Submitted:
30 August 2025
Posted:
03 September 2025
You are already at the latest version
Abstract
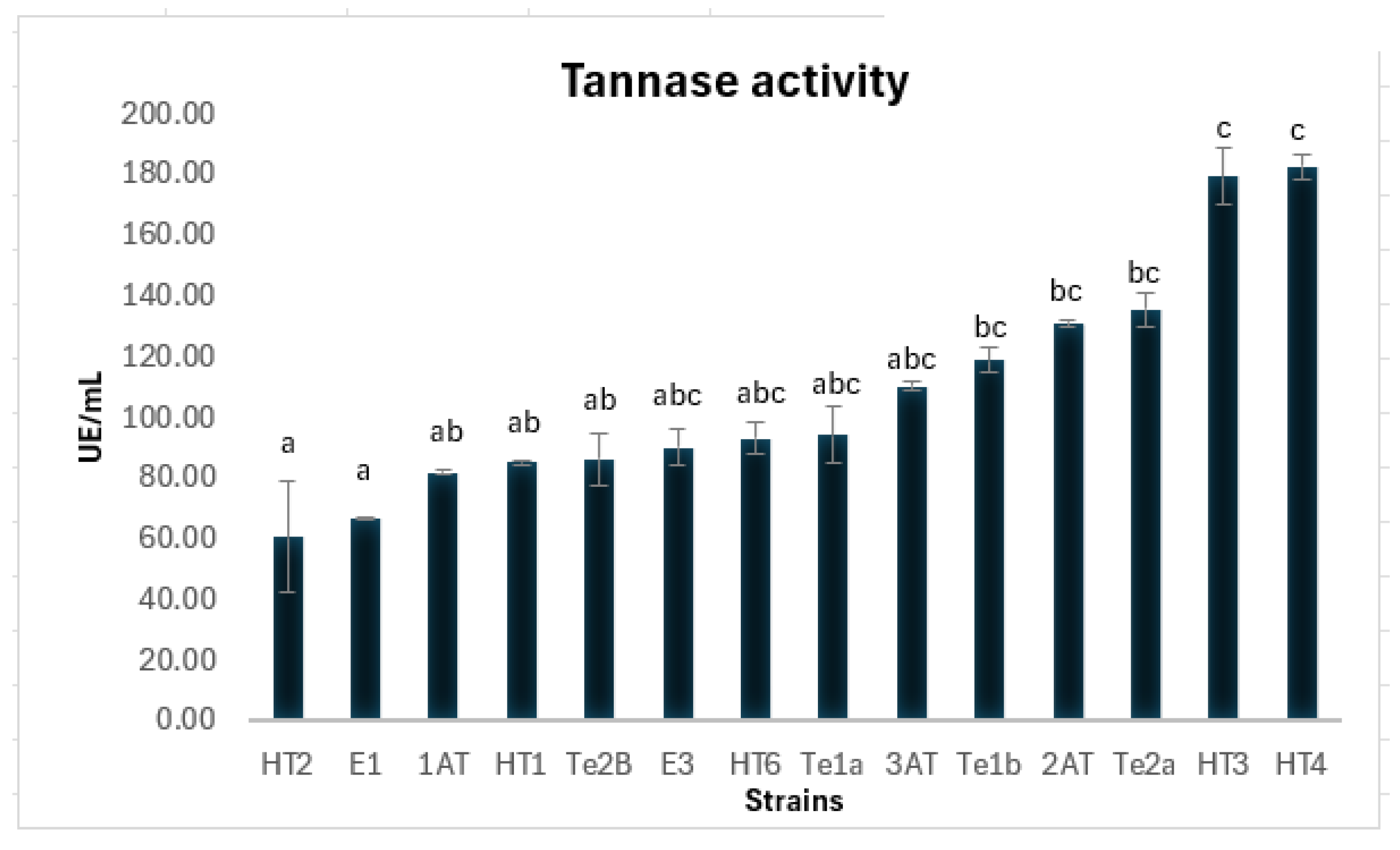
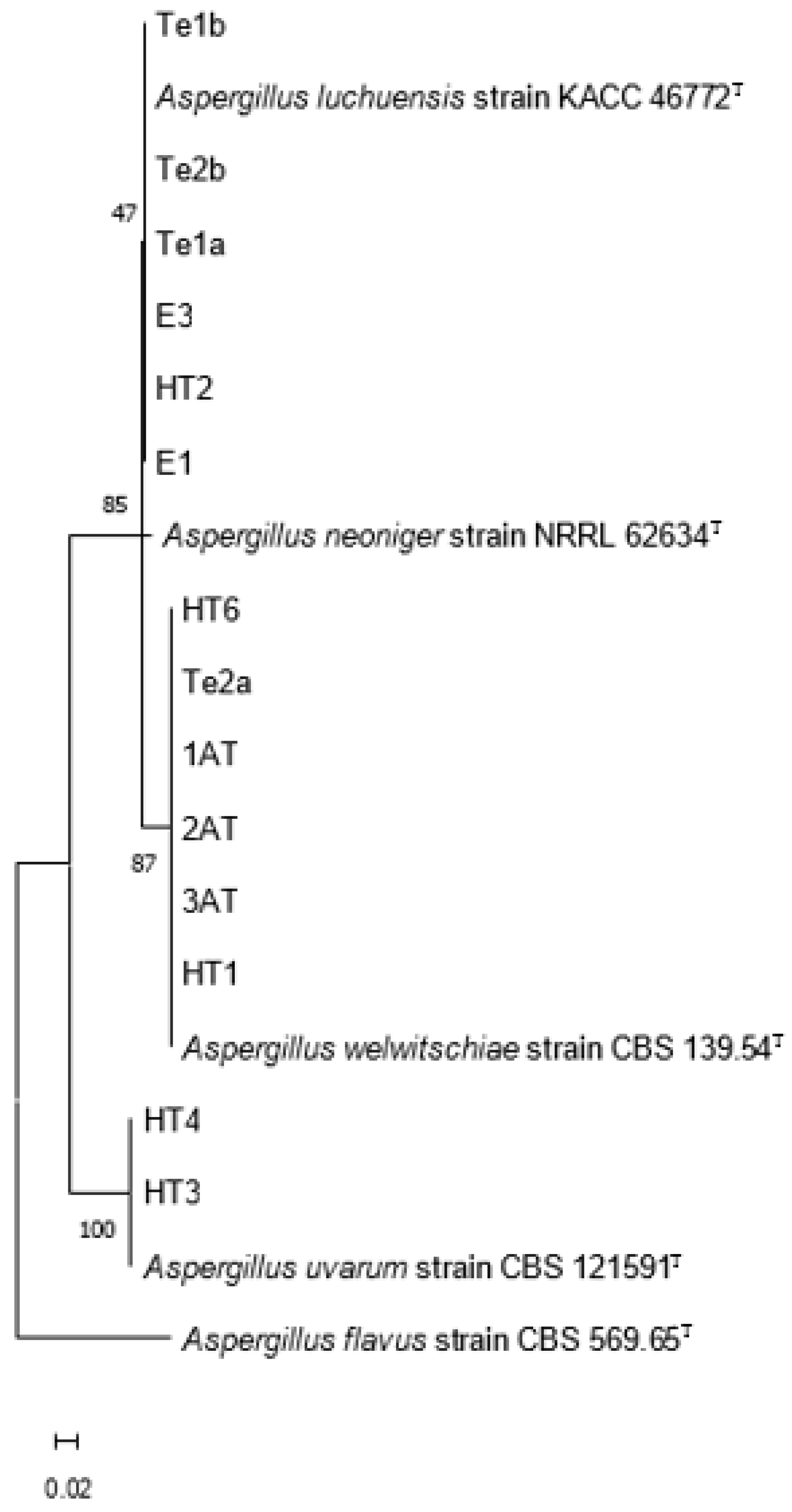
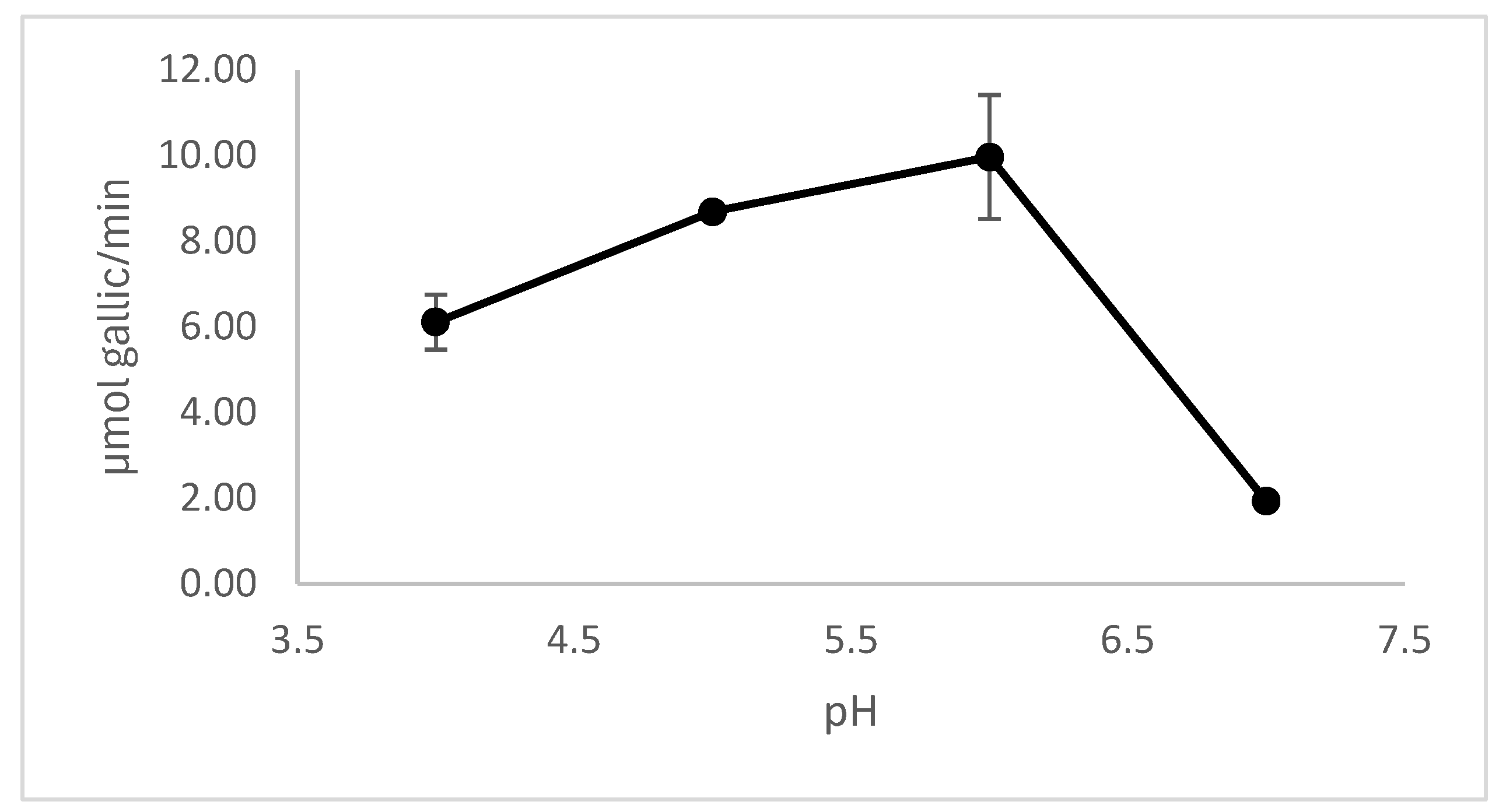
Tannases (tannin acyl hydrolases, EC 3.1.1.20) are enzymes of industrial interest due to their ability to hydrolyze hydrolyzable tannins into bioactive compounds like gallic acid. This study aimed to isolate and identify fungal strains capable of producing extracellular tannase, with a focus on their biotechnological potential. From tannin-rich substrates, 24 fungal isolates were obtained, of which 17 showed tannase activity. Molecular identification based on calmodulin gene sequencing identified three species of tannase-producing black aspergilli: Aspergillus luchuensis, A. welwitschiae, and A. uvarum. The isolate A. uvarum HT4 exhibited the highest extracellular tannase activity (182 U/mL) and was selected for further study. Whole-genome sequencing of HT4 revealed 15 putative tannase genes, most sharing high identity with A. uvarum CBS 121591. Two divergent genes appeared to be acquired