Submitted:
03 September 2025
Posted:
03 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RAD-Sequencing and SNP Statistics
2.3. Genetic Map Construction and Quality Evaluation
2.4. QTL Mapping and Screening of Candidate Genes
3. Results
3.1. RAD Sequencing Data Analysis
3.2. Genetic Map Construction
3.3. Quality Evaluation of the Genetic Map
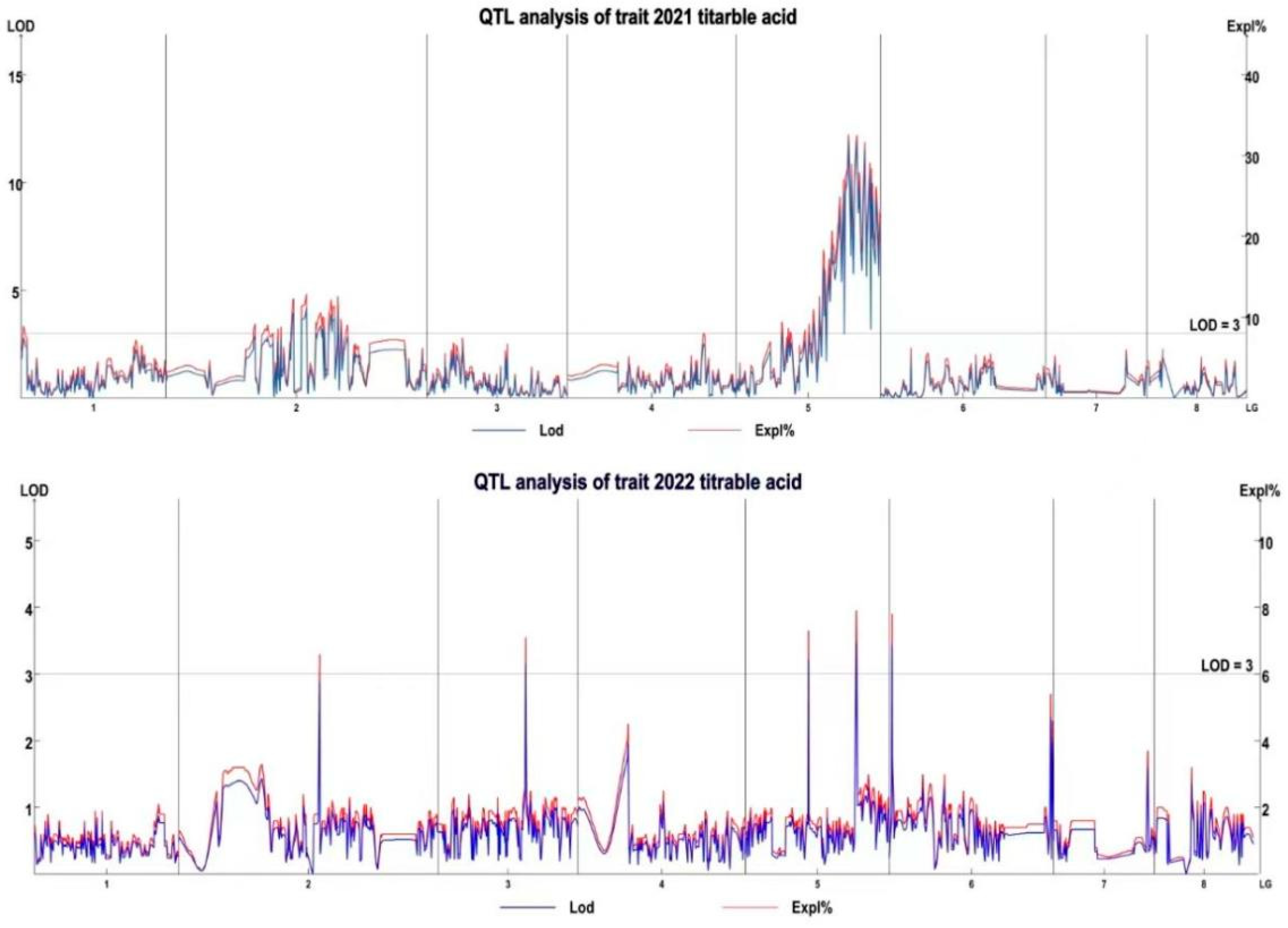
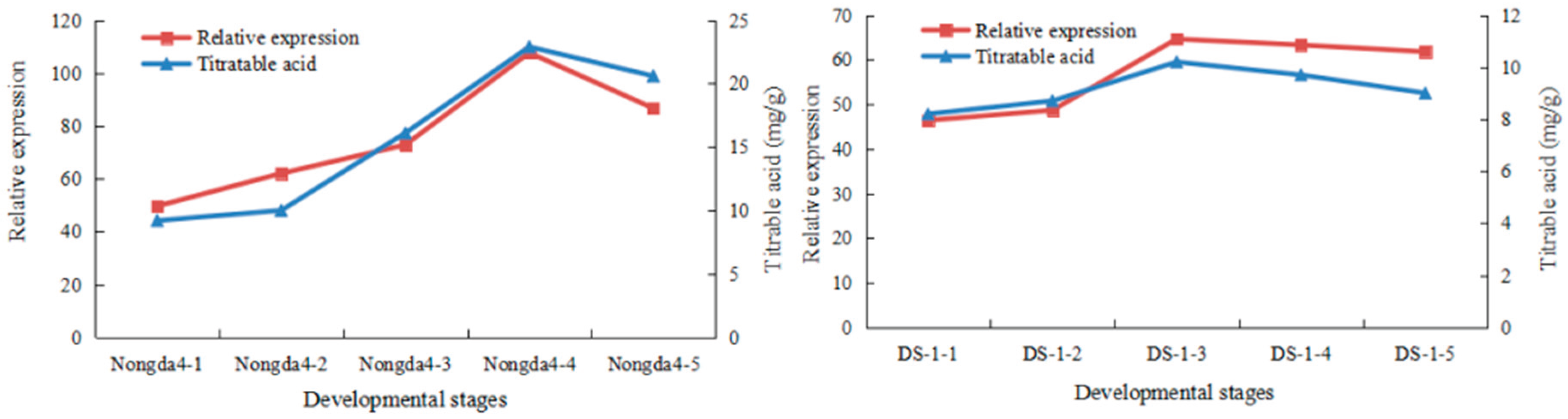
3.4. QTL Mapping and Candidate Gene Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RAD-seq | restriction site–associated DNA sequencing |
| SNP | single nucleotide polymorphism |
| QTL | quantitative trait locus |
| LOD | logarithm of the odds |
| PVE | phenotypic variation explained |
References
- Mo, C.; Li, W.D.; He, Y.X.; Ye, L.Q.; Zhang, Z.S.; Jin, J.S. Variability in the sugar and organic acid composition of the fruit of 57 genotypes of Chinese Dwarf Cherry [Cerasus humilis (Bge.) Sok]. J Hortic Sci Biotechnol. 2015, 90(4), 419–426. [Google Scholar] [CrossRef]
- Ren, J.; Sun, L.N.; Zhang, Q.Y.; Song, X.S. Drought tolerance is correlated with the activity of antioxidant enzymes in Cerasus humilis seedlings. Biomed Res. Int. 2016, 9851095. [Google Scholar] [CrossRef]
- Yang, R.; Yang, Y.; Hu, Y.; Yin, L.; Qu, P.; Wang, P.; Mu, X.; Zhang, S.; Xie, P.; Cheng, C.; et al. Comparison of bioactive compounds and antioxidant activities in differentially pigmented Cerasus humilis fruits. Molecules 2023, 28, 6272. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.Q.; Yang, C.X.; Li, W.D.; Hao, J.B.; Sun, M.; Zhang, J.R.; Zhang, Z.S. Evaluation of volatile compounds from Chinese Dwarf Cherry (Cerasus humilis (Bge.) Sok.) germplasms by headspace solid-phase micro-extraction and gas chromatography-mass spectrometry. Food Chem. 2017, 217, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.P.; Aryal, N.; Du, J.M.; Du, J.J. Oil content and fatty acid composition of the kernels of 31 different cultivars of Chinese dwarf cherry [Cerasus humilis (Bge.) Sok]. J Hortic Sci Biotechnol. 2015, 90, 525–529. [Google Scholar] [CrossRef]
