Submitted:
30 August 2025
Posted:
01 September 2025
You are already at the latest version
Abstract
Carnivorous plants survive in harsh habitats with limited nutrients and low pH. Much focus has been placed on carnivorous trap evolution as the primary mechanism to increase nutrient acquisition through insect digestion. Soil microbiome, however, may also play a pertinent role in nutrient acquisition influencing plant vigor and overall success. Dionaea muscipula, commonly known as the Venus’ Flytrap, is endemic to rims of the Carolina Bays located in southeast North Carolina and northeast South Carolina, where D. muscipula survives in nutrient poor soils with a vestigial root system. We utilized a combination of microscopy, plating, and metagenomics, to investigate the presence/ absence of fungal partners that may contribute to success and vigor of D. muscipula in its native habitat in order to further conservation of this carnivorous plant. Results support that D. muscipula forms both mycorrhizal and fungal endophytic associations, most likely to aid nutrient uptake from otherwise nutrient poor soils, as well as aid in stress defense. Several ectomycorrhizal, endophytic, and saprophytic fungal species were identified from the surrounding rhizosphere of D. muscipula roots presenting a first glimpse into fungal communities that may influence D. muscipula physiology and compose the microbiome of the Carolina Bays ecosystem.

Keywords:
1. Introduction
2. Materials and Methods
2.1. Collection of Roots and Soil
2.2. Microscopy
2.3. Fungal Cultures
2.4. Extraction of Fungal Isolates and Amplification of Fungal DNA
2.5. Rhizosphere Soil Samples
3. Results
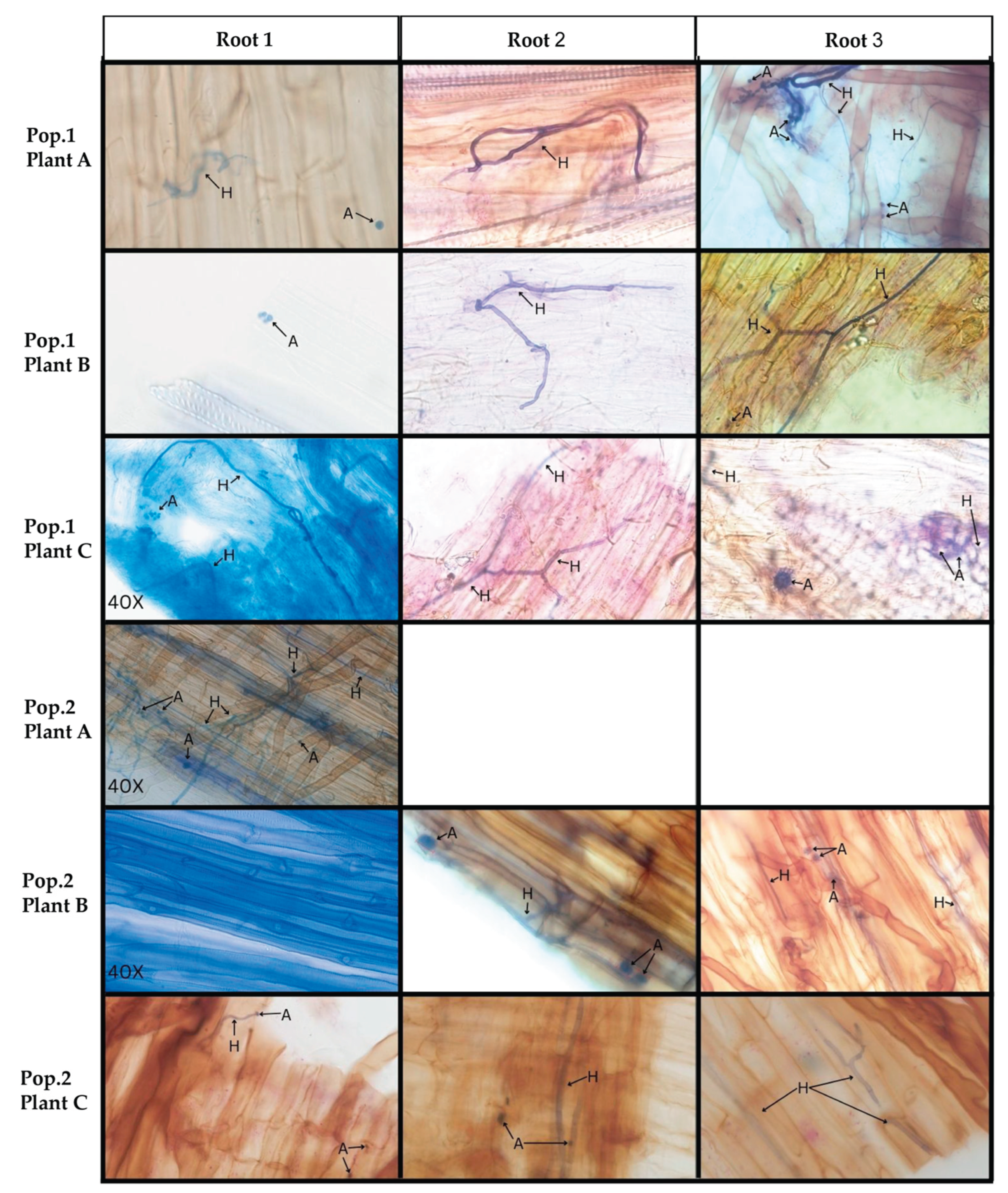
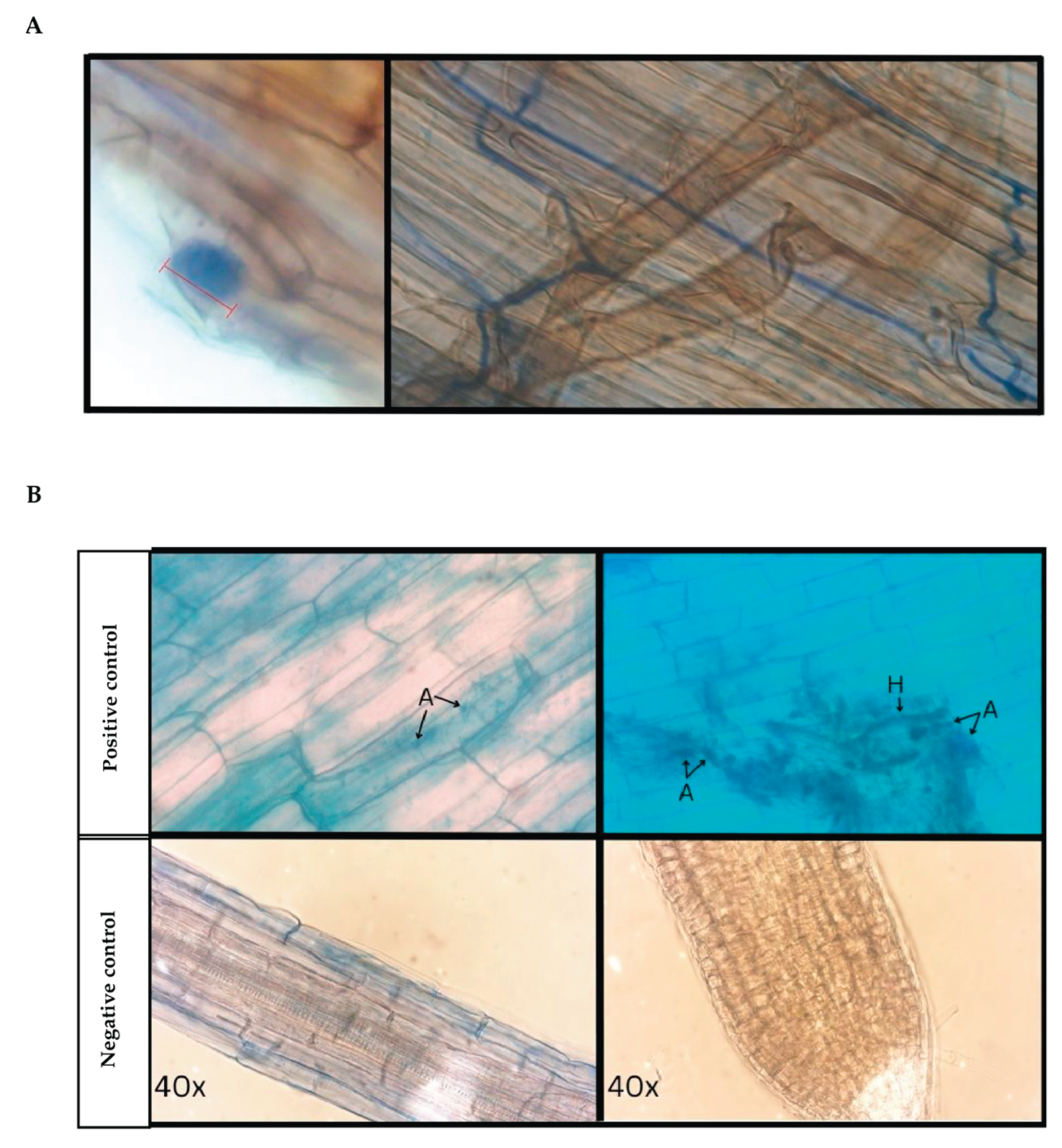
3.1. Microscopy of Trypan Blue Stained D. muscipula Root Tissues
3.2. Fungal Endophytes from D. muscipula Roots
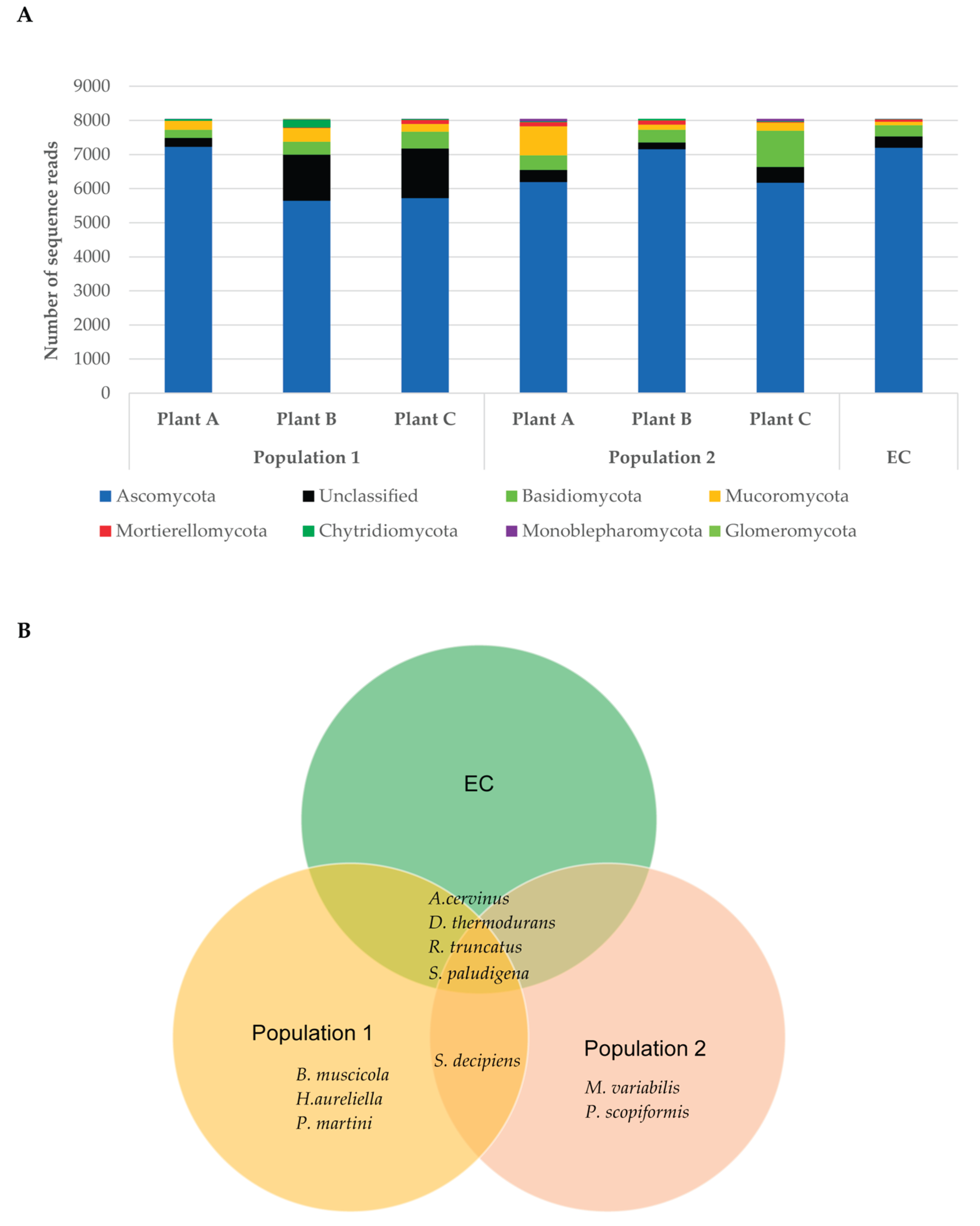
3.3. Fungal Rhizosphere Surrounding Native Venus’ Flytrap Roots of South Carolina
4. Discussion
4.1. Native D. Muscipula Roots Harbor Arbuscular Mycorrhizae
4.2. Fungal Endophytes of Native Venus’ Flytrap Roots
