Submitted:
22 May 2023
Posted:
23 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Identification of the NADP-ME Genes in Pepper (Capsicum annuum L.): Sequence and Cis-Regulatory Elements
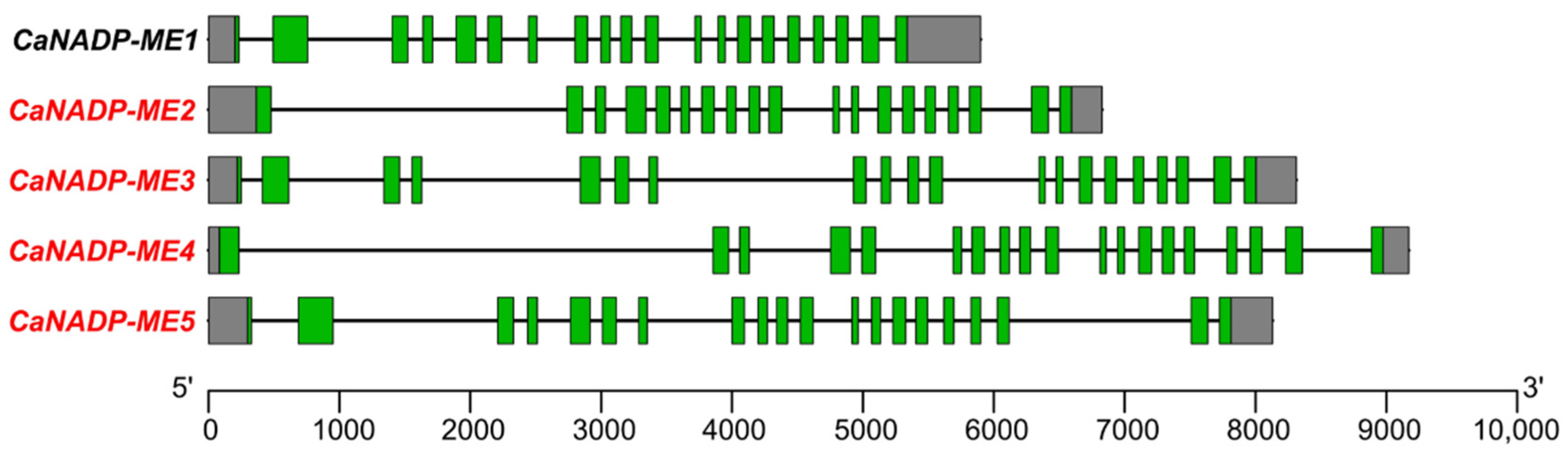
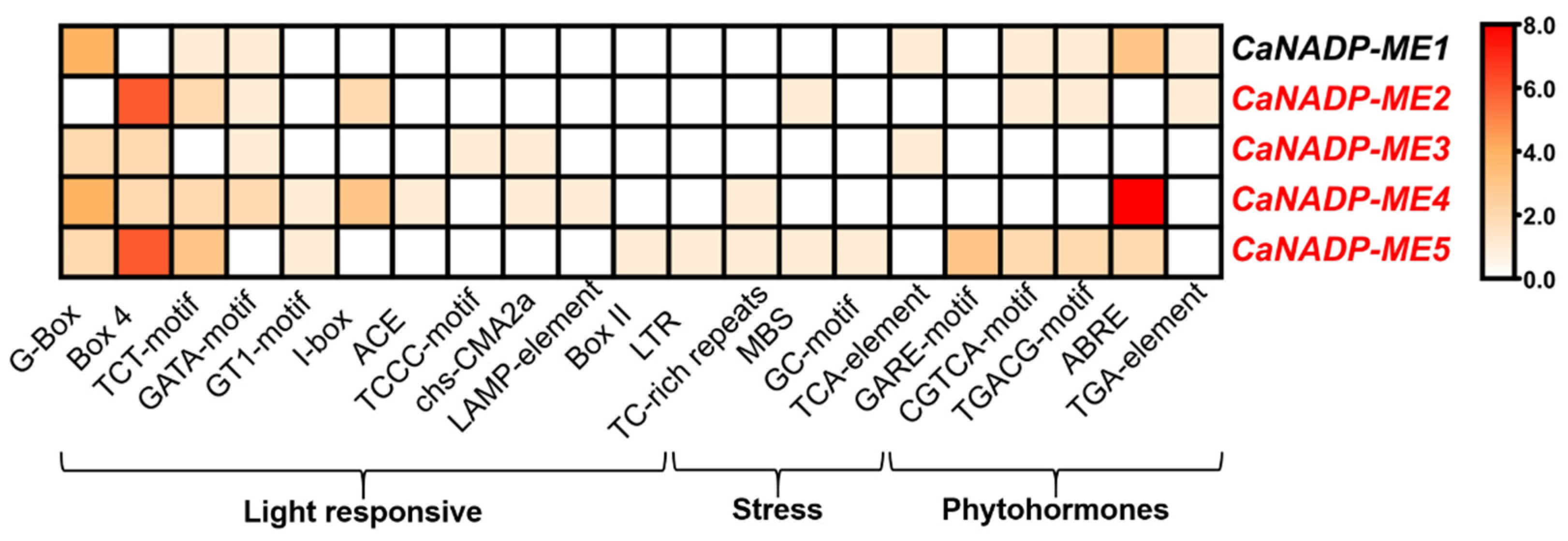
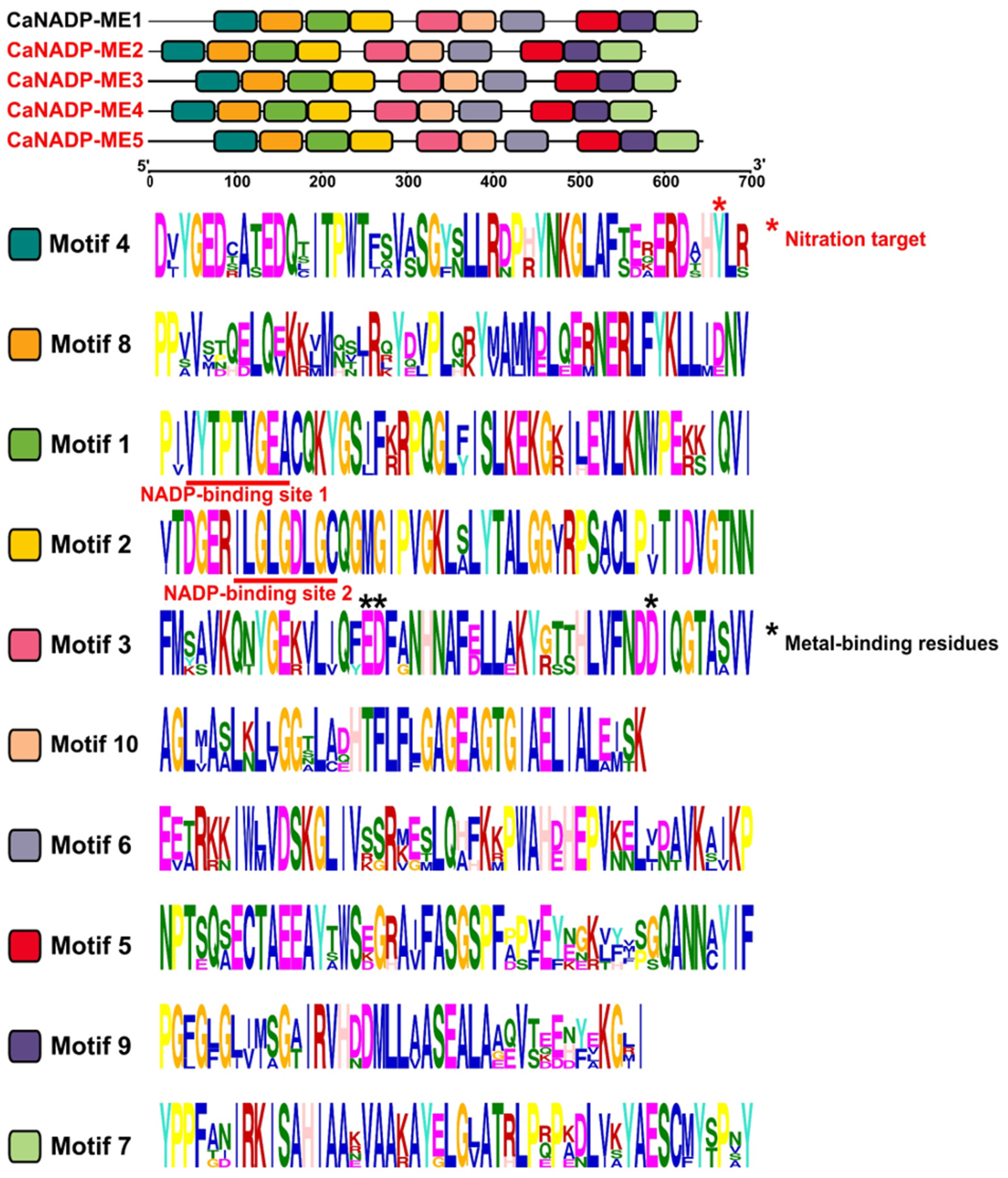
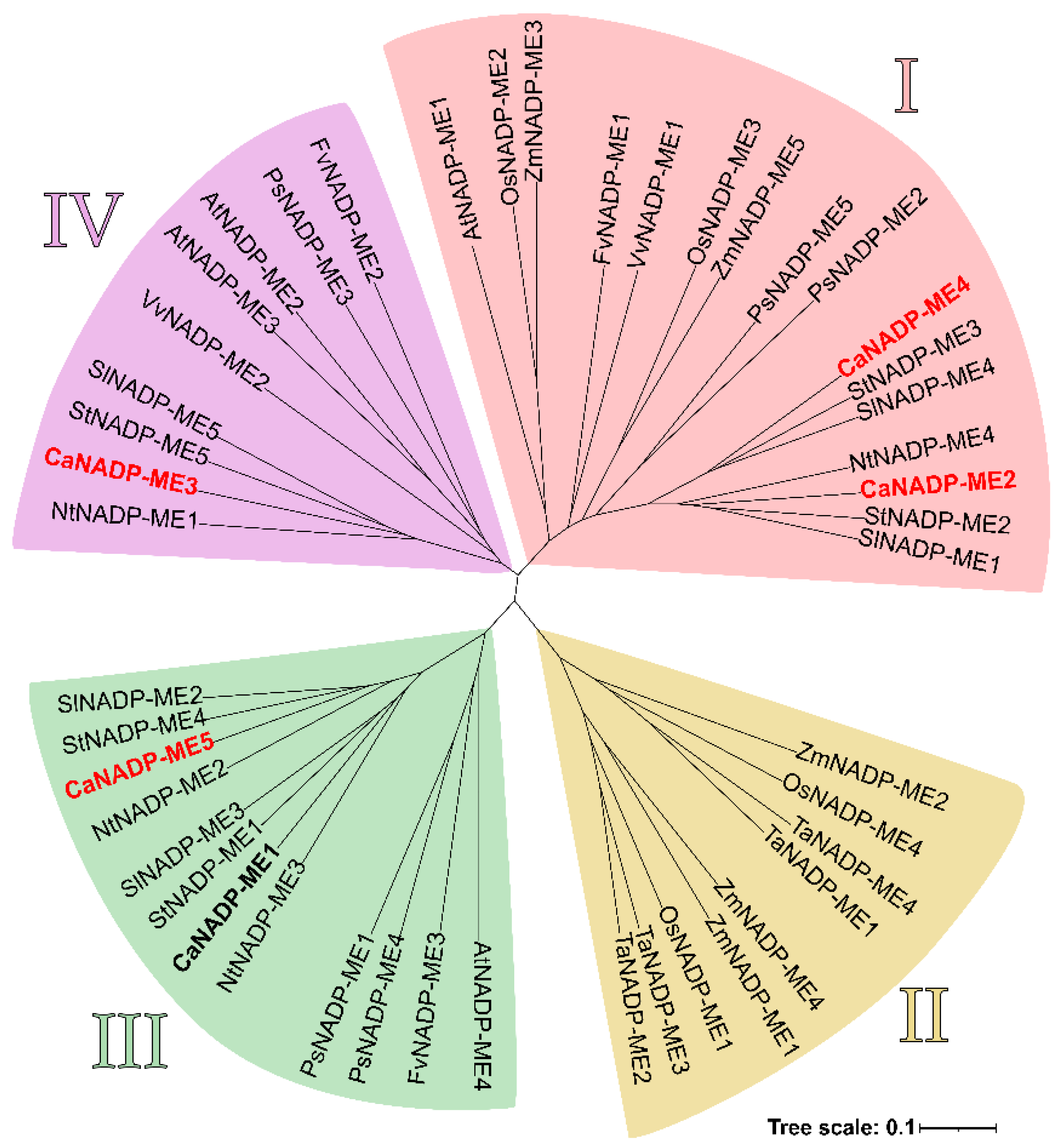
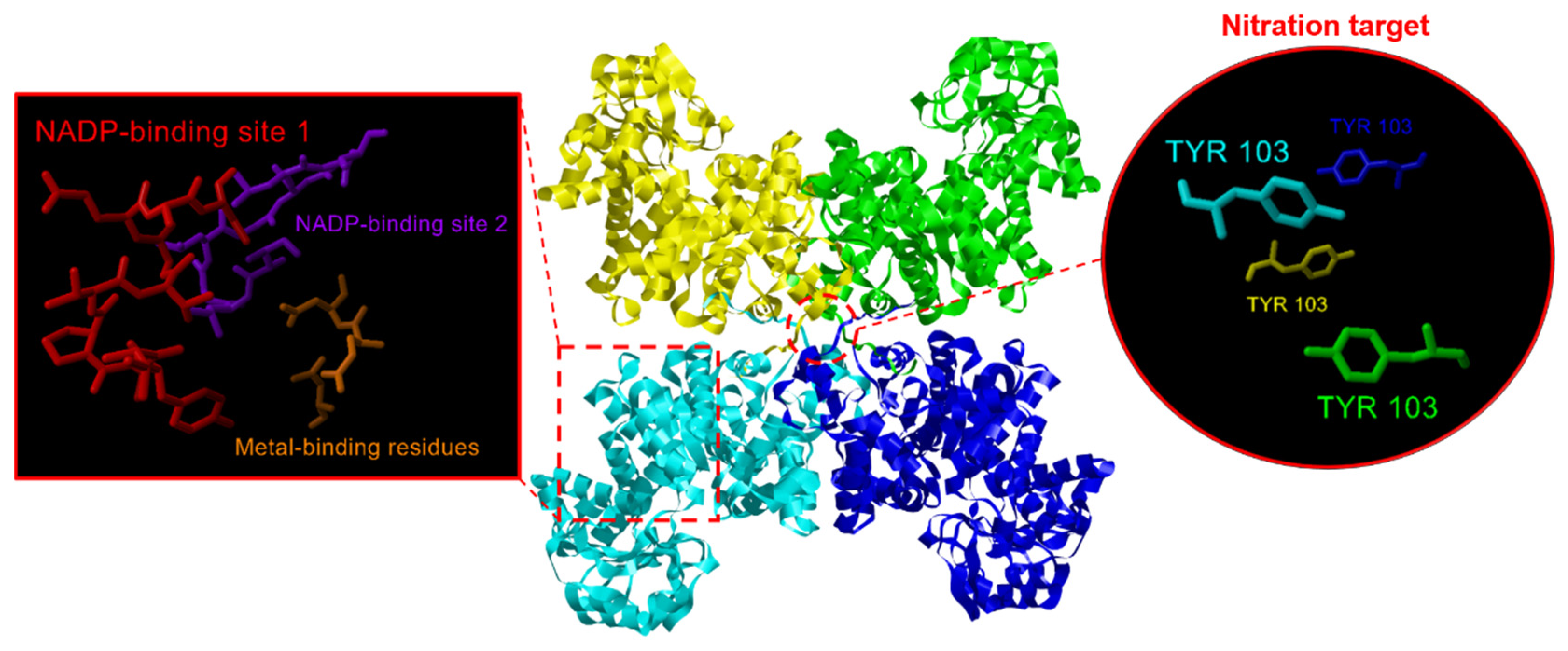
2.2. NADP-ME Proteins from Pepper: Sequence, Phylogenetic Analysis and Modeling
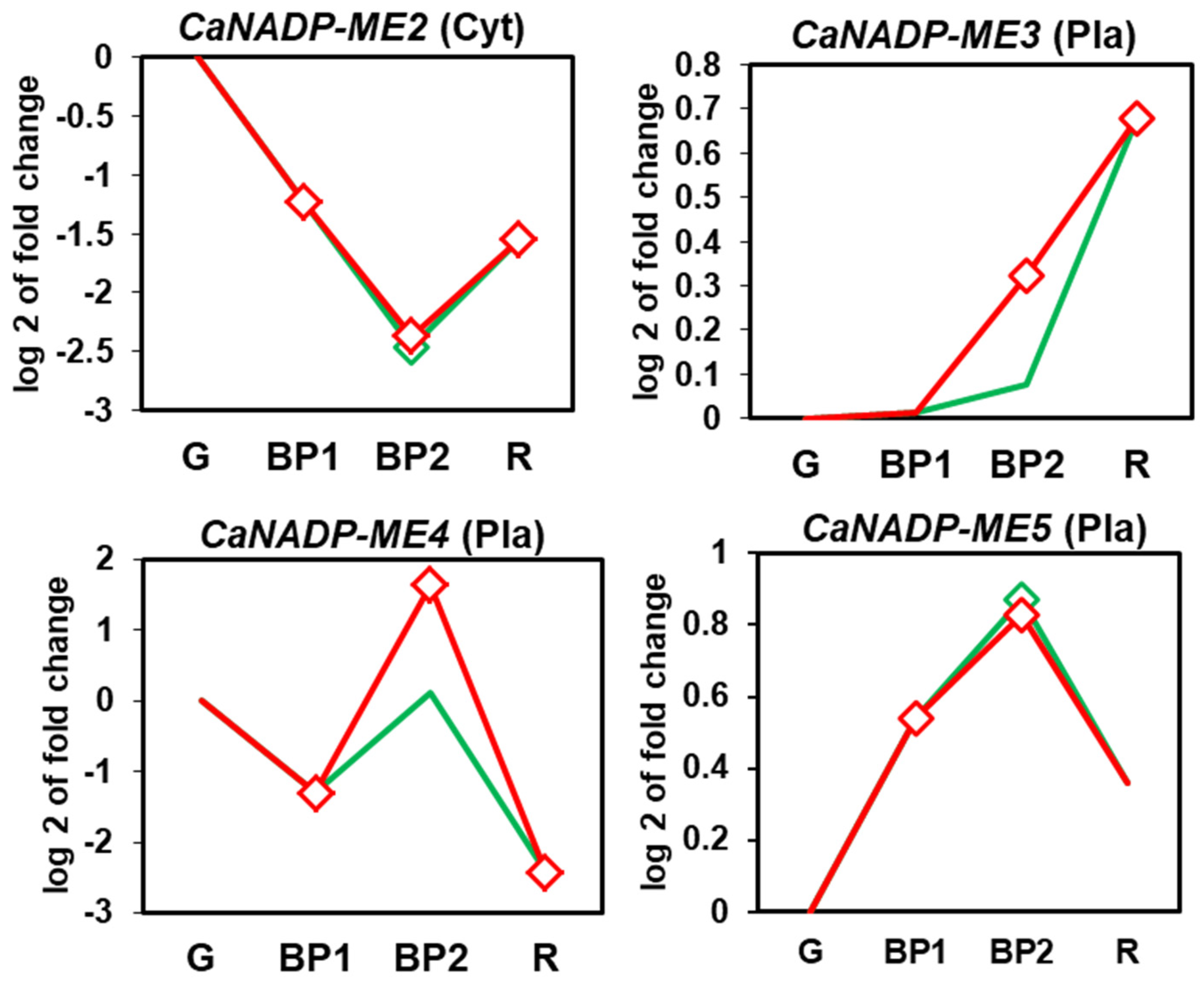
2.3. Fruit CaNADP-ME Genes: Expression during Ripening and by Exogenous NO Treatment
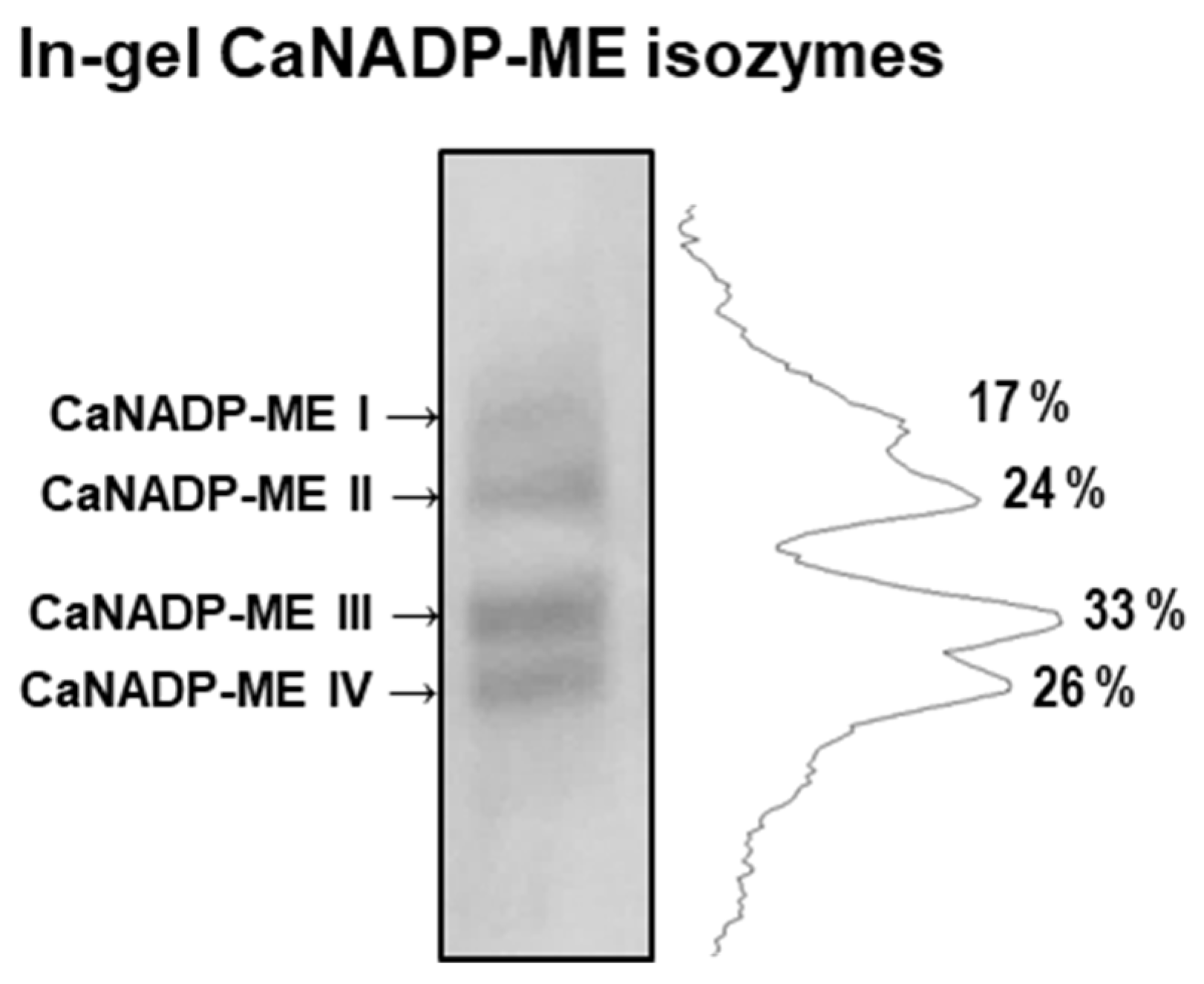
2.4. Identification of the CaNADP-ME Isozymes in Pepper Fruits
3. Discussion
3.1. Pepper Genome Contains Five CaNADP-ME Genes but only Four Genes (CaNADP-ME2 to ME5) Are Expressed in Fruits
3.2. During Fruit Ripening, the Expression of the CaNADP-ME2 and ME4 are Downregulated whereas CaNADP-ME3 Are Upregulated. Exogenous NO Gas Exerts a Negative Modulation of CaNADP-ME3 and ME4
4. Materials and Methods
4.1. Identification of the NADP-ME Genes in Pepper (Capsicum annuum L.), Chromosomal Location and Synteny Analysis
4.2. Phylogenetic and Conserved Motif Analyses of NADP-ME Protein Sequences
4.3. Introns/Exons and Cis-Regulatory Elements Analysis of the CaNADP-ME Genes
4.4. Plant Material and Exogenous Nitric Oxide (NO) Gas Treatment
4.5. Library Preparation and RNA-Sequencing
4.6. Fruit Extracts, Protein Assay, Protein Enrichment by Ammonium-Sulfate [(NH4)2SO4] Fractionation and In-Gel Isozyme Profile of NADP-ME Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corpas, F.J.; Barroso, J.B. NADPH-Generating Dehydrogenases: Their Role in the Mechanism of Protection against Nitro-Oxidative Stress Induced by Adverse Environmental Conditions. Frontiers in Environmental Science 2014, 2. [Google Scholar] [CrossRef]
- Habashi, R.; Hacham, Y.; Dhakarey, R.; Matityahu, I.; Holland, D.; Tian, L.; Amir, R. Elucidating the Role of Shikimate Dehydrogenase in Controlling the Production of Anthocyanins and Hydrolysable Tannins in the Outer Peels of Pomegranate. BMC Plant Biol 2019, 19, 476. [Google Scholar] [CrossRef]
- Meeks, K.R.; Tanner, J.J. Expression and Kinetic Characterization of PYCR3. Arch Biochem Biophys 2023, 733, 109468. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Production of Reactive Oxygen Species by Plant NADPH Oxidases. Plant Physiol 2006, 141, 336–340. [Google Scholar] [CrossRef]
- Kaya, H.; Takeda, S.; Kobayashi, M.J.; Kimura, S.; Iizuka, A.; Imai, A.; Hishinuma, H.; Kawarazaki, T.; Mori, K.; Yamamoto, Y.; et al. Comparative Analysis of the Reactive Oxygen Species-Producing Enzymatic Activity of Arabidopsis NADPH Oxidases. Plant J 2019, 98, 291–300. [Google Scholar] [CrossRef]