- Li, W.D.; Li, O.; Mo, C.; Jiang, Y.S.; He, Y.; Zhang, A.R.; Chen, L.M.; Jin, J.S. Mineral element composition of 27 Chinese dwarf cherry [Cerasus humilis (Bge.) Sok.] genotypes collected in China. J Hortic Sci Biotechnol. 2014, 89(6), 674–678. [Google Scholar] [CrossRef]
- Ji, X.L.; Zhang, M.Y.; Wang, D.; Li, Z.; Lang, S.Y.; Song, X.S. Genome-wide identification of WD40 superfamily in Cerasus humilis and functional characteristics of ChTTG1. Int J Biol Macromol. 2023, 225, 376–388. [Google Scholar] [CrossRef]
- Guo, C.Z.; Wang, P.F.; Zhang, J.C.; Guo, X.; Mu, X.P.; Du, J.J. Organic acid metabolism in Chinese dwarf cherry [Cerasus humilis (Bge.) Sok.] is controlled by a complex gene regulatory network. Front. Plant Sci. 2022, 13, 982112. [Google Scholar] [CrossRef]
- Mohd Shaha, F.R.; Liew, P.L.; Zaman, F.Q.; Nulit, R.; Barin, J.; Rolland, J.; Yong, H.Y.; Boon, S.H. Genotyping by sequencing for the construction of oil palm (Elaeis guineensis Jacq.) genetic linkage map and mapping of yield related quantitative trait loci. PeerJ. 2024, 12, e16570. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Z.; Qian, L.; Hang, S.; Niu, Y.; Shen, C.; Wei, Y.; Liu, B. Genetic mapping and validation of QTL controlling fruit diameter in cucumber. BMC Plant Biol. 2024, 24(1), 1271. [Google Scholar] [CrossRef]
- Kumar, R.; Das, S.P.; Choudhury, B.U.; Kumar, A.; Prakash, N.R.; Verma, R.; Chakraborti, M.; Devi, A.G.; Bhattacharjee, B.; Das, R.; et al. Advances in genomic tools for plant breeding: Harnessing DNA molecular markers, genomic selection, and genome editing. Biol Res. 2024, 57(1), 80. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12(7), 499–510. [Google Scholar] [CrossRef]