4.3. Soil Rhizome Surrounding Native D. muscipula Roots in South Carolina
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LOBHP | Lewis Ocean Bay Heritage Preserve |
| EC | Environmental Control |
| Pop1 | Population 1 |
| Pop2 | Population 2 |
| Pl. | Plant (A, B, C of each population) |
| PDA | Potato Dextrose Agar |
| AMF | Arbuscular mycorrhizal fungi |
| PEG | Polyethylene Glycol |
| OTU | Operational Taxonomic Unit |
References
- Roberts, P.R.; Oosting, H.J. Responses of Venus FlyTrap (Dionaea muscipula) to factors involved in its endemism. Ecol. Monogr. 1958, 28, 193–218. [Google Scholar] [CrossRef]
- Luken, J.O. Habitats of Dionaea muscipula (Venus’ FlyTrap) Droseraceae, associated with Carolina bays. SENA 2005, 4, 573–584. [Google Scholar] [CrossRef]
- Laliberte, L.; Luken, J.O.; Hutchens, J.J.Jr.; Godwin, K.S. , The ecological boundaries of six Carolina bays: Community compostion and ecotone distribution. Wetlands 2007, 27, 873–883. [Google Scholar] [CrossRef]
- Gray, J.B.; Wentworth, T.R.; Brownie, C. Extinction, colonization, and persistence of rare vascular flora in the longleaf pine- wiregrass ecosystem: Responses to fire frequency and population size. Nat. Areas J. 2003, 23, 210–219. [Google Scholar]
- SC SWAP. Chapter 2: South Carolina’s Priority Species (Species of Greatest Conservation Need). In South Carolina’s Priority Species SC, SC DNR: 2015.
- Garner, B. Successful protection and management efforts keep Venus Flytrap off the Endangered Species List. US Fish and Wildlife 2023.
- Margulies, J.D.; Trost, B.; Hamon, L.; Kerr, N.Z.; Kunz, M.; Randall, J.L.; Shew, R.D.; Shew, D.M.; Starke, L.; Suiter, D.; West, Z. , Expert assessment of illegal collecting impacts on Venus flytraps and priorities for research on illegal trade. Conserv. Biol. 2024, 38, e14320. [Google Scholar] [CrossRef]
- Luken, J.O. , Long-term outcomes of Venus Flytrap (Dionaea muscipula) establishment. Restor. Ecol. 2012, 20, 669–670. [Google Scholar] [CrossRef]
