Submitted:
18 August 2025
Posted:
19 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Experimental Strains and Culture Conditions
2.2. Stress Tolerance Analysis of L. Plantarum Strains
- Simulated wine A: 12% (v/v) ethanol, pH 3.60
- Simulated wine B: 10% (v/v) ethanol, pH 3.30
- Simulated wine C: 14% (v/v) ethanol, pH 3.80
2.3. Analysis of L-Malic Acid Content and Viable Bacterial Count
2.4. Analysis of Physicochemical Indices and Organic acid Contents
2.5. Analysis of Anthocyanin Contents and CIELAB Color Parameters
2.6. Analysis of Individual Phenolic Contents in Marselan Wine
2.7. Volatile Compound Analysis
2.8. Statistical Analysis
3. Results and Discussion
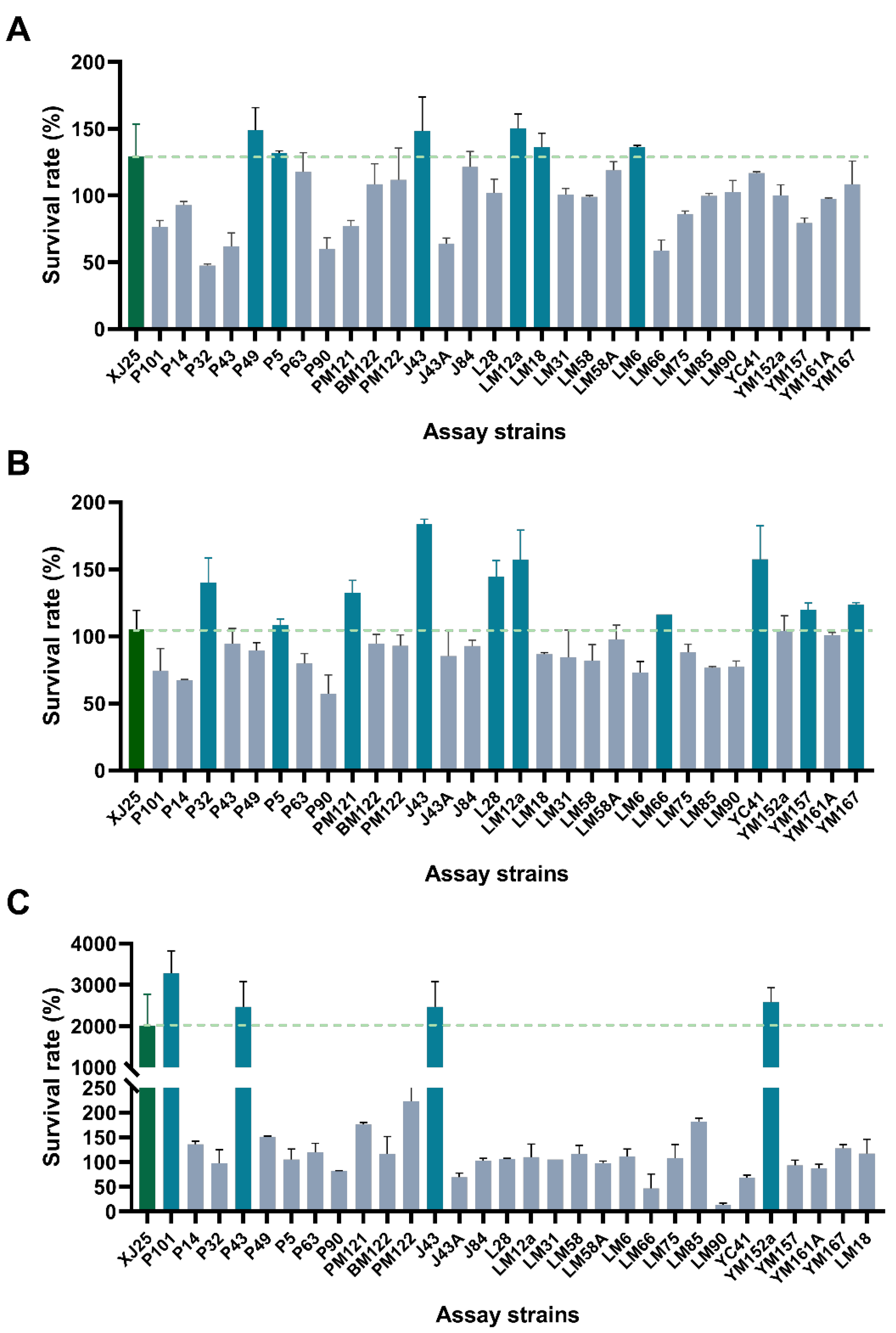
3.1. Combined Stress Tolerance Screening of L. plantarum Strains
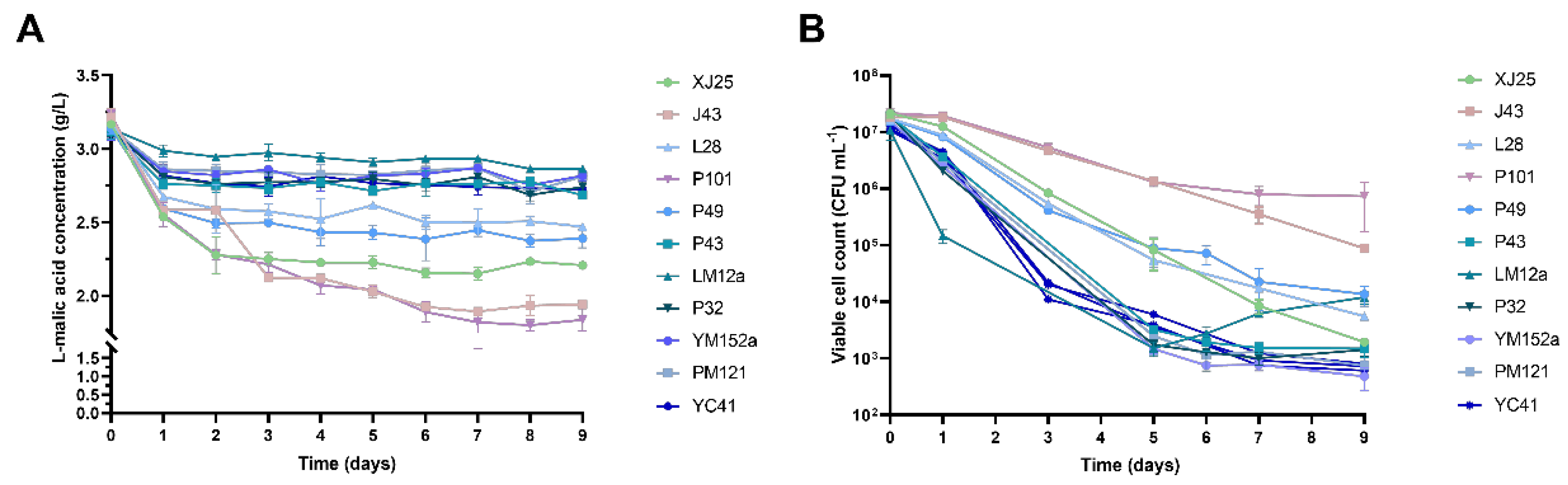
3.2. Viable Cell Counts and L-Malic Acid Consumption During MLF with Different L. plantarum Strains
3.3. Physicochemical Indices of Wines after MLF with Different L. plantarum Strains
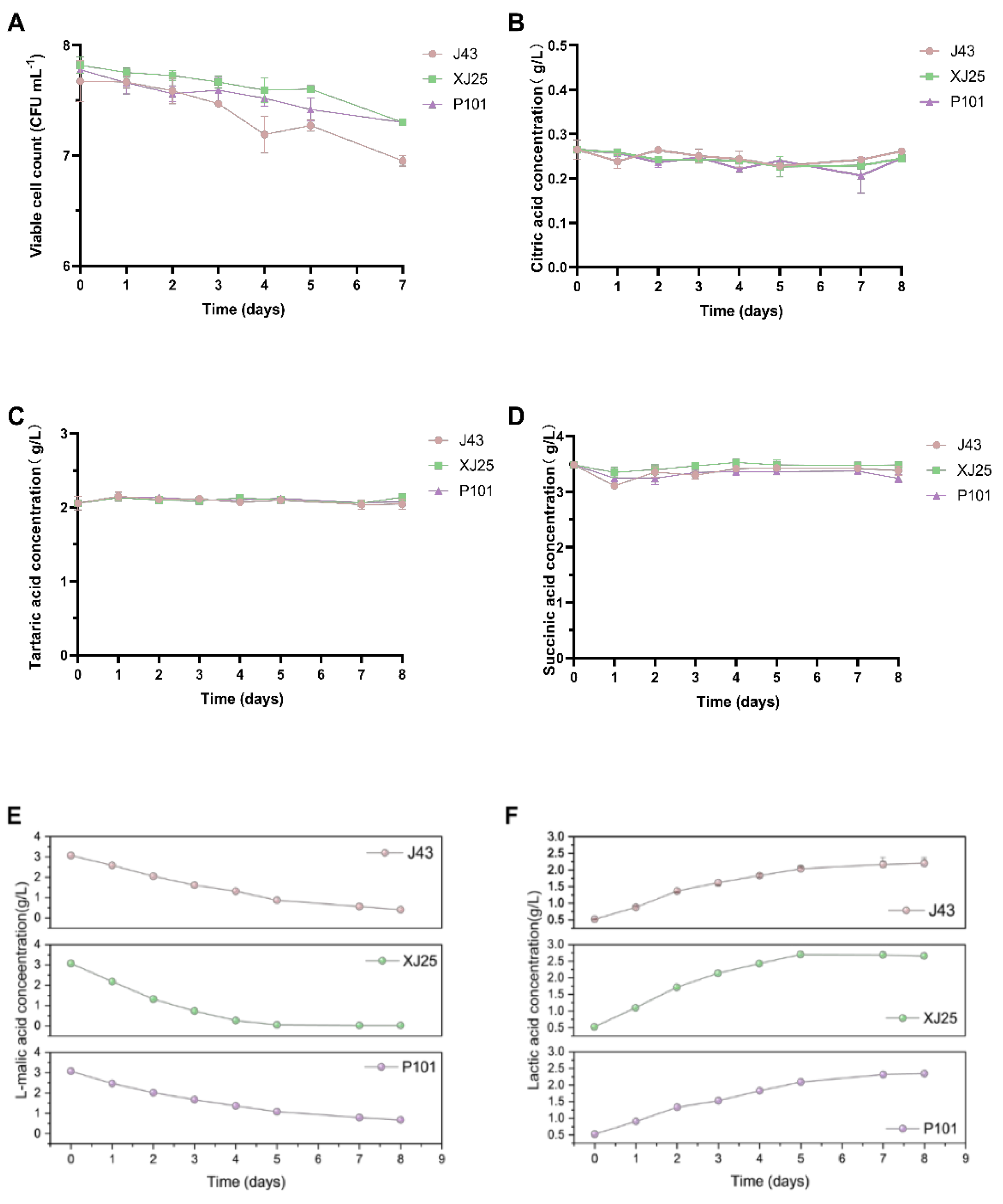
3.4. Changes in Viable L. plantarum Counts and Organic acid Contents in Marselan Wine
3.5. Effects of L. plantarum on Wine Color and Anthocyanin Contents Before and After MLF
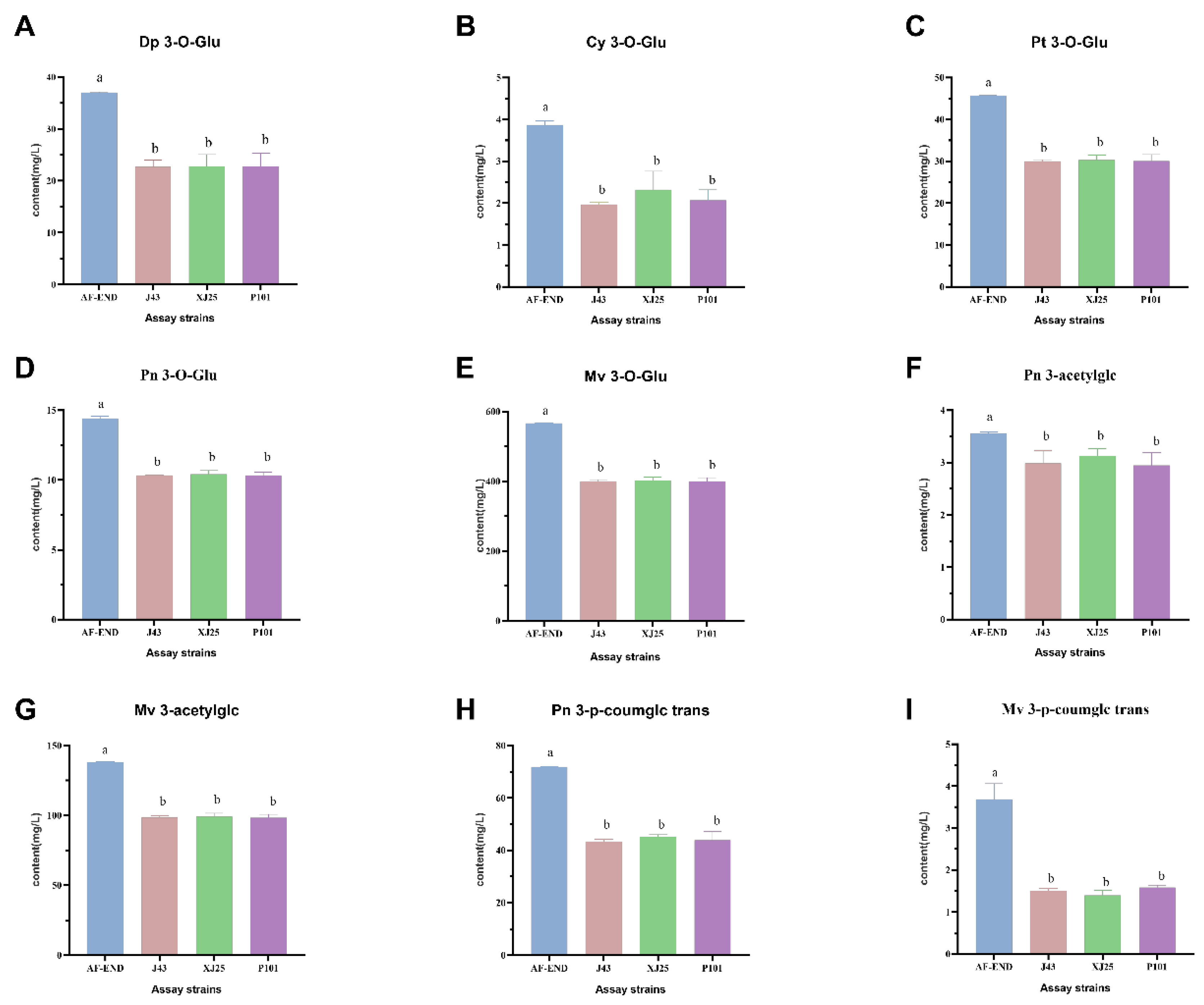
3.6. Individual Phenolic Contents During MLF with Different L. plantarum Strains
3.7. Volatile Compound and PCA Analysis of Marselan Wines Fermented with Different L. plantarum Strains
| Compounds | Aroma Concentration (μg/L) | Thresholds | OAV | Description | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AF-END | J43 | XJ25 | P101 | |||||||
| Esters (20) | ||||||||||
| Ethyl acetate | 31,039.50±187.59d | 33,513.88±331.35c | 37,143.24±107.61b | 39,475.58±2466.94a | 7500 | >1 | Banana, Strawberry | |||
| Ethyl butanoate | 110.39±1.74b | 101.86±1.36c | 111.98±2.62b | 122.11±5.25a | 400 | >0.1 | Strawberry, Banana, Pineapple | |||
| Ethyl 2-methylbutanoate | 1.44±0.33b | 6.71±0.07a | 6.02±0.91a | 0.00±0.00c | >1 | Apple | ||||
| N-propyl propionate | 0.00±0.00b | 0.00±0.00b | 0.00±0.00b | 6.69±0.60a | ||||||
| Ethyl 3-methylbutanoate | 0.70±0.02b | 0.94±0.02a | 0.93±0.03a | 0.92±0.06a | 3 | >0.1 | Strawberry, Sweet Fruity | |||
| Isoamyl acetate | 140.24±4.96a | 131.82±2.54a | 130.62±7.22a | 141.67±0.60a | 160 | >0.1 | Banana, Fruity | |||
| Isobutyl isovalerate | 86.09±1.00a | 85.31±0.04a | 87.45±4.63a | 88.51±3.63a | ||||||