- Antanaviciute, L.; Fernández-Fernández, F.; Jansen, J.; Banchi, E.; Evans, K.M.; Viola, R.; Velasco, R.; Dunwell, J.M.; Troggio, M.; Sargent, D.J. Development of a dense SNP-based linkage map of an apple rootstock progeny using the Malus Infinium whole genome genotyping array. BMC Genomics. 2012, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Su, K. , Guo, Y. S., Zhong, W. H., Lin, H., Liu, Z. D., Li, K., Li, Y. Y., & Guo, X. W. High-density genetic linkage map construction and white rot resistance quantitative trait loci mapping for genus Vitis based on restriction site-associated DNA sequencing. Phytopathology. 2021, 111(4), 659–670. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.H.; Feng, Y.; Chen, Q.; Zhao, F.K.; Zhou, S.J.; Ding, Y.; Song, X.L.; Li, P.; Wang, B.H. Genome-wide association analysis of salt tolerance QTLs with SNP markers in maize (Zea mays L.). Genes Genom. 2019, 41(10), 1135–1145. [Google Scholar] [CrossRef]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.G.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18(1), 324. [Google Scholar] [CrossRef]
- Barchi, L.; Lanteri, S.; Portis, E.; Valè, G.; Volante, A.; Pulcini, L.; Ciriaci, T.; Acciarri, N.; Barbierato, V.; Toppino, L.; et al. A RAD tag-derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS ONE. 2012, 7(8), e43740. [Google Scholar] [CrossRef]
- Jia, J.Z.; Zhao, S.C.; Kong, X.Y.; Li, Y.R.; Zhao, G.Y.; He, W.M.; Appels, R.; Pfeifer, M.; Tao, Y. ; Zhang, X; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013, 496, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Arce, N.; Rodríguez-Ezpeleta, N. Selecting RAD-seq data analysis parameters for population genetics: The more the better? Front Genet. 2019, 10, 533. [Google Scholar] [CrossRef]
- Monforte, A.J.; Diaz, A.; Caño-Delgado, A.; van der Knaap, E. The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J Exp Bot. 2014, 65, 4625–4637. [Google Scholar] [CrossRef]
- Argyris, J.M.; Díaz, A.; Ruggieri, V.; Fernández, M.; Jahrmann, T.; Gibon, Y.; Picó, B.; Martín-Hernández, A.M.; Monforte, A.J.; Garcia-Mas, J. QTL analyses in multiple populations employed for the fine mapping and identification of candidate genes at a locus affecting sugar accumulation in melon (Cucumis melo L.). Front Plant Sci. 2017, 8, 1679. [Google Scholar] [CrossRef]