- Humphreys, C.P.; Franks, P.J.; Rees, M.; Bidartondo, M.I.; Leake, J.R.; Beerling, D.J. , Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 2010, 1, 103. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, C.; Wang, G. , Inoculation with arbuscular mycorrhizal fungi improves plant biomass and nitrogen and phosphor rous nutrients: a meta-analysis. BMC Plant Biol. 2024, 24, 960. [Google Scholar] [CrossRef]
- Bermúdez-Contreras, A.I.; Monroy-Guzmán, C.; Pérez-Lucas, L.; Escutia-Sánchez, J.A.; Olmo-Ruiz, M.D.; Truong, C. , Mycor rhizal fungi associated with juniper and oak seedlins along a disturbance gradient in Central Mexico. Front. For. Glob. Change. 2022; 5, 736664. [Google Scholar]
- Leventis, G.; Tsiknia, M.; Feka, M.; Ladikou, C.V.; Papdakis, I.E.; Chatzipavlidis, I.; Papadopoulou, K.; Ehaliotis, C. , Arbuscular mycorrhizal fungi enhance growth of tomato under normal and drought conditions, via different water regulation mechanisms. Rhizosphere 2021, 19, 100394. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, Q.; Chen, Y.; Zhong, C.; Zhang, Y.; Chen, Z.; Pinyopusarerk, K.; Bush, D. , Arbuscular mycorrhizal fungi enhanced growth of Magnoia macclurei (Dandy) Figlar Seedling grown under glasshouse conditions. For. Sci. 2017, 63, 441–448. [Google Scholar]
- Cosme, M.; Fernádez, I.; Van der Heijden, M.G.G.; Pieterse, C.M.J. , Non-mycorrhizal plants: The exceptions that prove the rule. Trends Plant Sci. 2018, 23, 577–587. [Google Scholar] [CrossRef]
- Chandrasekaran, M. , Arbuscular mycorrhizal fungi mediated alleviation of drought stress via non-enzymatic antioxidants: A Meta-analysis. Plants 2022, 11, 2448. [Google Scholar] [CrossRef]
- Stratton, C.A.; Ray, S.; Bradley, B.A.; Kaye, J.P.; Ali, J.G.; Murrell, E.G. , Nutrition vs association: plant defenses are altered by arbuscular mycorrhizal fungi association not by nutritional provisioning alone. BMC Plant Biol. 2022, 22, 400. [Google Scholar] [CrossRef]
- Kawahara, A.; An, G-H. ; Miyakawa, S.; Sonoda, J.; Ezawa, T., Nestedness in arbuscular mycorrhizal fungal communities along soil pH gradients in early primary succession: Acid-tolerant fungi are pH generalists. PLoS One 2016, 11, e0165034. [Google Scholar] [CrossRef]