- Wu, B.; Li, P.; Hong, X.; Xu, C.; Wang, R.; Liang, Y. The Receptor-like Cytosolic Kinase RIPK Activates NADP-Malic Enzyme 2 to Generate NADPH for Fueling ROS Production. Mol Plant 2022, 15, 887–903. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. NO Source in Higher Plants: Present and Future of an Unresolved Question. Trends Plant Sci 2022, 27, 116–119. [Google Scholar] [CrossRef]
- Edwards, G.E.; Andreo, C.S. NADP-Malic Enzyme from Plants. Phytochemistry 1992, 31, 1845–1857. [Google Scholar] [CrossRef]
- Drincovich, M.F.; Casati, P.; Andreo, C.S. NADP-Malic Enzyme from Plants: A Ubiquitous Enzyme Involved in Different Metabolic Pathways. FEBS Lett 2001, 490, 1–6. [Google Scholar] [CrossRef]
- Yazdanpanah, F.; Maurino, V.G.; Mettler-Altmann, T.; Buijs, G.; Bailly, M.N.; Karimi Jashni, M.; Willems, L.; Sergeeva, L.I.; Rajjou, L.C.; Hilhorst, H.W.M.; et al. NADP-MALIC ENZYME 1 Affects Germination after Seed Storage in Arabidopsis Thaliana. Plant Cell Physiol 2019, 60, 318–328. [Google Scholar] [CrossRef]
- Airaki, M.; Leterrier, M.; Valderrama, R.; Chaki, M.; Begara-Morales, J.C.; Barroso, J.B.; del Río, L.A.; Palma, J.M.; Corpas, F.J. Spatial and Temporal Regulation of the Metabolism of Reactive Oxygen and Nitrogen Species during the Early Development of Pepper (Capsicum Annuum) Seedlings. Ann Bot 2015, 116, 679–693. [Google Scholar] [CrossRef]
- Goodenough, P.W.; Prosser, I.M.; Young, K. NADP-Linked Malic Enzyme and Malate Metabolism in Ageing Tomato Fruit. Phytochemistry 1985, 24, 1157–1162. [Google Scholar] [CrossRef]
- Franke, K.E.; Adams, D.O. Cloning of a Full-Length CDNA for Malic Enzyme (EC 1.1.1.40) from Grape Berries. Plant Physiol 1995, 107, 1009–1010. [Google Scholar] [CrossRef]
- Famiani, F.; Walker, R.P.; Técsi, L.; Chen, Z.H.; Proietti, P.; Leegood, R.C. An Immunohistochemical Study of the Compartmentation of Metabolism during the Development of Grape (Vitis Vinifera L.) Berries. J Exp Bot 2000, 51, 675–683. [Google Scholar]
- Famiani, F.; Casulli, V.; Baldicchi, A.; Battistelli, A.; Moscatello, S.; Walker, R.P. Development and Metabolism of the Fruit and Seed of the Japanese Plum Ozark Premier (Rosaceae). J Plant Physiol 2012, 169, 551–560. [Google Scholar] [CrossRef]
- Osorio, S.; Vallarino, J.G.; Szecowka, M.; Ufaz, S.; Tzin, V.; Angelovici, R.; Galili, G.; Fernie, A.R. Alteration of the Interconversion of Pyruvate and Malate in the Plastid or Cytosol of Ripening Tomato Fruit Invokes Diverse Consequences on Sugar but Similar Effects on Cellular Organic Acid, Metabolism, and Transitory Starch Accumulation. Plant Physiol 2013, 161, 628–643. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, B.; Ding, H.; Zhang, J.; Li, S. Review: The Role of NADP-Malic Enzyme in Plants under Stress. Plant Sci 2019, 281, 206–212. [Google Scholar] [CrossRef]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Gómez-Rodríguez, M.V.; Chaki, M.; Pedrajas, J.R.; Fernández-Ocaña, A.; Del Río, L.A.; Barroso, J.B. The Dehydrogenase-Mediated Recycling of NADPH Is a Key Antioxidant System against Salt-Induced Oxidative Stress in Olive Plants. Plant Cell Environ 2006, 29, 1449–1459. [Google Scholar] [CrossRef]