- Zeng, Y.L.; Wang, M.Y.; Hunter, D.C.; Matich, A.J.; McAtee, P.A.; Knäbel, M.; Hamiaux, C.; Popowski, E.A.; Jaeger, S.R.; Nieuwenhuizen, N.J.; et al. Sensory-directed genetic and biochemical characterization of volatile terpene production in kiwifruit. Plant Physiol. 2020, 183(1), 51–66. [Google Scholar] [CrossRef]
- Diaz-Garcia, L. , Schlautman, B., Covarrubias-Pazaran, G., Maule, A., Johnson-Cicalese, J., Grygleski, E., Vorsa, N., & Zalapa, J. Massive phenotyping of multiple cranberry populations reveals novel QTLs for fruit anthocyanin content and other important chemical traits. Mol Genet Genomics. 2018, 293(6), 1379–1392. [Google Scholar] [CrossRef]
- Tang, H.X. , Pei, G.Y., Zhang, Q., Wang, Z.T. QTL mapping analysis of jujube fruit-related traits. Acta Hortic Sin. 2023, 50(4), 754–764. [Google Scholar] [CrossRef]
- Jiang, F.C. , Yang, L., Zhang, J.H., Zhang, M.L., Yu, W.J., Sun, H.Y. QTL mapping and screening of major-effect genes regulating organic acid accumulation in apricot fruit. Acta Hortic Sin. 2025, 52(4), 846–856. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kimura, T.; Shoda, M.; Imai, T.; Saito, T.; Sawamura, Y.; Kotobuki, K.; Hayashi, T.; Matsuta, N. Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet. 2002, 106, 9–18. [Google Scholar] [CrossRef]
- Gangadhara Rao, P.; Behera, T.K.; Gaikwad, A.B.; Munshi, A.D.; Srivastava, A.; Boopalakrishnan, G.; Vinod. Genetic analysis and QTL mapping of yield and fruit traits in bitter gourd (Momordica charantia L.). Sci Rep. 2021, 11(1), 4109. [Google Scholar] [CrossRef]