| Ethyl caproate | 183.10±1.95a | 126.95±1.86c | 124.48±1.25c | 145.04±5.67b | 14 | >1 | Green apple, Strawberry | |||
| Ethyl lactate | 5,696.61±109.69c | 8,452.46±72.65b | 10,514.85±890.43a | 10,125.87±617.30a | 14000 | >0.1 | Milk, Butter | |||
| Methyl caprylate | 41.82±1.17a | 0.21±0.04b | 0.01±0.00b | 0.44±0.06b | ||||||
| Ethyl caprylate | 806.04±4.23a | 483.09±10.60c | 408.13±17.75d | 549.00±18.75b | 5 | >1 | Pineapple, Pear, Floral | |||
| Ethyl 3-hydroxybutanoate | 259.54±0.03b | 282.64±3.83a | 278.41±11.78ab | 294.31±14.40a | 200000 | <0.1 | ||||
| Ethyl pelargonate | 12.95±0.15a | 9.89±0.24b | 7.31±0.62c | 9.11±0.37b | 200 | <0.1 | Fruity | |||
| Ethyl caprate | 551.25±10.49a | 417.11±4.64b | 339.74±17.62c | 426.03±8.34b | 200 | >1 | Fruity | |||
| Diethyl succinate | 101.82±1.08b | 112.89±3.12a | 105.02±0.91b | 111.31±0.06a | 6000 | <0.1 | Fruity, Melon | |||
| Ethyl benzoate | 3.99±0.00a | 3.98±0.01a | 3.95±0.01b | 3.96±0.00b | ||||||
| Ethyl 9-decanoate | 0.00±0.00c | 1.09±0.12a | 0.18±0.00bc | 0.32±0.03b | ||||||
| Phenylethyl acetate | 12.41±0.16a | 12.21±0.39ab | 10.97±0.34c | 11.41±0.13bc | 250 | <0.1 | Rose, Sweet | |||
| Ethyl laurate | 9.11±0.00ab | 9.32±0.35a | 8.43±0.31b | 9.37±0.27a | 1500 | <0.1 | Sweet, Beeswax | |||
| Ethyl nonacosanoate | 5.53±0.02a | 5.66±0.07a | 5.64±0.11a | 5.65±0.06a | ||||||
| Alcohols (13) | ||||||||||
| 1-Propanol | 24,180.88±210.70ab | 21,199.51±567.61b | 26,965.99±2592.89a | 26,482.49±1510.11a | 306000 | <0.1 | Mello, Mature fruity, Floral and Green | |||
| Isobutanol | 20,490.93±572.41b | 21,429.31±98.03a | 21,817.10±117.27a | 21,755.57±438.80a | 40000 | >0.1 | Chemical | |||
| 1-Butanol | 1,008.94±5.06b | 1,007.35±0.65b | 1,043.25±15.06a | 1,043.41±3.64a | 150000 | <0.1 | Fruity, Green, Malt, Chemical, Alcohol | |||
| 4-Methyl-2-pentanol | 2,066.00±0.00a | 2,066.00±0.00a | 2,066.00±0.00a | 2,066.00±0.00a | ||||||
| Isoamylol | 59,476.92±258.64a | 50,966.47±129.62c | 26,789.52±816.49d | 57,480.29±1242.33b | 30000 | >1 | Caramel, Lipid | |||
| 4-Methyl-1-pentanol | 20.08±0.04ab | 18.84±0.31c | 19.93±0.16b | 20.47±0.09a | 50000 | <0.1 | ||||
| 3-Methyl-1-pentanol | 16.08±0.13a | 13.59±0.24b | 15.62±0.23a | 16.43±0.75a | 500 | <0.1 | ||||
| Hexyl alcohol | 837.77±1.03b | 766.70±8.22c | 817.96±7.20b | 844.34±14.77a | 8000 | <0.1 | ||||
| 1-Heptanol | 28.59±0.08a | 27.23±0.15b | 27.49±0.03b | 27.74±0.22b | 2500 | <0.1 | ||||