- Manai, J.; Gouia, H.; Corpas, F.J. Redox and Nitric Oxide Homeostasis Are Affected in Tomato (Solanum Lycopersicum) Roots under Salinity-Induced Oxidative Stress. J. Plant Physiol. 2014, 171, 1028–1035. [Google Scholar] [CrossRef]
- Bouthour, D.; Kalai, T.; Chaffei, H.C.; Gouia, H.; Corpas, F.J. Differential Response of NADP-Dehydrogenases and Carbon Metabolism in Leaves and Roots of Two Durum Wheat (Triticum Durum Desf.) Cultivars (Karim and Azizi) with Different Sensitivities to Salt Stress. J. Plant Physiol. 2015, 179, 56–63. [Google Scholar] [CrossRef]
- Signorelli, S.; Corpas, F.J.; Borsani, O.; Barroso, J.B.; Monza, J. Water Stress Induces a Differential and Spatially Distributed Nitro-Oxidative Stress Response in Roots and Leaves of Lotus Japonicus. Plant Sci. 2013, 201–202, 137–146. [Google Scholar] [CrossRef]
- Doubnerová Hýsková, V.; Miedzińska, L.; Dobrá, J.; Vankova, R.; Ryšlavá, H. Phosphoenolpyruvate Carboxylase, NADP-Malic Enzyme, and Pyruvate, Phosphate Dikinase Are Involved in the Acclimation of Nicotiana Tabacum L. to Drought Stress. J Plant Physiol 2014, 171, 19–25. [Google Scholar] [CrossRef]
- Houmani, H.; Rodríguez-Ruiz, M.; Palma, J.M.; Corpas, F.J. Mechanical Wounding Promotes Local and Long Distance Response in the Halophyte Cakile Maritima through the Involvement of the ROS and RNS Metabolism. Nitric Oxide 2018, 74, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Aguayo-Trinidad, S.; Ogawa, T.; Yoshimura, K.; Shigeoka, S. Activation of NADPH-Recycling Systems in Leaves and Roots of Arabidopsis Thaliana under Arsenic-Induced Stress Conditions Is Accelerated by Knock-out of Nudix Hydrolase 19 (AtNUDX19) Gene. J Plant Physiol 2016, 192, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Ruíz-Torres, C.; Feriche-Linares, R.; Rodríguez-Ruíz, M.; Palma, J.M.; Corpas, F.J. Arsenic-Induced Stress Activates Sulfur Metabolism in Different Organs of Garlic (Allium Sativum L.) Plants Accompanied by a General Decline of the NADPH-Generating Systems in Roots. J Plant Physiol 2017, 211, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ruiz, M.; Aparicio-Chacón, M.V.; Palma, J.M.; Corpas, F.J. Arsenate Disrupts Ion Balance, Sulfur and Nitric Oxide Metabolisms in Roots and Leaves of Pea (Pisum Sativum L.) Plants. Environmental and Experimental Botany 2019, 161, 143–156. [Google Scholar] [CrossRef]