- Qin, M.F.; Li, L.T.; Singh, J.; Sun, M.Y.; Bai, B.; Li, S.W.; Ni, J.P.; Zhang, J.Y.; Zhang, X.; Wei, W.L.; et al. Construction of a high-density bin-map and identification of fruit quality-related quantitative trait loci and functional genes in pear. Hortic Res. 2022, 9, uhac141. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Xu, Z.; Zhang, S.Y.; Wang, X.J.; Ma, X.F.; Zheng, J.C.; Xing, L.B.; Zhang, D.; Ma, J.J.; Han, M.Y.; Zhao, C.P. Construction of a high-density SNP-based genetic map and identification of fruit-related QTLs and candidate genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2020, 20(1), 438. [Google Scholar] [CrossRef] [PubMed]
- Battistoni, B.; Salazar, J.; Vega, W.; Valderrama-Soto, D.; Jiménez-Muñoz, P.; Sepúlveda-González, A.; Ahumada, S.; Cho, I.; Gardana, C.S.; Morales, H. An upgraded, highly-saturated linkage map of Japanese plum (Prunus salicina Lindl.), and identification of a new major locus controlling the flavan-3-ol composition in fruits. Front Plant Sci. 2022, 13, 805744. [Google Scholar] [CrossRef]
- Oh, S.; Ahn, S.; Han, H.; Kim, K.; Kim, S.A.; Kim, D. Genetic linkage maps and QTLs associated with fruit skin color and acidity in apple (Malus×domestica). Hortic. Environ. Biotechnol. 2023, 64(2), 299–310. [Google Scholar] [CrossRef]
- Wang, P.F.; Yi, S.K.; Mu, X.P.; Zhang, J.C.; Du, J.J. Chromosome-level genome assembly of Cerasus humilis Using PacBio and Hi-C Technologies. Front. Genet. 2020, 11, 956. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.H.; Zhou, X. Statistical methods for SNP heritability estimation and partition: A review. Comput Struct Biotechnol J. 2020, 18, 1557–1568. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kimura, T.; Shoda, M.; Imai, T.; Saito, T.; Sawamura, Y.; Kotobuki, K.; Hayashi, T.; Matsuta, N. Genetic linkage maps constructed by using an interspecific cross between Japanese and European pears. Theor Appl Genet. 2002, 106(1), 9–18. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and genotyping loci de novo from short-read sequences. G3 (Bethesda). 2011, 1(3), 171–182. [Google Scholar] [CrossRef]