via horizontal gene transfer from Aspergillus brunneoviolaceus and Penicillium angulare. Proteomic analysis of the secretome confirmed the expression of two extracellular tannases. The enzyme showed optimal activity at pH 5.0–6.0 and 40–50 °C. Secretome analysis revealed hydrolytic enzymes typical of saprophytic fungi in lignocellulose-rich environments. Importantly, no biosynthetic gene clusters of major mycotoxins were detected, supporting the biosafety of HT4 for industrial applications.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Isolation of Tannase-Producing Fungi
2.3. Selection of Fungi with Extracellular Tannase Activity
2.3.1. Extracellular Tannase Production in Liquid Medium
2.3.2. Tannase Activity Assay
2.4. Molecular Identification of Tannase Producers
2.4.1. DNA Extraction
2.4.2. Calmodulin Gene Amplification
2.4.3. Phylogenetic Analysis of Calmodulin Sequences
2.5. Whole Genome Sequencing, Assembly, and Annotation of the Selected Isolate
2.5.1. Identification and Phylogenetic Analysis of Tannase Sequences
2.6. Characterization of TE Produced by the Selected Isolate
2.6.1. Protein Determination
2.6.2. Effect of pH and Temperature on Tannase Activity
2.6.3. Verification of Tannase Activity by Zymogram
2.6.4. Protein Identification by Nano LC-MS/MS
2.6.5. Secretory Proteins in the HT4 Enzymatic Extract from HT4
3. Results
3.1. Isolation and Screening of Tannase-Producing Fungi
3.2. Extracellular Tannase Production in Liquid Medium
3.3. Molecular Identification of Tannase Producers
3.4. Genome Analysis of the Selected Strain
3.4.1. Tannase Sequences in HT4 Genome
3.4.2. Identification of Secondary Metabolite Gene Clusters
3.5. Characterization of TE Produced by the Selected Isolate
3.5.1. Protein Determination
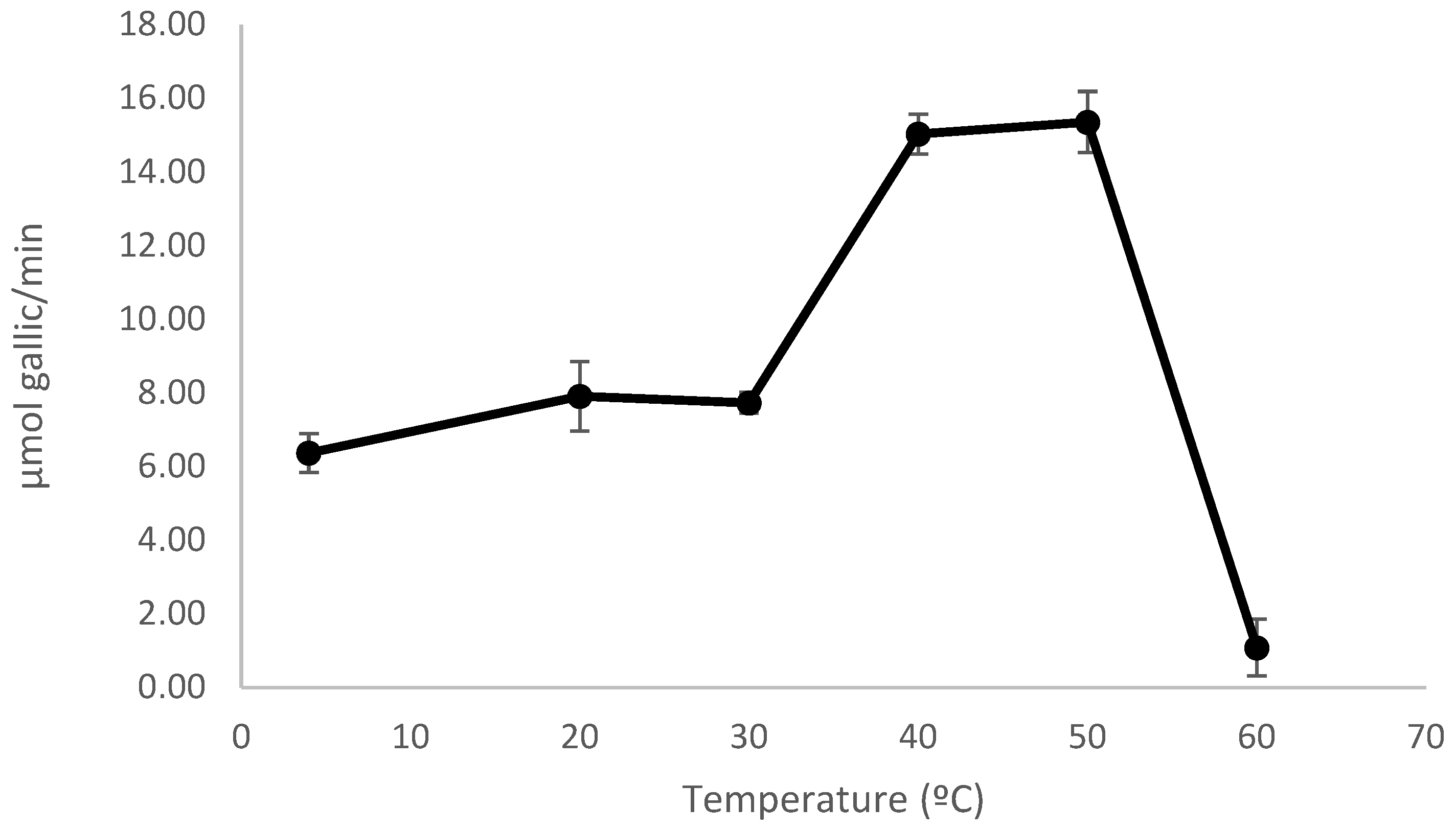
3.5.2. Effect of pH and Temperature on Enzyme Activity
3.5.3. Tannase Verification by Zymogram:
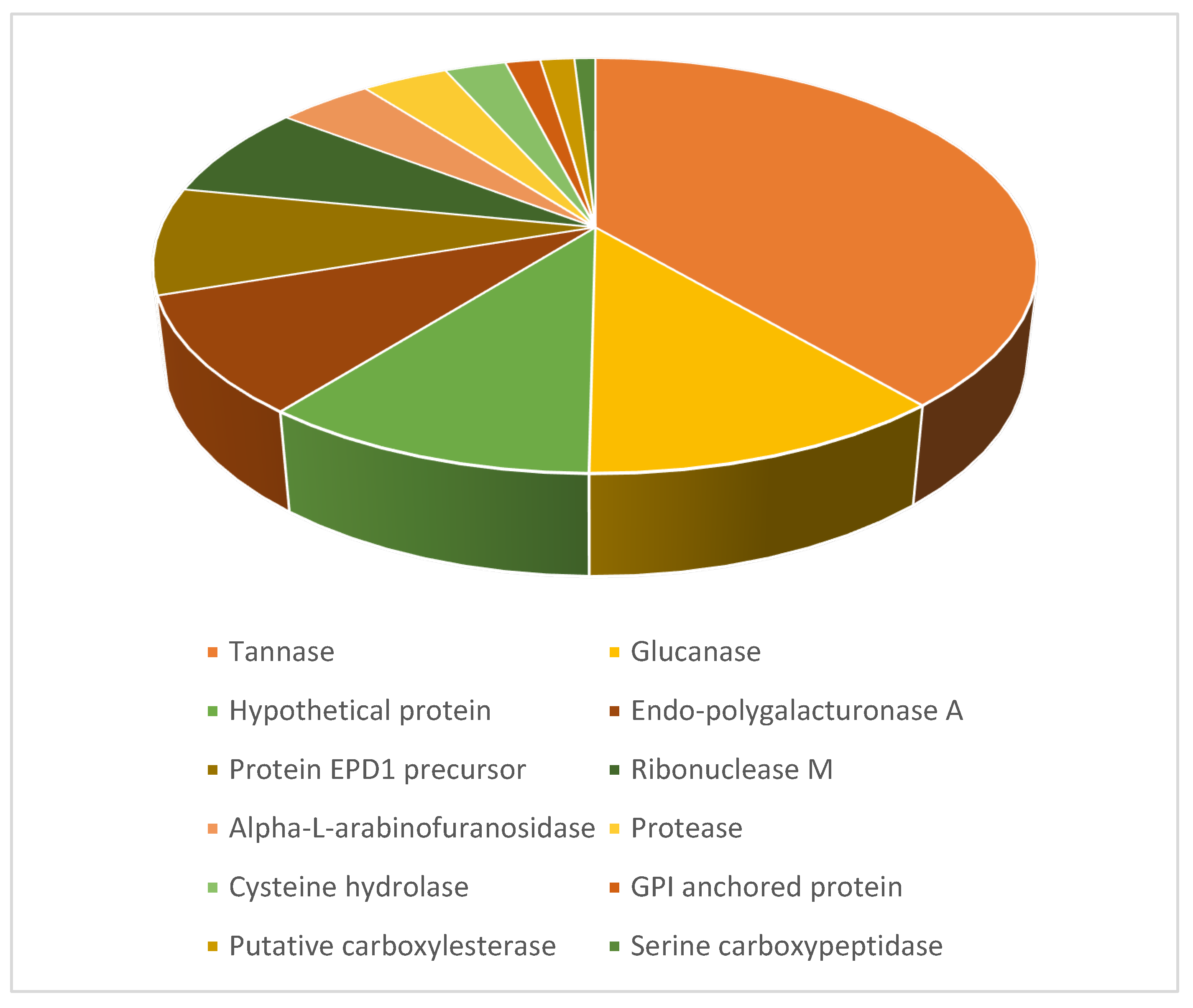
3.6. Protein Identification by Nano LC-MS/MS
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TE | Tannase extract |
| BGCs | Biosynthetic gene clusters |
References