- Kharbech, O.; Houmani, H.; Chaoui, A.; Corpas, F.J. Alleviation of Cr(VI)-Induced Oxidative Stress in Maize (Zea Mays L.) Seedlings by NO and H2S Donors through Differential Organ-Dependent Regulation of ROS and NADPH-Recycling Metabolisms. J. Plant Physiol. 2017, 219, 71–80. [Google Scholar] [CrossRef]
- de Freitas-Silva, L.; Rodríguez-Ruiz, M.; Houmani, H.; da Silva, L.C.; Palma, J.M.; Corpas, F.J. Glyphosate-Induced Oxidative Stress in Arabidopsis Thaliana Affecting Peroxisomal Metabolism and Triggers Activity in the Oxidative Phase of the Pentose Phosphate Pathway (OxPPP) Involved in NADPH Generation. J Plant Physiol 2017, 218, 196–205. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Gómez-Rodríguez, M.V.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; López-Jaramillo, J.; Corpas, F.J.; Barroso, J.B. Short-Term Low Temperature Induces Nitro-Oxidative Stress That Deregulates the NADP-Malic Enzyme Function by Tyrosine Nitration in Arabidopsis Thaliana. Antioxidants (Basel) 2019, 8, 448. [Google Scholar] [CrossRef]
- Sánchez-McSweeney, A.; González-Gordo, S.; Aranda-Sicilia, M.N.; Rodríguez-Rosales, M.P.; Venema, K.; Palma, J.M.; Corpas, F.J. Loss of Function of the Chloroplast Membrane K+/H+ Antiporters AtKEA1 and AtKEA2 Alters the ROS and NO Metabolism but Promotes Drought Stress Resilience. Plant Physiol Biochem 2021, 160, 106–119. [Google Scholar] [CrossRef]
- Houmani, H.; Debez, A.; Freitas-Silva, L. de; Abdelly, C.; Palma, J.M.; Corpas, F.J. Potassium (K+) Starvation-Induced Oxidative Stress Triggers a General Boost of Antioxidant and NADPH-Generating Systems in the Halophyte Cakile Maritima. Antioxidants (Basel) 2022, 11, 401. [Google Scholar] [CrossRef]
- Guevara, L.; Domínguez-Anaya, M.Á.; Ortigosa, A.; González-Gordo, S.; Díaz, C.; Vicente, F.; Corpas, F.J.; Pérez Del Palacio, J.; Palma, J.M. Identification of Compounds with Potential Therapeutic Uses from Sweet Pepper (Capsicum Annuum L.) Fruits and Their Modulation by Nitric Oxide (NO). Int J Mol Sci 2021, 22, 4476. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Bautista, R.; Claros, M.G.; Cañas, A.; Palma, J.M.; Corpas, F.J. Nitric Oxide-Dependent Regulation of Sweet Pepper Fruit Ripening. J Exp Bot 2019, 70, 4557–4570. [Google Scholar] [CrossRef]
- González-Gordo, S.; Rodríguez-Ruiz, M.; Palma, J.M.; Corpas, F.J. Superoxide Radical Metabolism in Sweet Pepper (Capsicum Annuum L.) Fruits Is Regulated by Ripening and by a NO-Enriched Environment. Front Plant Sci 2020, 11, 485. [Google Scholar] [CrossRef]