- Rastas, P.; Paulin, L.; Hanski, I.; Lehtonen, R.; Auvinen, P. Lep-MAP: Fast and accurate linkage map construction for large SNP datasets. Bioinformatics. 2013, 29(24), 3128–3134. [Google Scholar] [CrossRef]
- Guo, C.Z. Genetic analysis of fruit organic acids, screening and functional verification of key organic acid metabolism regulating genes in Chinese dwarf cherry. Ph.D. Dissertation, Shanxi Agricultural University, Shanxi, 2023. [Google Scholar]
- Jiang, Y.M.; Dong, L.; Li, H.Q.; Liu, Y.N.; Wang, X.D.; Liu, G.Q. Genetic linkage map construction and QTL analysis for plant height in proso millet (Panicum miliaceum L.). Theor Appl Genet. 2024, 137(4), 78. [Google Scholar] [CrossRef]
- Shi, P.; Xu, Z.; Zhang, S.Y.; Wang, X.J.; Ma, X.F.; Zheng, J.C.; Xing, L.B.; Zhang, D.; Ma, J.J.; Han, M.Y.; Zhao, C.P. Construction of a high-density SNP-based genetic map and identification of fruit-related QTLs and candidate genes in peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2020, 20(1), 438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Georgi, L.L.; Reighard, G.L.; Scorza, R.; Abbott, A.G. Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch]. J Hered. 2002, 93(5), 352–358. [Google Scholar] [CrossRef]
- Wang, Y.; Georgi, L.L.; Zhebentyayeva, T.N.; Reighard, G.L.; Scorza, R.; Abbott, A.G. High-throughput targeted SSR marker development in peach (Prunus persica). Genome 2002, 45(2), 319–328. [Google Scholar] [CrossRef]
- Sun, R. , Chang, Y., Yang, F., Wang, Y., Li, H., Zhao, Y., Chen, D., Wu, T., Zhang, X., & Han, Z. A dense SNP genetic map constructed using restriction site-associated DNA sequencing enables detection of QTLs controlling apple fruit quality. BMC Genom. 2015, 16(1), 747. [Google Scholar] [CrossRef]
- Li, H. , Chen, A., Tang, H., & Luan, M. High-density genetic map construction and QTL analysis of the first flower node in kenaf using RAD-seq. BMC Plant Biol. 2024, 24(1), 1191. [Google Scholar] [CrossRef]