- Fernádez, I.; Cosme, M.; Stringlis, I.A.; Yu, K.; de Jonge, R.; van Wees, S.M.; Pozo, M.J.; Pieterse, C.M.J.; van der Heijden, M.G.A. , Molecular dialogue between arbuscular mycorrhizal fungi and the nonhost plant Arabidopsis thaliana switches from initial detection to antagonism. New Phytol. 2019, 223, 867–881. [Google Scholar] [CrossRef]
- Adamec, L.; Pavlovič, A. , Mineral nutrition of terrestrial carnivorous plants. In Carnivorous Plants: Physiology, ecology, and evolution, Aaron Ellison, L. A., Ed. Oxford, 2017; online edn, Oxford Academic 2018: 2017; Chapter 17.
- Adamec, L. , Leaf absorption of mineral nutrients in carnivorous plants stimulates root nutrient uptake. New Phytol. 2002, 155, 89–100. [Google Scholar] [CrossRef]
- Adlassnig, W.; Peroutka, M.; Lambers, H.; Lichtscheidl, I.K. , The roots of carnivorous plants. Plant Soil. 2005, 274, 127–140. [Google Scholar] [CrossRef]
- Gao, P.; Loeffler, T.S.; Honsel, A.; Kruse, J.; Krol, E.; Scherzer, S.; Kreuzer, I.; Bemm, F.; Buegger, F.; Burzlaff, T.; Hedrich, R.; Rennenberg, H. , Integration of trap- and root-derived nitrogen nutrition of carnivorous Dionaea muscipula. New Phytol. 2015, 205, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Santiago, Y.; Darnowski, D.W. , Mycorrhizal formation by various carnivorous plants. Carniv. Plant Newsl. 2012, 41, 4–7. [Google Scholar] [CrossRef]
- Gange, A.C.; Eschen, R.; Wearn, J.A.; Thawer, A.; Sutton, B.C. , Differential effects of foliar endophytic fungi on insect herbivores attacking a herbaceous plant. Oecologia 2012, 168, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Cosme, M.; Lu, J.; Erb, M.; Stout, M.J.; Franken, P.; Wurst, S. , A fungal endophyte helps plants to tolerate root herbivory through changes in gibberellin and jasmonate signaling. New Phytol. 2016, 211, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; Lu, Y.; Zhou, L.; Li, N.; Li, Q.; Zhang, Z. , Endophytic Beauveria bassiana promotes plant biomass growth and suppresses pathogen damage by directional recruitment. Front. Microbiol. 2023, 14, 1227269. [Google Scholar] [CrossRef] [PubMed]