- González-Gordo, S.; Rodríguez-Ruiz, M.; López-Jaramillo, J.; Muñoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. Nitric Oxide (NO) Differentially Modulates the Ascorbate Peroxidase (APX) Isozymes of Sweet Pepper (Capsicum Annuum L.) Fruits. Antioxidants (Basel) 2022, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Cañas, A.; Muñoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. Lipoxygenase (LOX) in Sweet and Hot Pepper (Capsicum Annuum L.) Fruits during Ripening and under an Enriched Nitric Oxide (NO) Gas Atmosphere. Int J Mol Sci 2022, 23, 15211. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Palma, J.M.; Corpas, F.J. Small Heat Shock Protein (SHSP) Gene Family from Sweet Pepper (Capsicum Annuum L.) Fruits: Involvement in Ripening and Modulation by Nitric Oxide (NO). Plants (Basel) 2023, 12, 389. [Google Scholar] [CrossRef]
- Palma, J.M.; Terán, F.; Contreras-Ruiz, A.; Rodríguez-Ruiz, M.; Corpas, F.J. Antioxidant Profile of Pepper (Capsicum Annuum L.) Fruits Containing Diverse Levels of Capsaicinoids. Antioxidants (Basel) 2020, 9, 878. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, M.; Mioto, P.; Palma, J.M.; Corpas, F.J. S-Nitrosoglutathione Reductase (GSNOR) Activity Is down-Regulated during Pepper (Capsicum Annuum L.) Fruit Ripening. Nitric Oxide 2017, 68, 51–55. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, M.; González-Gordo, S.; Cañas, A.; Campos, M.J.; Paradela, A.; Corpas, F.J.; Palma, J.M. Sweet Pepper (Capsicum Annuum L.) Fruits Contain an Atypical Peroxisomal Catalase That Is Modulated by Reactive Oxygen and Nitrogen Species. Antioxidants (Basel) 2019, 8, 374. [Google Scholar] [CrossRef]
- Mateos, R.M.; Bonilla-Valverde, D.; del Río, L.A.; Palma, J.M.; Corpas, F.J. NADP-Dehydrogenases from Pepper Fruits: Effect of Maturation. Physiol Plant 2009, 135, 130–139. [Google Scholar] [CrossRef]
- Muñoz-Vargas, M.A.; González-Gordo, S.; Palma, J.M.; Corpas, F.J. Inhibition of NADP-Malic Enzyme Activity by H2 S and NO in Sweet Pepper (Capsicum Annuum L.) Fruits. Physiol Plant 2020, 168, 278–288. [Google Scholar] [CrossRef]
- Rothermel, B.A.; Nelson, T. Primary Structure of the Maize NADP-Dependent Malic Enzyme. J Biol Chem 1989, 264, 19587–19592. [Google Scholar] [CrossRef] [PubMed]
- Börsch, D.; Westhoff, P. Primary Structure of NADP-Dependent Malic Enzyme in the Dicotyledonous C4 Plant Flaveria Trinervia. FEBS Lett 1990, 273, 111–115. [Google Scholar] [CrossRef]
- Tronconi, M.A.; Andreo, C.S.; Drincovich, M.F. Chimeric Structure of Plant Malic Enzyme Family: Different Evolutionary Scenarios for NAD- and NADP-Dependent Isoforms. Front Plant Sci 2018, 9, 565. [Google Scholar] [CrossRef] [PubMed]
- Detarsio, E.; Alvarez, C.E.; Saigo, M.; Andreo, C.S.; Drincovich, M.F. Identification of Domains Involved in Tetramerization and Malate Inhibition of Maize C4-NADP-Malic Enzyme. J Biol Chem 2007, 282, 6053–6060. [Google Scholar] [CrossRef]
