20, 36. [Google Scholar] [CrossRef]
- Wang, X.; Chi, Y.; Song, S. Important soil microbiota's effects on plants and soils: a comprehensive 30-year systematic literature review. Front Microbiol. 2024, 15, 1347745. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Bianchelli, S.; Candela, M.; Dell’Anno, A.; Gambi, C.; Rastelli, E.; Varrella, S.; Danovaro, R. Microbiome-assisted restoration of degraded marine habitats: a new nature-based solution? Front. Mar. Sci. 2023, 10, 1227560. [Google Scholar] [CrossRef]
- Farrer, E.C.; Van Bael, S.A.; Clay, K.; et al. Plant-Microbial Symbioses in Coastal Systems: Their Ecological Importance and Role in Coastal Restoration. Estuaries and Coasts 2022, 45, 1805–1822. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Voolstra, C.R.; Sweet, M.; et al. Harnessing the microbiome to prevent global biodiversity loss. Nat Microbiol 2022, 7, 1726–1735. [Google Scholar] [CrossRef]

 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