- Wang, D.S. , Cheng, B.B., & Zhang, J.J. High-density genetic map and quantitative trait loci map of skin color in hawthorn (Crataegus pinnatifida bge. Var. major N.E.Br.). Front Genet. 2024, 15, 1405604. [Google Scholar] [CrossRef] [PubMed]
- Powder, K.E. Quantitative Trait Loci (QTL) mapping. In: Methods in Molecular Biology (Ed.) Clifton, N.J. 2020, pp. 211–229. [CrossRef]
- Rawandoozi, Z.J. , Hartmann, T.P., Carpenedo, S., Gasic, K., Linge, C.d.S., Cai, L., Van de Weg, E., Byrne, D.H. Identification and characterization of QTLs for fruit quality traits in peach through a multi-family approach. BMC Genom. 2020, 21(1), 522. [Google Scholar] [CrossRef] [PubMed]
- Dondini, L. , Domenichini, C., Dong, Y., Gennari, F., Bassi, D., Foschi, S., Lama, M., Adami, M., De Franceschi, P., Cervellati, C., Bergonzoni, L., Alessandri, S., Tartarini, S. Quantitative trait loci mapping and identification of candidate genes linked to fruit acidity in apricot (Prunus armeniaca L.). Front Plant Sci. 2022, 13, 838370. [Google Scholar] [CrossRef]
- Sun, R.; Chang, Y.; Yang, F.; Wang, Y.; Li, H.; Zhao, Y.; Chen, D.; Wu, T.; Zhang, X.; Han, Z. A dense SNP genetic map constructed using restriction site-associated DNA sequencing enables detection of QTLs controlling apple fruit quality. BMC Genom. 2015, 16, 747. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.J.; You, Q.B.; Duan, G.Z.; Ren, J.J.; Chu, S.S.; Zhao, J.Q.; Li, X.; Zhou, X.N.; & Jiao, Y.Q. Quantitative trait loci analysis of seed oil content and composition of wild and cultivated soybean. BMC Plant Biol. 2020, 20, 51. [Google Scholar] [CrossRef]
- Wu, S.; Li, M.; Zhang, C.; Tan, Q.; Yang, X.; Sun, X.; Pan, Z.; Deng, X.; Hu, C. Effects of phosphorus on fruit soluble sugar and citric acid accumulations in citrus. Plant Physiol Biochem. 2021, 160, 73–81. [Google Scholar] [CrossRef]
- Dong, B.; Meng, D.; Song, Z.; Cao, H.; Du, T.; Qi, M.; Wang, S.; Xue, J.; Yang, Q.; Fu, Y. CcNFYB3-CcMATE35 and LncRNA CcLTCS-CcCS modules jointly regulate the efflux and synthesis of citrate to enhance aluminium tolerance in pigeon pea. Plant Biotechnol J. 2024, 22, 181–199. [Google Scholar] [CrossRef]