- Saigo, M.; Alvarez, C.E.; Andreo, C.S.; Drincovich, M.F. Plastidial NADP-Malic Enzymes from Grasses: Unraveling the Way to the C4 Specific Isoforms. Plant Physiol Biochem 2013, 63, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, Y.; Lyu, M.-J.A.; Zhu, X.-G. Two Major Metabolic Factors for an Efficient NADP-Malic Enzyme Type C4 Photosynthesis. Plant Physiol 2022, 189, 84–98. [Google Scholar] [CrossRef]
- Honda, H.; Akagi, H.; Shimada, H. An Isozyme of the NADP-Malic Enzyme of a CAM Plant, Aloe Arborescens, with Variation on Conservative Amino Acid Residues. Gene 2000, 243, 85–92. [Google Scholar] [CrossRef]
- Laporte, M.M.; Shen, B.; Tarczynski, M.C. Engineering for Drought Avoidance: Expression of Maize NADP-Malic Enzyme in Tobacco Results in Altered Stomatal Function. J Exp Bot 2002, 53, 699–705. [Google Scholar] [CrossRef]
- Arias, C.L.; Andreo, C.S.; Drincovich, M.F.; Gerrard Wheeler, M.C. Fumarate and Cytosolic PH as Modulators of the Synthesis or Consumption of C(4) Organic Acids through NADP-Malic Enzyme in Arabidopsis Thaliana. Plant Mol Biol 2013, 81, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Badia, M.B.; Arias, C.L.; Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovich, M.F.; Wheeler, M.C.G. Enhanced Cytosolic NADP-ME2 Activity in A. Thaliana Affects Plant Development, Stress Tolerance and Specific Diurnal and Nocturnal Cellular Processes. Plant Sci 2015, 240, 193–203. [Google Scholar] [CrossRef]
- Aghdam, M.S.; Palma, J.M.; Corpas, F.J. NADPH as a Quality Footprinting in Horticultural Crops Marketability. Trends in Food Science & Technology 2020, 103, 152–161. [Google Scholar] [CrossRef]
- Maurino, V.G.; Saigo, M.; Andreo, C.S.; Drincovich, M.F. Non-Photosynthetic “malic Enzyme” from Maize: A Constituvely Expressed Enzyme That Responds to Plant Defence Inducers. Plant Mol Biol 2001, 45, 409–420. [Google Scholar] [CrossRef]
- Walter, M.H.; Grima-Pettenati, J.; Feuillet, C. Characterization of a Bean (Phaseolus Vulgaris L.) Malic-Enzyme Gene. Eur J Biochem 1994, 224, 999–1009. [Google Scholar] [CrossRef]
- Wheeler, M.C.G.; Tronconi, M.A.; Drincovich, M.F.; Andreo, C.S.; Flügge, U.-I.; Maurino, V.G. A Comprehensive Analysis of the NADP-Malic Enzyme Gene Family of Arabidopsis. Plant Physiol 2005, 139, 39–51. [Google Scholar] [CrossRef]
- Jo, B.-S.; Choi, S.S. Introns: The Functional Benefits of Introns in Genomes. Genomics Inform 2015, 13, 112–118. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Lampe, M.-K.; Sultan, L.D.; Ostersetzer-Biran, O. Organellar Maturases: A Window into the Evolution of the Spliceosome. Biochim Biophys Acta 2015, 1847, 798–808. [Google Scholar] [CrossRef]