| Octanol | 5.21±0.20a | 3.14±0.01c | 2.84±0.04c | 3.50±0.21b | 40 | >0.1 | Floral | |||
| 1-Decanol | 28.41±2.12c | 52.91±1.00a | 46.23±1.94b | 56.45±2.10a | 400 | >0.1 | Orange, Fatty | |||
| Benzyl alcohol | 537.12±2.66c | 589.82±11.65b | 630.14±23.13a | 618.40±20.07ab | 200000 | <0.1 | Roast, Fruity | |||
| Phenethyl alcohol | 12,456.63±35.15a | 12,143.24±1000.30a | 11,895.77±395.40a | 12,682.42±867.55a | 400 | >1 | Orange, Fatty | |||
| Terpenes (14) | ||||||||||
| Limonene | 0.88±0.07a | 0.30±0.00b | 0.58±0.04ab | 0.77±0.16ab | 10 | >0.1 | Sweet, Citrus, lemon | |||
| Linalool | 21.66±0.01c | 21.83±0.03a | 21.74±0.01b | 21.87±0.03a | 25 | >0.1 | Floral, Citrus | |||
| 1-Octen-3-ol | 15.11±0.10a | 13.56±0.11c | 13.30±0.23c | 14.12±0.34b | ||||||
| (E)-3-Hexenol | 38.01±1.45a | 33.42±0.03b | 35.02±0.37b | 35.19±0.65b | ||||||
| (Z)-Rose oxide | 3.35±0.00a | 3.34±0.00c | 3.35±0.00d | 3.35±0.00b | ||||||
| (Z)-3-Hexenol | 1.64±0.02d | 20.21±0.10b | 17.85±0.56c | 24.47±1.86a | ||||||
| α-Terpineol | 12.36±0.12b | 19.96±6.09a | 12.13±0.03b | 12.70±0.11b | 250 | <0.1 | Lilac | |||
| Nerol | 127.68±1.17c | 145.60±10.33ab | 148.60±2.52a | 133.38±0.52b | 400 | >0.1 | Floral, Green | |||
| Ethyl geranyl ether | 12.12±0.02a | 12.02±0.03a | 12.09±0.10a | 12.16±0.11a | ||||||
| Terpinen-4-ol | 9.88±0.02b | 12.39±1.68a | 10.38±0.05ab | 10.38±0.03ab | ||||||
| (E)-Beta-farnesene | 3,219.34±38.03a | 2,256.72±40.30c | 1,935.72±64.89d | 2,417.84±61.92b | ||||||
| Citronellol | 8.40±0.35a | 6.87±0.69b | 5.95±0.16b | 6.50±0.09b | 100 | <0.1 | Green, Lilac, Rose | |||
| Nerolido | 9.83±0.02a | 9.95±0.21a | 9.97±0.27a | 9.83±0.02a | 400 | <0.1 | Green, Floral | |||
| Dodecatrienol | 81.28±0.00b | 83.38±1.62ab | 85.22±0.95a | 82.61±0.35b | ||||||
| Aldehydes and Ketones (5) | ||||||||||
| 2-Hexenal | 401.26±1.77a | 244.66±11.87c | 237.31±2.94c | 291.43±16.09b | ||||||
| Nonanal | 0.02±0.01a | 0.00±0.00b | 0.00±0.00b | 0.01±0.01b | 2.5 | <0.1 | Citrus | |||
| α-Ionone | 10.54±0.25a | 9.96±0.19ab | 8.74±0.10c | 9.40±0.67bc | ||||||
| (E)-5-Butyldihydro-4-methyl-2(3H)-furanone | 227.48±0.01c | 227.84±0.15a | 227.46±0.12c | 227.69±0.09ab | ||||||
| Ionone | 9.45±0.01a | 9.47±0.01a | 9.45±0.01a | 9.45±0.00a | ||||||
| Volatile Phenols(4) | ||||||||||
| Phenol | 16.16±0.01a | 15.48±0.73ab | 15.07±0.10b | 15.27±0.01b | 30 | >0.1 | ||||
| 4-Ethylguaiacol | 125.70±0.03b | 125.56±0.02c | 125.62±0.03c | 125.79±0.04a | ||||||