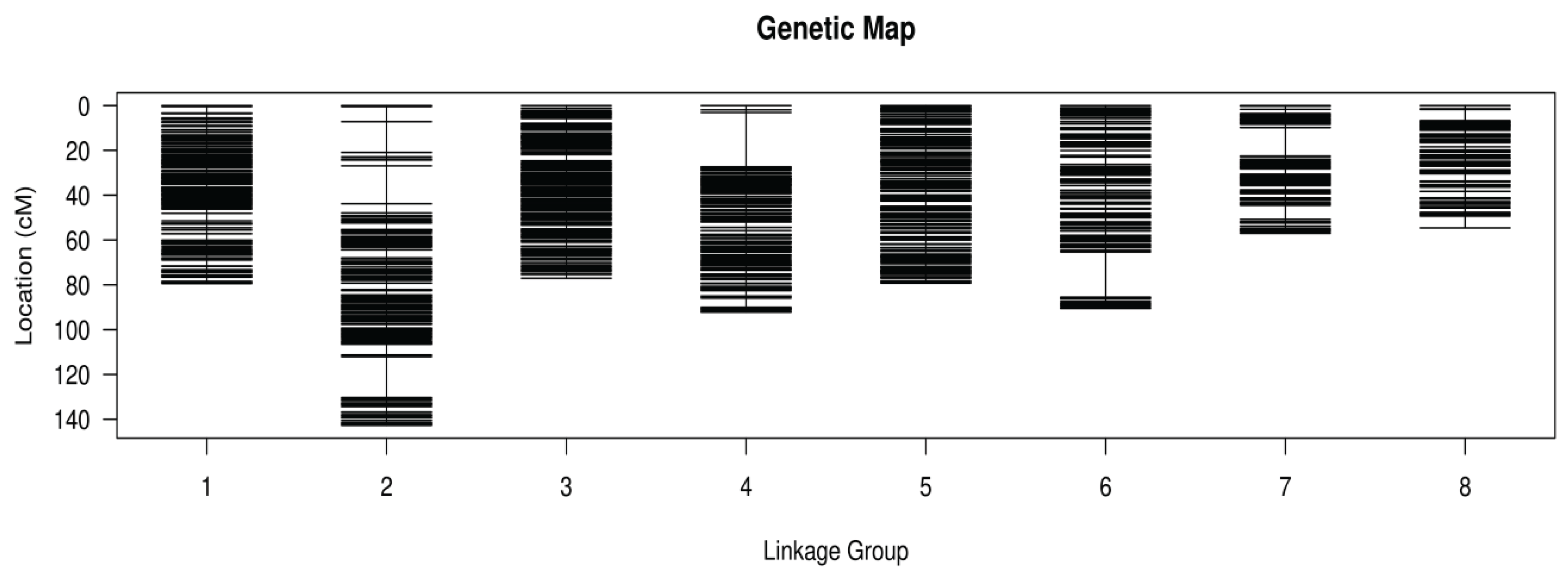
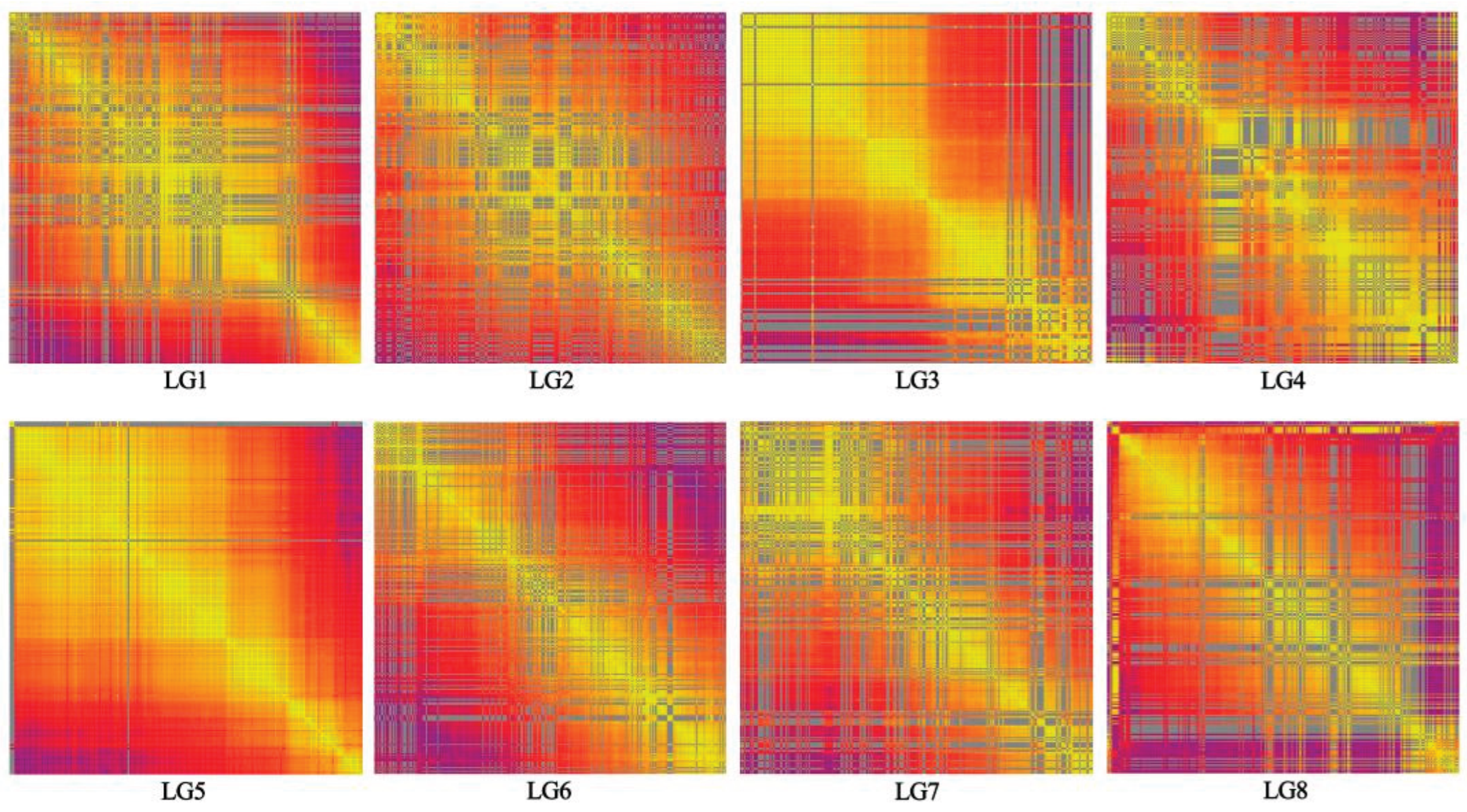
| Linkage group | SNP number | Total distance (cM) | Average distance (cM) | Max Gap (cM) | Gaps <5 cM (%) |
|---|---|---|---|---|---|
| LG1 | 465 | 79.45 | 0.17 | 3.35 | 100 |
| LG2 | 389 | 142.71 | 0.37 | 18.23 | 98.97 |
| LG3 | 384 | 77.04 | 0.2 | 2.87 | 100 |
| LG4 | 333 | 92.18 | 0.28 | 24.22 | 99.7 |
| LG5 | 305 | 79.2 | 0.26 | 2.39 | 100 |
| LG6 | 265 | 90.52 | 0.34 | 20.17 | 99.62 |
| LG7 | 197 | 57.01 | 0.29 | 12.71 | 98.98 |
| LG8 | 153 | 54.6 | 0.36 | 5.28 | 98.68 |
| Total | 2491 | 672.71 | - | - | - |
| Average | 311.38 | 84.09 | 0.27 | 11.15 | 99.49 |
| Year | Linkage Group | QTL | Confidence interval (cM) | Mark number | LOD | PVE (%) |
|---|---|---|---|---|---|---|
| 2021 | LG2 | 21TA-1 | 69.433–69.912 | 6 | 3.87 | 12.03 |
| LG2 | 21TA-2 | 74.218–77.568 | 16 | 3.86 | 12.01 | |
| LG2 | 21TA-3 | 84.51–85.467 | 4 | 3.33 | 10.45 | |
| LG2 | 21TA-4 | 86.663–87.381 | 6 | 3.25 | 10.19 | |
| LG2 | 21TA-5 | 89.056–90.97 | 13 | 3.41 | 10.69 | |
| LG2 | 21TA-6 | 91.209–92.405 | 10 | 3.68 | 11.45 | |
| LG2 | 21TA-7 | 94.08–94.319 | 3 | 3.91 | 12.13 | |
| LG5 | 21TA-8 | 42.349–42.588 | 3 | 3.47 | 10.83 | |
| LG5 | 21TA-9 | 45.7–45.94 | 3 | 3.56 | 11.13 | |
| LG5 | 21TA-10 | 48.093–48.811 | 8 | 5.43 | 16.44 | |
| LG5 | 21TA-11 | 50.007–59.338 | 35 | 6.29 | 18.71 | |
| LG5 | 21TA-12 | 60.056–61.971 | 10 | 11.28 | 31.13 | |
| LG5 | 21TA-13 | 63.407–79.197 | 55 | 8.36 | 24.06 | |
| 2022 | LG1 | 22TA-1 | 29.432 | 3 | 3.86 | 8.7 |
| LG2 | 22TA-2 | 60.336 | 1 | 3.81 | 8.6 | |
| LG3 | 22TA-3 | 48.332 | 1 | 3.14 | 7.1 | |
| LG5 | 22TA-4 | 34.693 | 1 | 3.23 | 7.3 | |
| LG6 | 22TA-5 | 1.436 | 1 | 3.45 | 7.8 |
| Correlation coefficient | Phosphatidate phosphatase gene (FPKM) |
|---|---|
| Titratable acid content | 0.93** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