- Fu, Z.-Y.; Zhang, Z.-B.; Hu, X.-J.; Shao, H.-B.; Ping, X. Cloning, Identification, Expression Analysis and Phylogenetic Relevance of Two NADP-Dependent Malic Enzyme Genes from Hexaploid Wheat. C R Biol 2009, 332, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Guo, W.; Li, B.; Wang, W.; Yue, Z.; Lei, J.; Zhao, Y.; Zhong, X. Genome-Wide Identification, Classification, and Analysis of NADP-ME Family Members from 12 Crucifer Species. Mol Genet Genomics 2016, 291, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.C.G.; Arias, C.L.; Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovitch, M.F. Arabidopsis Thaliana NADP-Malic Enzyme Isoforms: High Degree of Identity but Clearly Distinct Properties. Plant Mol Biol 2008, 67, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Centeno, D.C.; Osorio, S.; Nunes-Nesi, A.; Bertolo, A.L.F.; Carneiro, R.T.; Araújo, W.L.; Steinhauser, M.-C.; Michalska, J.; Rohrmann, J.; Geigenberger, P.; et al. Malate Plays a Crucial Role in Starch Metabolism, Ripening, and Soluble Solid Content of Tomato Fruit and Affects Postharvest Softening. Plant Cell 2011, 23, 162–184. [Google Scholar] [CrossRef]
- Ruffner, H.P.; Possner, D.; Brem, S.; Rast, D.M. The Physiological Role of Malic Enzyme in Grape Ripening. Planta 1984, 160, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, F.; Palma, F.; Jamilena, M.; Garrido, D. Preconditioning Treatment Induces Chilling Tolerance in Zucchini Fruit Improving Different Physiological Mechanisms against Cold Injury. Annals of Applied Biology 2015, 166, 340–354. [Google Scholar] [CrossRef]
- Batista Silva, W.; Cosme Silva, G.M.; Santana, D.B.; Salvador, A.R.; Medeiros, D.B.; Belghith, I.; da Silva, N.M.; Cordeiro, M.H.M.; Misobutsi, G.P. Chitosan Delays Ripening and ROS Production in Guava (Psidium Guajava L.) Fruit. Food Chem 2018, 242, 232–238. [Google Scholar] [CrossRef]
- Cherian, S.; Figueroa, C.R.; Nair, H. 'Movers and Shakers' in the Regulation of Fruit Ripening: a Cross-Dissection ff Climacteric Versus Non-Climacteric Fruit. Journal of Experimental Botay 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [PubMed]
- Jarret, R.L.; Berke, T.; Baldwin, E.A.; Antonious, G.F. Variability for Free Sugars and Organic Acids in Capsicum Chinense. Chem Biodivers 2009, 6, 138–145. [Google Scholar] [CrossRef]
- de Ï Vila Silva, L.; Condori-Apfata, J.A.; Costa, P.M. de A.; Martino, P.B.O.; Tavares, A.C.A.; Marcelino, M.M.; Raimundi, S.B.C.J.R.; Picoli, E.A. de T.; Araï Jo, W.L.; Zsï Gï N, A.; et al. Source Strength Modulates Fruit Set by Starch Turnover and Export of Both Sucrose and Amino Acids in Pepper. Plant Cell Physiol 2019, 60, 2319–2330. [Google Scholar] [CrossRef]
- Selinski, J.; Scheibe, R. Malate Valves: Old Shuttles with New Perspectives. Plant Biol (Stuttg) 2019, 21 Suppl 1, 21–30. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, H.; Zhou, J.-M.; Smith, S.M.; Li, J. Malate Circulation: Linking Chloroplast Metabolism to Mitochondrial ROS. Trends Plant Sci 2020, 25, 446–454. [Google Scholar] [CrossRef]