- Rodriquez, R.J.; Henson, J.; Vokenburgh, E.V.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.-O.; Redman, R.S. , Stress tolerance in plants via habitat-adapted symbiosis. The ISME Journal: Multidisciplinary Microb. Ecol. 2008, 2, 404–416. [Google Scholar] [CrossRef]
- Quilliam, R.S.; Jones, D.L. , Fungal root endophytes of the carnivorous plant Drosera rotundifolia. Mycorrhiza 2010, 20, 341–348. [Google Scholar] [CrossRef]
- Naseem, F.; Kayang, H. , Endophytic fungal diversity of endemic carnivorous plant Nepenthes khasians in Meghalaya, India. Stud. Fungi 2021, 6, 138–150. [Google Scholar] [CrossRef]
- Rueda-Almazán, J.E.; Hernández, V.M.; Alcalá-Martínez, J.R.; Fernández-Duque, A.; Ruiz-Aguilar, M.; Alcalá, R.E. , Spatial and temporal differences in the community structure of endophytic fungi in the carnivorous plant Pinguicula moranensis (Lentibular iaceae). Fungal Ecol. 2021, 53, 101087. [Google Scholar] [CrossRef]
- Shaw, B. J.P.; Ryves, D.B.; Glanville, H.; Young, E.B.; Millett, J. , Culturable endophytes from carnivorous plant traps in the UK: commonality of endophyte species across host species and sites. Mycol. Prog. 2024. [Google Scholar]
- Sun, P-F. ; Lu, M.R.; Liu, Y-C.; Shaw, B.J. P.; Lin, C-P.; Chen, H-W.; Lin, Y-f.; Hoh, D.Z.; Ke, H-M.; Wang, I-F.; Lu, M-Y.J.; Young, E.B.; Millett, J.; Kirschner, R.; Lin, Y-C.J.; Chen, Y-L.; Tsai, I.J., An acidophilic fungus promotes prey digestion in a carnivorous plant. Nat. Microbiol. 2024, 9, 2522–2537. [Google Scholar]
- Hawkins, H.-J.; Cargill, R. I. M.; Nuland, M. E. V.; Hagen, S. C.; Field, K. J.; Sheldrake, M.; Soudzilovskaia, N. A.; Kiers, E. T. , Mycorrhizal mycelium as a global carbon pool. Curr. Biol. 2023, 33, R560–R573. [Google Scholar] [CrossRef]
- Moukarzel, R.; Ridgway, H.J.; Guerin-Laguette, A.; Jones, E.E. , An improved clearing and staining protocol for evaluation of arbuscular mycorrhizal colonisation in darkly pigmented woody roots. N. Z. Plant Prot. 2020, 73, 33–39. [Google Scholar] [CrossRef]
- Morawetz, J.J. , A clearing protocol for whole tissues: An example using haustoria of Orobanchaceae. Appl. Plant Sci. 2013; 1, 1200361. [Google Scholar]