- Magaya, R.P; Mutibvu, T; Ncube, S; Nyahangare, E. T; Mapfumo, P; Mtambanengwe, F; Nyakudya, E; Nhamo, A. Effects of Phytase and Tannase Enzyme Supplementation on in Vitro Nutrient Digestibility and Tannin Degradation of Sorghum-Based Broiler Diets. International Journal of Life Science and Agriculture Research. (2024), 3(5), 346-354. [CrossRef]
- Lekshmi, R., Arif Nisha, S., Thirumalai Vasan, P., & Kaleeswaran, B. A comprehensive review on tannase: Microbes associated production of tannase exploiting tannin rich agro-industrial wastes with special reference to its potential environmental and industrial applications. Environmental Research (2021), 201, 111625. [CrossRef]
- da Silva, G. R., dos Santos, E. S., de Souza, A. P., de Oliveira, R. A., & de Macedo, G. R. Production of lignocellulolytic enzymatic complex using pretreated carnauba straw as carbon source and application on sugarcane bagasse hydrolysis. Biomass Conversion and Biorefinery (2020), 12(1), 85–95. [CrossRef]
- Cavalcanti, R. M. F., Jorge, J. A., & Guimarães, L. H. S. Characterization of Aspergillus fumigatus CAS-21 tannase with potential for propyl gallate synthesis and treatment of tannery effluent from leather industry. 3 Biotech (2018), 8(6), 270. [CrossRef]
- Núñez Sellés, A. J., Vélez Castro, H. T., Agüero-Agüero, J., González-González, J., Naddeo, F., De Simone, F., & Rastrelli, L. Isolation and quantitative analysis of phenolic antioxidants, free sugars, and polyols from mango (Mangifera indica L.) stem bark aqueous decoction used in Cuba as a nutritional supplement. Journal of Agricultural and Food Chemistry (2002), 50(4), 762-766. [CrossRef]
- Bradoo, S., Gupta, R., & Saxena, R. K. Screening of extracellular tannase-producing fungi: Development of a rapid and simple plate assay. Journal of General and Applied Microbiology (1996), 42(4), 325–329. [CrossRef]
- Sharma, S., Bhat, T. K., & Dawra, R. K. A spectrophotometric method for assay of tannase using rhodanine. Analytical biochemistry (2000), 279(1), 85-89. [CrossRef]
- Di Rienzo, J. A., Casanoves, F., Balzarini, M. G., González, L., Tablada, M., & Robledo, C. W. (2015). InfoStat (versión 2015). Grupo InfoStat, Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, Argentina. Recuperado de http://www.infostat.com.ar.
- Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory manual (2ª ed.). Cold Spring Harbor Laboratory Press.