| 4-Ethyl-pheno | 66.10±0.02a | 66.16±0.09a | 66.12±0.04a | 66.21±0.03a | ||||||
| 2,4-Di-t-butylphenol | 177.36±0.10a | 179.34±5.71a | 185.39±8.94a | 181.06±2.40a | ||||||
| Fatty acids (5) | ||||||||||
| Propanoic acid | 8,232.04±120.08b | 8,853.37±137.58a | 8,869.48±261.78a | 8,880.14±243.06a | ||||||
| Isovaleric acid | 80.37±65.62c | 196.47±16.12b | 302.09±15.53a | 202.13±16.33b | 3000 | <0.1 | Sour, Cheese | |||
| Octanoic acid | 353.73±7.14b | 498.00±65.80a | 435.19±14.94ab | 501.68±30.48a | 500 | >1 | Sour, Cheese, Fatty | |||
| Decanoic acid | 188.28±2.28c | 245.13±17.88ab | 232.35±5.00b | 255.88±5.69a | 1000 | >0.1 | Sour, Fatty | |||
| Benzoic acid | 23,418.69±3.04a | 23,416.63±5.84a | 23,423.12±9.65a | 23,433.27±9.87a | ||||||
| Other (1) | ||||||||||
| 2-Methoxy-3-isobutyl pyrazine | 0.94±0.00a | 0.93±0.00b | 0.93±0.00b | 0.93±0.00b | ||||||
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daeschel, M.A.; Jung, D.S.; Watson, B.T. Controlling Wine Malolactic Fermentation with Nisin and Nisin-Resistant Strains of Leuconostoc oenos. Appl Environ Microbiol 1991, 57, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.C.; Richelieu, M. Control of flavor development in wine during and after malolactic fermentation by Oenococcus oeni. Appl Environ Microbiol 1999, 65, 740–745. [Google Scholar] [CrossRef]
- Sun, J.; Ge, Y.; Gu, X.; Li, R.; Ma, W.; Jin, G. Identification and Characterization of Malolactic Bacteria Isolated from the Eastern Foothills of Helan Mountain in China. Foods 2022, 11. [Google Scholar] [CrossRef]
- Capozzi, V.; Tufariello, M.; De Simone, N.; Fragasso, M.; Grieco, F. Biodiversity of Oenological Lactic Acid Bacteria: Species- and Strain-Dependent Plus/Minus Effects on Wine Quality and Safety. Fermentation 2021, 7(1), 24. [Google Scholar] [CrossRef]
- Virdis, C.; Sumby, K.; Bartowsky, E.; Jiranek, V. Lactic Acid Bacteria in Wine: Technological Advances and Evaluation of Their Functional Role. Front Microbiol 2020, 11, 612118. [Google Scholar] [CrossRef]
- Arevalo-Villena, M.; Bartowsky, E.J.; Capone, D.; Sefton, M.A. Production of indole by wine-associated microorganisms under oenological conditions. Food Microbiol 2010, 27, 685–690. [Google Scholar] [CrossRef]
- Tofalo, R.; Battistelli, N.; Perpetuini, G.; Valbonetti, L.; Rossetti, A.P.; Perla, C.; Zulli, C.; Arfelli, G. Oenococcus oeni Lifestyle Modulates Wine Volatilome and Malolactic Fermentation Outcome. Front Microbiol 2021, 12, 736789. [Google Scholar] [CrossRef]