- Delaux, P-M. ; Varala, K.; Edger, P.P.; Coruzzi, G.M.; Pires, J.C.; Ané, J-M., Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014, (7), e1004487. [Google Scholar]
- Sané, A.K.; Diallo, B.; Kane, A.; Ngom, M.; Cissoko, M.; Sy, M.O. , Response to inoculation with arbuscular mycorrhizal fungi of two tomato (Solanum lycopersicum L.) varieties subjected to water stress under semi-controlled conditions. Agric. Sci. 2022, 13, 790–819. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. , High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLOS One 2012, (7), e40863. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Young, J.P.W. , Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T. L. , BLAST+: architecture and applications. BMC Bioinformatics 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J., Chapter 1: The symbionts forming arbuscular mycorrhizas. In Mycorrhizal Symbiosis, 3rd ed.; Smith, S. E.; Read, D., Eds. Academic Press: London, UK, 2008; pp 13-41.
- Vilela, L. A.F.; Damásio, M.M. , Chapter 41: Molecular and cellular changes of arbuscular mycorrhizal fungi-plant interaction in pesticide contamination. In Handbook of Bioremediation: Physiological, Molecular and Biotechnological Interventions, Prasad, M.H. a. M.N.V., Ed. Academic Press: 2020; pp 649-656.
- Ziane, H.; Hamza, N.; Meddad-Hamza, A. , Arbuscular mycorrhizal fungi and fertilization rates optimize tomato (Solanum lycopersicum L.) growth and yield in a Mediterranean agroecosystem. J. Saudi Soc. Agric. Sci. 2021, 20, 454–458. [Google Scholar] [CrossRef]
- Toghueo, R.M.K.; Boyom, F.F. , Endophytic Penicillium species and their agricultural, biotechnological, and pharmaceutical applications. 3 Biotech 2020, 8, 107. [Google Scholar] [CrossRef]
- MacDougal, D.T. , Symbiotic Saprophytism. Ann. Bot. London 1899, 13, 1–46. [Google Scholar] [CrossRef]
- Venugopal, N.; Devi, K.R. An interesting observation on the mycorrhizal symbiosis in the insectivorous plant, Drosera peltata Sm., in Meghalaya, North-East India. CPN 2007, 36, 9-13.