- Garmendia, G., & Vero, S. Occurrence and biodiversity of Aspergillus section Nigri on ‘Tannat’grapes in Uruguay. International Journal of Food Microbiology (2016)., 216, 31-39. [CrossRef]
- Bolger, A. M., Lohse, M., & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics (2014), 30(15), 2114–2120. [CrossRef]
- Bankevich, A., Nurk, S., Antipov, D., Gurevich, A.A., Dvorkin, M., Kulikov, A.S., Lesin, V.M., Nikolenko, S.I., Pham, S., Prjibelski, A.D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M.A., Pevzner, P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. (2012) 19, 455–477. [CrossRef]
- Simão, F.A., Waterhouse, R.M., Ioannidis, P., Kriventseva, E. V., Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics (2015) 31, 3210–3212. [CrossRef]
- Stanke, M., Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. (2005) 33, W465–W467. [CrossRef]
- Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., Maslen, J., Mitchell, A., Nuka, G., Pesseat, S., Quinn, A. F., Sangrador-Vegas, A., Scheremetjew, M., Yong, S.-Y., Lopez, R., & Hunter, S. InterProScan 5: Genome-scale protein function classification. Bioinformatics (2014), 30(9), 1236–1240. [CrossRef]
- El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., Qureshi, M., Richardson, L. J., Salazar, G. A., Smart, A., Sonnhammer, E. L. L., Hirsh, L., Paladin, L., Piovesan, D., Tosatto, S. C. E., & Finn, R. D. The Pfam protein families database in 2019. Nucleic Acids Research (2019), 47(D1), D427–D432. [CrossRef]
- Smith, P. K., Khron, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J., & Klenk, D. C. Analytical Biochemistry (1985), 150, 76–85. [CrossRef]
- Neuhoff, V., Arold, N., Taube, D., & Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis (1988), 9(6), 255-262. [CrossRef]
- Neuhoff, V., Stamm, R., Pardowitz, I., Arold, N., Ehrhardt, W., & Taube, D. Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis(1990), 11(2), 101-117. [CrossRef]
- Rossello, J., Lima, A., Gil, M., Rodríguez Duarte, J., Correa, A., Carvalho, P. C., Kierbel, A. & Durán, R. The EAL-domain protein FcsR regulates flagella, chemotaxis and type III secretion system in Pseudomonas aeruginosa by a phosphodiesterase independent mechanism. Scientific Reports (2017), 7(1), 10281. [CrossRef]
- Santos, M. D., Lima, D. B., Fischer, J. S., Clasen, M. A., Kurt, L. U., Camillo-Andrade, A. C., ... & Carvalho, P. C. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nature protocols (2022)., 17(7), 1553-1578. [CrossRef]
- Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., von Heijne, G. & Nielsen, H. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature biotechnology, 37(4), 420-423. [CrossRef]
- Godio, R. P., & Martín, J. F. (2009). Modified oxidosqualene cyclases in the formation of bioactive secondary metabolites: biosynthesis of the antitumor clavaric acid. Fungal Genetics and Biology, 46(3), 232-242. [CrossRef]
- Zheng, Y. Y., Liang, Z. Y., Shen, N. X., Liu, W. L., Zhou, X. J., Fu, X. M., Chen, M. & Wang, C. Y. (2019). New naphtho-γ-pyrones isolated from marine-derived fungus Penicillium sp. HK1-22 and their antimicrobial activities. Marine Drugs, 17(6), 322. [CrossRef]
- Wang, X., Gong, X., Li, P., Lai, D., & Zhou, L. (2018). Structural diversity and biological activities of cyclic depsipeptides from fungi. Molecules, 23(1), 169. [CrossRef]
- Matsuda, K. (2018). Okaramines and other plant fungal products as new insecticide leads. Current Opinion in Insect Science, 30, 67-72. [CrossRef]
- Paper de Frisvad JC, Thrane U, Samson RA et al. (2006) Important mycotoxins and the fungi which produce them. In: Pitt J, Samson RA, Thrane U (eds) Advances in food mycology. Springer, New York, pp 3–31.
- Egea, M. B., Dantas, L. A., Sousa, T. L. D., Lima, A. G., & Lemes, A. C. (2023). The potential, strategies, and challenges of Monascus pigment for food application. Frontiers in Sustainable Food Systems, 7, 1141644. [CrossRef]
- Biswas, I., Mitra, D., & Mohapatra, P. K. D. (2022). Structural and catalytic advancement of fungal tannase: A proteomic contribution in industrial applicability. Bioresource Technology Reports, 19, 101103. [CrossRef]
- Aracri, F. M., Cavalcanti, R. M. F., & Guimarães, L. H. S. Extracellular tannase from Aspergillus ochraceus: influence of the culture conditions on biofilm formation, enzyme production, and application. J. Microbiol. Biotechnol (2019), 29(11), 1749-1759. [CrossRef]
- Cruz, R., de Lima, J. S., Fonseca, J. C., dos Santos Fernandes, M. J., Lima, D. M. M., Duda, G. P., ... & de Souza Motta, C. M. Diversity of filamentous fungi of area from Brazilian Caatinga and high-level tannase production using mango (Mangifera indica L.) and surinam cherry (Eugenia uniflora L.) leaves under SSF. Advances in Microbiology (2013), 3(8), 52-60. [CrossRef]
- Thakur, N., Nath, A. K., & Sharma, A. Optimization of production conditions, isolation, purification, and characterization of tannase from filamentous fungi. Folia Microbiologica (2024), 1-13. [CrossRef]
- Lal, D., & Gardner, J. J. (2012). Production, characterization and purification of tannase from Aspergillus niger. European Journal of Experimental Biology, 2(5), 1430-1438.