- E, G.A.; López, I.; Ruiz, J.I.; Sáenz, J.; Fernández, E.; Zarazaga, M.; Dizy, M.; Torres, C.; Ruiz-Larrea, F. High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 2004, 230, 53–61. [Google Scholar] [CrossRef]
- Chen, Q.; Hao, N.; Zhao, L.; Yang, X.; Yuan, Y.; Zhao, Y.; Wang, F.; Qiu, Z.; He, L.; Shi, K.; et al. Comparative functional analysis of malate metabolism genes in Oenococcus oeni and Lactiplantibacillus plantarum at low pH and their roles in acid stress response. Food Res Int 2022, 157, 111235. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhao, H.; Zhang, C.; Yu, J.; Lu, Z. Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. J Agric Food Chem 2013, 61, 11676–11682. [Google Scholar] [CrossRef]
- Knoll, C.; Divol, B.; du Toit, M. Genetic screening of lactic acid bacteria of oenological origin for bacteriocin-encoding genes. Food Microbiol 2008, 25, 983–991. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl Environ Microbiol 2004, 70, 5715–5731. [Google Scholar] [CrossRef]
- Mtshali, P.S.; Divol, B.; van Rensburg, P.; du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J Appl Microbiol 2010, 108, 1389–1397. [Google Scholar] [CrossRef]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Implications of new research and technologies for malolactic fermentation in wine. Appl Microbiol Biotechnol 2014, 98, 19, 8111–32. [Google Scholar] [CrossRef]
- Hu, L.; Chen, X.; Cao, Y.; Gao, P.; Xu, T.; Xiong, D.; Zhao, Z. Lactiplantibacillus plantarum exerts strain-specific effects on malolactic fermentation, antioxidant activity, and aroma profile of apple cider. Food Chem: X 2024, 23, 101575. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Cai, H.; Kou, J.; Xie, Y.; Yao, X.; Li, J.; Zhou, P.; He, F.; Duan, C.; Pan, Q.; et al. Variety-specific flavor characteristics in the Shandong region: Interaction between fermentation and variety. Food Chem 2025, 478, 143707. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, X.; Ren, Y.; Xuan, X.; Pervaiz, T.; Shangguan, L.; Fang, J. Geographical location influence 'Cabernet Franc' fruit quality in Shandong province. Sci Rep 2024, 14, 2382. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Benavent-Gil, Y.; Navascués, E.; Calvo, A.; Albors, C.; Pardo, I.; Ferrer, S. Lowering histamine formation in a red Ribera del Duero wine (Spain) by using an indigenous O. oeni strain as a malolactic starter. Int J Food Microbiol 2017, 244, 11–18. [Google Scholar] [CrossRef]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Curilén, Y.; Delfederico, L.; Caballero, A.; Semorile, L.; Pozo-Bayón, M.; Tymczyszyn, E.E. Advantages of Using Blend Cultures of Native L. plantarum and O. oeni Strains to Induce Malolactic Fermentation of Patagonian Malbec Wine. Front Microbiol 2018, 9, 2109. [Google Scholar] [CrossRef]
- Franquès, J.; Araque, I.; El Khoury, M.; Lucas, P.M.; Reguant, C.; Bordons, A. Selection and characterization of autochthonous strains of Oenococcus oeni for vinification in Priorat (Catalonia, Spain). OENO One 2018, 52. [Google Scholar] [CrossRef]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J Appl Microbiol 2015, 118, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Romero, J.; Ilabaca, C.; Ruiz, M.; Jara, C. Oenococcus oeni in Chilean Red Wines: Technological and Genomic Characterization. Front Microbiol 2018, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Selection of autochthonous Oenococcus oeni strains according to their oenological properties and vinification results. Int J Food Microbiol 2010, 137, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, N.; Perpetuini, G.; Perla, C.; Arfelli, G.; Zulli, C.; Rossetti, A.P.; Tofalo, R. Characterization of natural Oenococcus oeni strains for Montepulciano d’Abruzzo organic wine production. Eur Food Res Technol 2020, 246, 1031–1039. [Google Scholar] [CrossRef]
- Liu, X.; Fu, J.; Ma, W.; Jin, G. Screening and evaluation of high stress tolerance, high esterase activity and safety of Oenococcus oeni strains adapt to challenging conditions in Northwest China wine. LWT--Food Sci Technol 2024, 213, 116975. [Google Scholar] [CrossRef]
- Meng, Q.; Yuan, Y.; Li, Y.; Wu, S.; Shi, K.; Liu, S. Optimization of Electrotransformation Parameters and Engineered Promoters for Lactobacillus plantarum from Wine. ACS Synth Biol 2021, 10, 1728–1738. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, D.; Liu, H.; Shen, J.; Zhang, J.; He, L.; Li, J.; Zhou, P.; Guan, X.; Liu, S.; et al. Impact of indigenous Oenococcus oeni and Lactiplantibacillus plantarum species co-culture on Cabernet Sauvignon wine malolactic fermentation: Kinetic parameters, color and aroma. Food Chem: X 2024, 22, 101369. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, S.; He, L.; Tian, Y. Draft Genome Sequence of Lactobacillus plantarum XJ25 Isolated from Chinese Red Wine. G enome announc 2016, 4. [Google Scholar] [CrossRef]