- Harikumar, V.S. , Are there arbuscular mycorrhizal associations in carnivorous plants Drosera burmanii and D. indica? BOT SERB 2013, 37, 13–19. [Google Scholar]
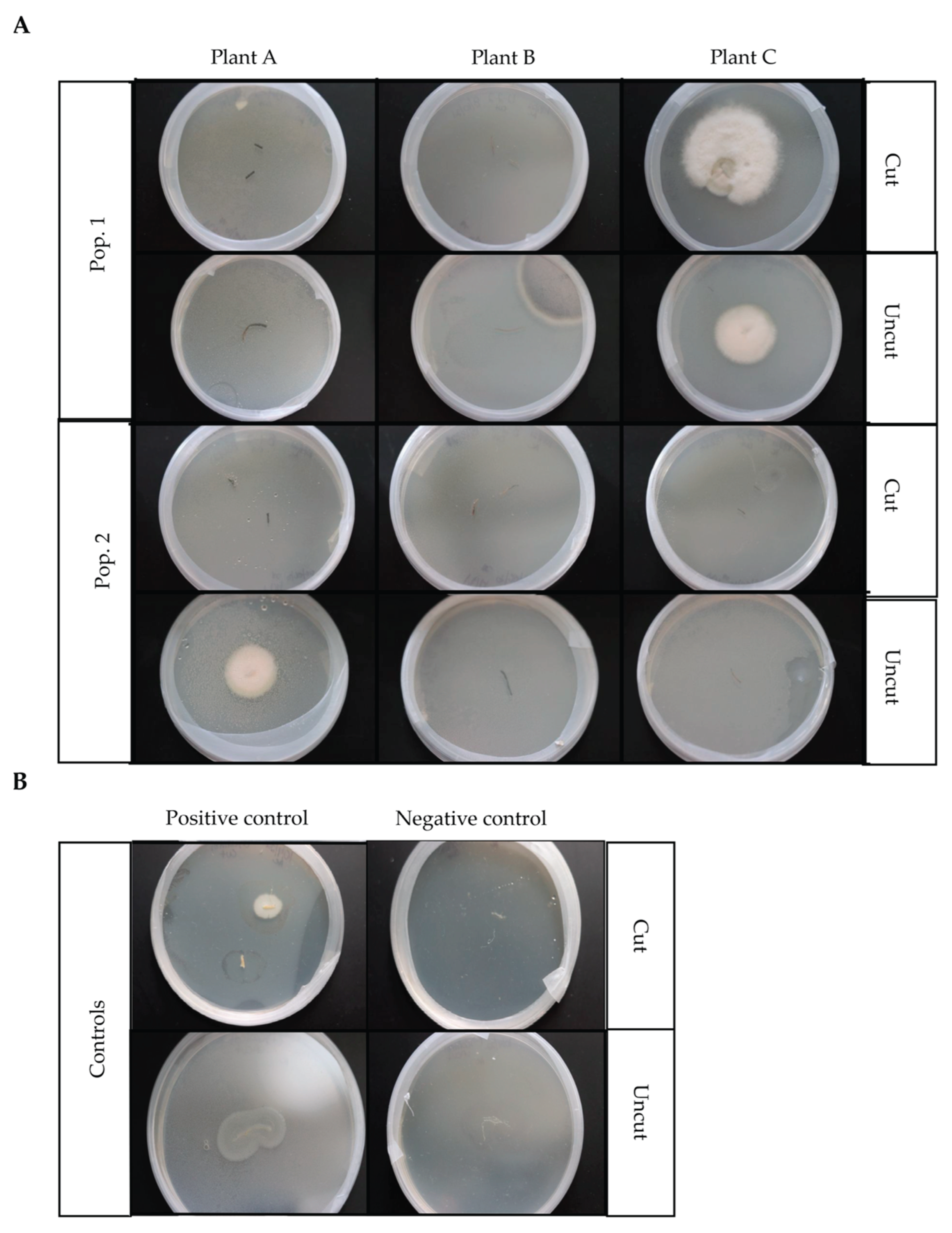
| Plant | Morphotype | Description |
|---|---|---|
| Pop. 1 Plant C cut root | I | Solid, white, fuzzy, circular formation with circles connecting in line, slightly raised |
| Pop. 1 Plant C cut root | II | Branching, transparent white fuzzy circular formation, with little to no center growth, very slightly raised, formed in connecting clusters |
| Pop. 1 Plant C cut root | III | Solid, white, fuzzy, uneven circular formation, raised |