- Sharma, K. P., & John, P. J. Purification and characterization of tannase and tannase gene from Enterobacter sp. Process Biochemistry (2011), 46(1), 240–244. [CrossRef]
- Yao, J., Guo, G. S., Ren, G. H., & Liu, Y. H. Production, characterization and applications of tannase. Journal of Molecular Catalysis B: Enzymatic (2014), 101, 137–147. [CrossRef]
- Batra, A., & Saxena, R. K. Potential tannase producers from the genera Aspergillus and Penicillium. Process Biochemistry . (2005), 40(5), 1553–1557. [CrossRef]
- Osipov, D. O., Matys, V. Y., Nemashkalov, V. A., Rozhkova, A. M., Shashkov, I. A., Satrutdinov, A. D., ... & Sinitsyn, A. P. Cloning, Isolation, and Properties of a New Recombinant Tannase from the Aspergillus niger Fungus. Applied Biochemistry and Microbiology (2022), 58(9), 958-965. [CrossRef]
- Zhursinali, A. B., & Kurmanbaev, A. A. Biologically active substances of fungus Aspergillus niger. Eurasian Journal of Applied Biotechnology (2020), (1). [CrossRef]
- Busk, P. K., Lange, M., Pilgaard, B., & Lange, L. Several genes encoding enzymes with the same activity are necessary for aerobic fungal degradation of cellulose in nature. PLoS One. (2014), 9(12), e114138. [CrossRef]
- Alfaro, M., Oguiza, J. A., Ramírez, L., & Pisabarro, A. G. (2014). Comparative analysis of secretomes in basidiomycete fungi. Journal of Proteomics, 102, 28-43. [CrossRef]

| Identifier | Length (AA) | Identity (%) | Homolog source |
|---|---|---|---|
| NODE_1.g376 | 512 | 95.20 | Aspergillus uvarum CBS 121591 XP_025490290.1 |
| NODE_3.g1172 | 537 | 100 | Aspergillus uvarum CBS 121591 XP_025488689.1 |
| NODE_9.g2866 | 543 | 97.18 |
Aspergillus uvarum CBS 121591 XP_025492021.1 |
| NODE_11.g3384 | 1477 | 99.60 |
Aspergillus uvarum CBS 121591 XP_025487606.1 |
| NODE_11.g3515 | 548 | 100 | Aspergillus uvarum CBS 121591 XP_025485684.1 |
| NODE_16.g4469 | 588 | 99.83 | Aspergillus uvarum CBS 121591 XP_025491148 |
| NODE_16.g4509 | 572 | 99.48 | Aspergillus uvarum CBS 121591 XP_025491187 |
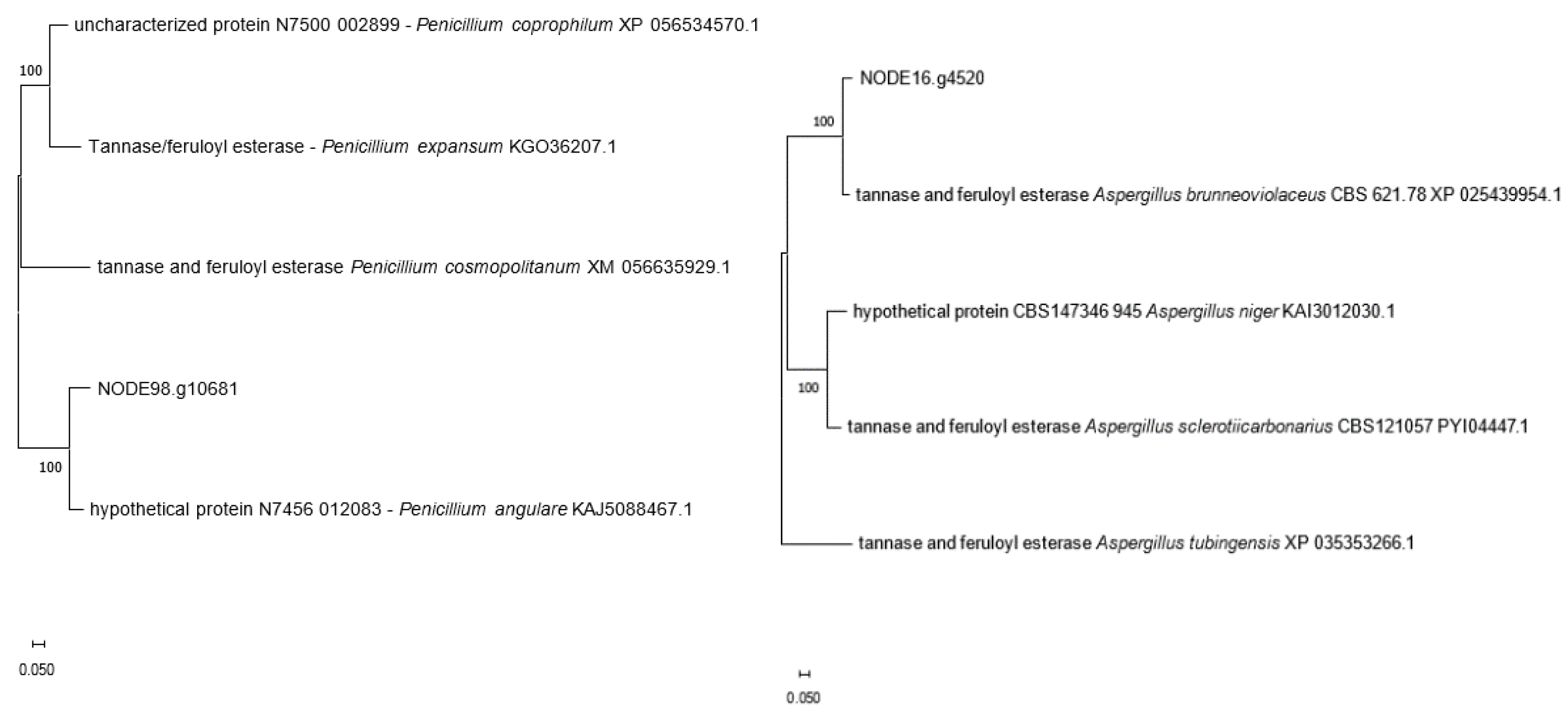
| NODE_16.g4520 | 585 | 92.31 | Aspergillus brunneoviolaceus CBS 621.78 XP_025439954 |
| NODE_18.g4952 | 739 | 99.30 | Aspergillus uvarum CBS121591 XP_025493482 |