- Betteridge, A.; Grbin, P.; Jiranek, V. Improving Oenococcus oeni to overcome challenges of wine malolactic fermentation. Trends Biotechnol 2015, 33, 547–553. [Google Scholar] [CrossRef]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; Valdes La Hens, D.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron J Biotechnol 2019, 38, 10–18. [Google Scholar] [CrossRef]
- du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: the Next Generation of Malolactic Fermentation Starter Cultures—an Overview. Food Bioprocess Technol 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Davis, C.R.; Wibowo, D.; Fleet, G.H.; Lee, T.H. Properties of Wine Lactic Acid Bacteria: Their Potential Enological Significance. Am J Enol Vitic 1988, 39, 137. [Google Scholar] [CrossRef]
- Sauer, M.; Russmayer, H.; Grabherr, R.; Peterbauer, C.K.; Marx, H. The Efficient Clade: Lactic Acid Bacteria for Industrial Chemical Production. Trends Biotechnol 2017, 35, 756–769. [Google Scholar] [CrossRef]
- Pereira, R.; Mohamed, E.T.; Radi, M.S.; Herrgård, M.J.; Feist, A.M.; Nielsen, J.; Chen, Y. Elucidating aromatic acid tolerance at low pH in Saccharomyces cerevisiae using adaptive laboratory evolution. Proc Natl Acad Sci 2020, 117, 27954–27961. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Pian, R.; Xing, Y.; Zhou, W.; Yang, F.; Chen, X.; Zhang, Q. Effects of citric acid on fermentation characteristics and bacterial diversity of Amomum villosum silage. Bioresour Technol 2020, 307, 123290. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Zhao, H.; Gu, P.; Chen, Y.; Zhang, B.; Zhu, B. Acetaldehyde released by Lactobacillus plantarum enhances accumulation of pyranoanthocyanins in wine during malolactic fermentation. Food Res Int 2018, 108, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, D.; Liu, H.; Shen, J.; Zhang, J.; He, L.; Li, J.; Zhou, P.; Guan, X.; Liu, S.; et al. Impact of indigenous Oenococcus oeni and Lactiplantibacillus plantarum species co-culture on Cabernet Sauvignon wine malolactic fermentation: Kinetic parameters, color and aroma. Food Chem: X 2024, 22, 101369. [Google Scholar] [CrossRef]
- He, F.; Liang, N.N.; Mu, L.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Anthocyanins and their variation in red wines I. Monomeric anthocyanins and their color expression. Molecules 2012, 17, 1571–1601. [Google Scholar] [CrossRef]