- Rödiger, A.; Agne, B.; Dobritzsch, D.; Helm, S.; Müller, F.; Pötzsch, N.; Baginsky, S. Chromoplast Differentiation in Bell Pepper (Capsicum Annuum) Fruits. The Plant Journal 2021, 105, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Palma, J.M.; Corpas, F.J. Peroxisomal Proteome Mining of Sweet Pepper (Capsicum Annuum L.) Fruit Ripening Through Whole Isobaric Tags for Relative and Absolute Quantitation Analysis. Front Plant Sci 2022, 13, 893376. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Rodríguez-Ruiz, M.; Paradela, A.; Ramos-Fernández, A.; Corpas, F.J.; Palma, J.M. Mitochondrial Protein Expression during Sweet Pepper (Capsicum Annuum L.) Fruit Ripening: ITRAQ-Based Proteomic Analysis and Role of Cytochrome c Oxidase. J Plant Physiol 2022, 274, 153734. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Rodríguez-Ruiz, M.; Muñoz-Vargas, M.A.; Palma, J.M. Thiol-Based Oxidative Posttranslational Modifications (OxiPTMs) of Plant Proteins. Plant Cell Physiol 2022, 63, 889–900. [Google Scholar] [CrossRef]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-Specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol 2015, 167, 1731–1746. [Google Scholar] [CrossRef]
- Aroca, A.; Benito, J.M.; Gotor, C.; Romero, L.C. Persulfidation Proteome Reveals the Regulation of Protein Function by Hydrogen Sulfide in Diverse Biological Processes in Arabidopsis. J Exp Bot 2017, 68, 4915–4927. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Research 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc Int Conf Intell Syst Mol Biol 1994, 2, 28–36. [Google Scholar] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-MSubP: A Computational Framework for the Prediction of Single- and Multi-Target Protein Subcellular Localization Using Integrated Machine-Learning Approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Gayte, I.G.; Moreno, R.B.; Zonjic, P.S.; Claros, M.G. DEgenes Hunter - A Flexible R Pipeline for Automated RNA-Seq Studies in Organisms without Reference Genome. Genomics and Computational Biology 2017, 3, 31. [Google Scholar] [CrossRef]
| Gene Name | Gene ID | Chr. | Genomic Location | Protein ID | Length (aa) | Da | Theoretical pI | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| CaNADP-ME1 | 107861795 | 3 | 267357814-267362949 | XP_047266087.1 | 643 | 70,840 | 5.94 | Plastid |
| CaNADP-ME2 | 107843507 | 5 | 158139423-158145651 | XP_016543302.1 | 578 | 64,245 | 5.80 | Cytosol |
| CaNADP-ME3 | 107847755 | 8 | 26364857-26372640 | XP_016547705.1 | 618 | 68,468 | 6.57 | Plastid |
| CaNADP-ME4 | 107842298 | 9 | 199489875-199498766 | XP_016541547.1 | 590 | 65,360 | 6.13 | Plastid |
| CaNADP-ME5 | 107850438 | 12 | 97276100-97283614 | XP_016550447.1 | 644 | 70,709 | 6.28 | Plastid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