| Pop. 1 Plant C uncut root | I | Solid, white, fuzzy, circular formation with circles connecting in line, slightly raised |
| Pop. 2 Plant A uncut root | IV | Solid, yellowish, fuzzy semicircular formation with translucent outer rim surround the circular formation, slight yellow indent center in circle formation |
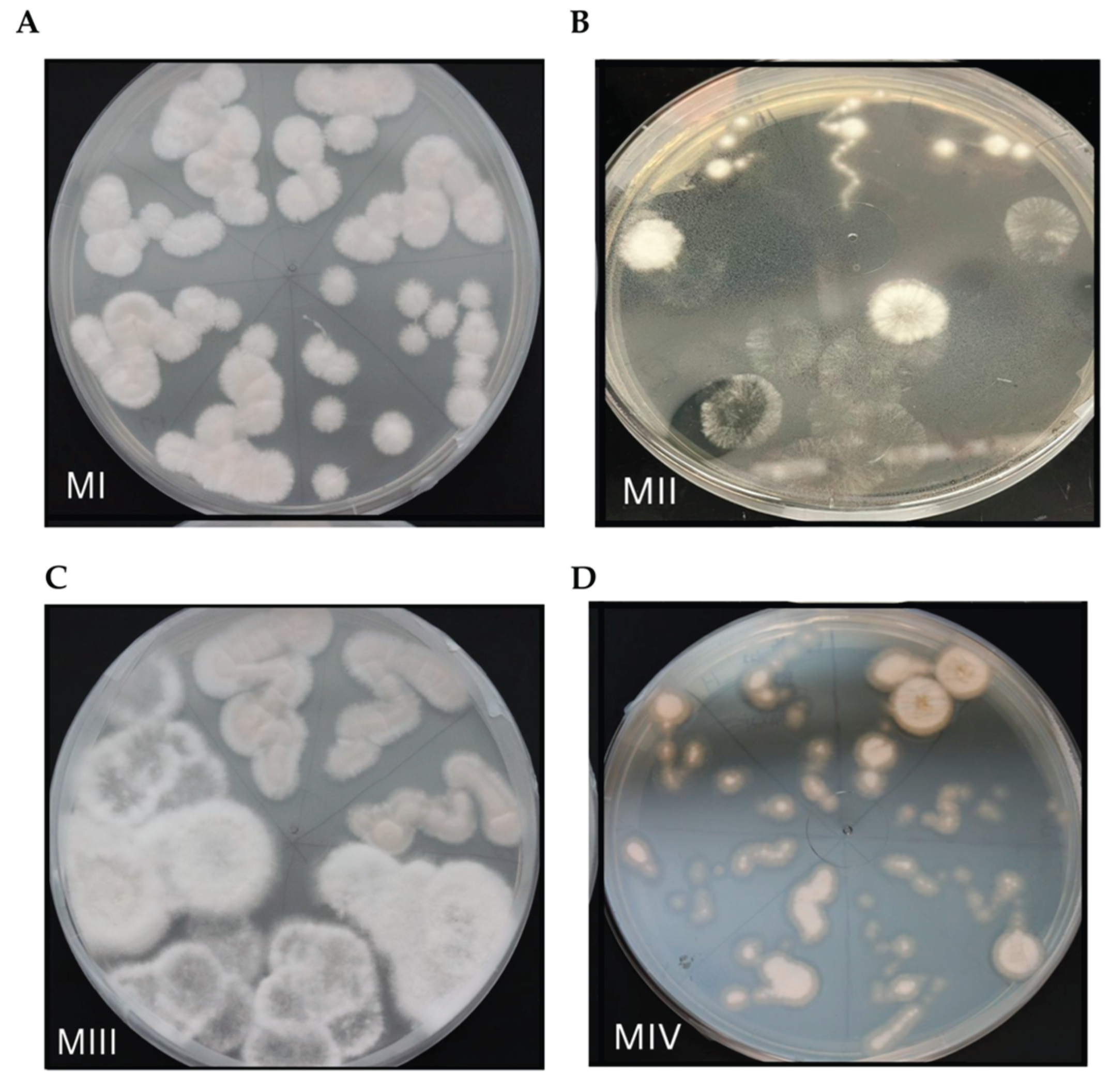
| Morphotype | Species | Phylum | % Identity1 | Region | |
|---|---|---|---|---|---|
| MI | Penicillium rolfsii | Ascomycota | 99.4 | ITS | |
| MII | Neopestalotiopsis sp. | Ascomycota | 96-99.7 | ITS | |
| MIII | Neopestalotiopsis sp. | Ascomycota | 99.4-99.7 | ITS | |
| MIV | Penicillium limosum | Ascomycota | 97.3 | 18s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