| NODE_55.g9244 | 578 | 97.75 | Aspergillus uvarum CBS 121591 XP_025487768.1 |
| NODE_57.g9409 | 545 | 100 | Aspergillus uvarum CBS 121591 XP_025487823 |
| NODE_60.g9637 | 584 | 100 | Aspergillus uvarum CBS 121591 XP_025496649.1 |
| NODE_70.g10053 | 522 | 100 | Aspergillus uvarum CBS 121591 XP_025489644.1 |
| NODE_71.g10109 | 532 | 96.75 | Aspergillus uvarum CBS 121591 XP_025495855.1 |
| NODE_98.g10681 | 505 | 88.20 | Penicillium angulare N7456_012083 KAJ5088467 |
| Region | Type | Metabolite | Reported Bioactivity | Potential Application |
| 3.1 | Terpene | Clavaric acid | Farnesyltransferase inhibition [23] | Anticancer therapeutics |
| 3.2 | T1PKS | Naphtho-γ-pyrone | Antimicrobial [24] | Food preservatives, antibiotics |
| 6.1 | NRPS | Cyclic depsipeptides | Insecticidal, antimicrobial, anticancer [25] | Biopesticides, drug discovery |
| 10.1 | Indole | Okaramine D | Insecticidal [26] | Agricultural biocontrol |
| 21.4 | Indole, NRPS, NRPS-like (híbrido) | histidyltryptophanyldiketopiperazine/ roquefortine C/ roquefortine D / meleagrine/ glandicoline A/B/ | Neurotoxicity (roquefortines), antimicrobial, cytotoxic, acetylcholinesterase inhibition (meleagrine, glandicolines) [27] | Natural product scaffolds for drug discovery; neuropharmacology; antimicrobial agents (limited due to toxicity) |
| 59.1 | T1PKS | Monascorubrin | Pigmentation, antioxidant [28] | Natural food colorants |
| Identifier | # Unique peptides | # Sequence counts | # Spectrum count | % coverage | Protein Score |
|---|---|---|---|---|---|
| NODE_18_length_653725_cov_92.2.g4952 | 17 | 17 | 93 | 21.4 | 46.99 |
| NODE_60_length_181120_cov_91.6.g9637 | 13 | 13 | 41 | 15.4 | 35.50 |
| NODE_43_length_316552_cov_92.1.g8261 | 11 | 11 | 30 | 16.2 | 25.57 |
| NODE_58_length_211462_cov_89.3.g9479 | 8 | 8 | 14 | 24.2 | 23.12 |
| NODE_44_length_299260_cov_90.2.g8317 | 1 | 1 | 12 | 3.9 | 3.94 |
| NODE_25_length_475153_cov_90.6.g6245 | 4 | 4 | 11 | 6.6 | 11.21 |
| NODE_23_length_521226_cov_91.4.g5957 | 4 | 4 | 9 | 6.5 | 6.95 |
| NODE_86_length_77309_cov_91.69.g10522 | 2 | 2 | 3 | 5.7 | 4.28 |
| NODE_16_length_703737_cov_86.9.g4509 | 1 | 1 | 2 | 2.3 | 3.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