- Sacchi, K.L.; Bisson, L.F.; Adams, D.O. A Review of the Effect of Winemaking Techniques on Phenolic Extraction in Red Wines. Am J Enol Vitic 2005, 56, 197. [Google Scholar] [CrossRef]
- Ginjom, I.R.; D'Arcy, B.R.; Caffin, N.A.; Gidley, M.J. Phenolic contents and antioxidant activities of major Australian red wines throughout the winemaking process. J Agric Food Chem 2010, 58, 10133–10142. [Google Scholar] [CrossRef]
- Philippe, C.; Chaïb, A.; Jaomanjaka, F.; Cluzet, S.; Lagarde, A.; Ballestra, P.; Decendit, A.; Petrel, M.; Claisse, O.; Goulet, A.; et al. Wine Phenolic Compounds Differently Affect the Host-Killing Activity of Two Lytic Bacteriophages Infecting the Lactic Acid Bacterium Oenococcus oeni. Viruses 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, D.; Ivanov, Y.; Chengolova, Z.; Godjevargova, T. Antimicrobial Potential, Antioxidant Activity, and Phenolic Content of Grape Seed Extracts from Four Grape Varieties. Microorganisms 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Míguez, M.J.; Cacho, J.F.; Ferreira, V.; Vicario, I.M.; Heredia, F.J. Volatile components of Zalema white wines. Food Chem 2007, 100, 1464–1473. [Google Scholar] [CrossRef]
- Aznar, M.; Arroyo, T. Analysis of wine volatile profile by purge-and-trap-gas chromatography-mass spectrometry. Application to the analysis of red and white wines from different Spanish regions. J Chromatogr A 2007, 1165, 151–157. [Google Scholar] [CrossRef]
- Rocha, S.l.M.; Rodrigues, F.; Coutinho, P.; Delgadillo, I.; Coimbra, M.A. Volatile composition of Baga red wine: Assessment of the identification of the would-be impact odourants. Anal Chim Acta 2004, 513, 257–262. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Liu, X.; Bian, X.; Li, J.; Meng, N.; Liu, M.; Huang, M.; Sun, B.; Li, J. Flavor Interactions in Wine: Current Status and Future Directions From Interdisplinary and Crossmodal Perspectives. Compr Rev Food Sci Food Saf 2025, 24, e70199. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Yan, X.; Wang, Q.; Zhang, Y.; Tao, Y. Performance of selected P. fermentans and its excellular enzyme in co-inoculation with S. cerevisiae for wine aroma enhancement. LWT--Food Sci Technol 2017, 86, 361–370. [Google Scholar] [CrossRef]
- GB/T 15038-2006; Analytical Methods ofWine and Fruit Wine. China Standard Publishing House: Beijing, China, 2006.
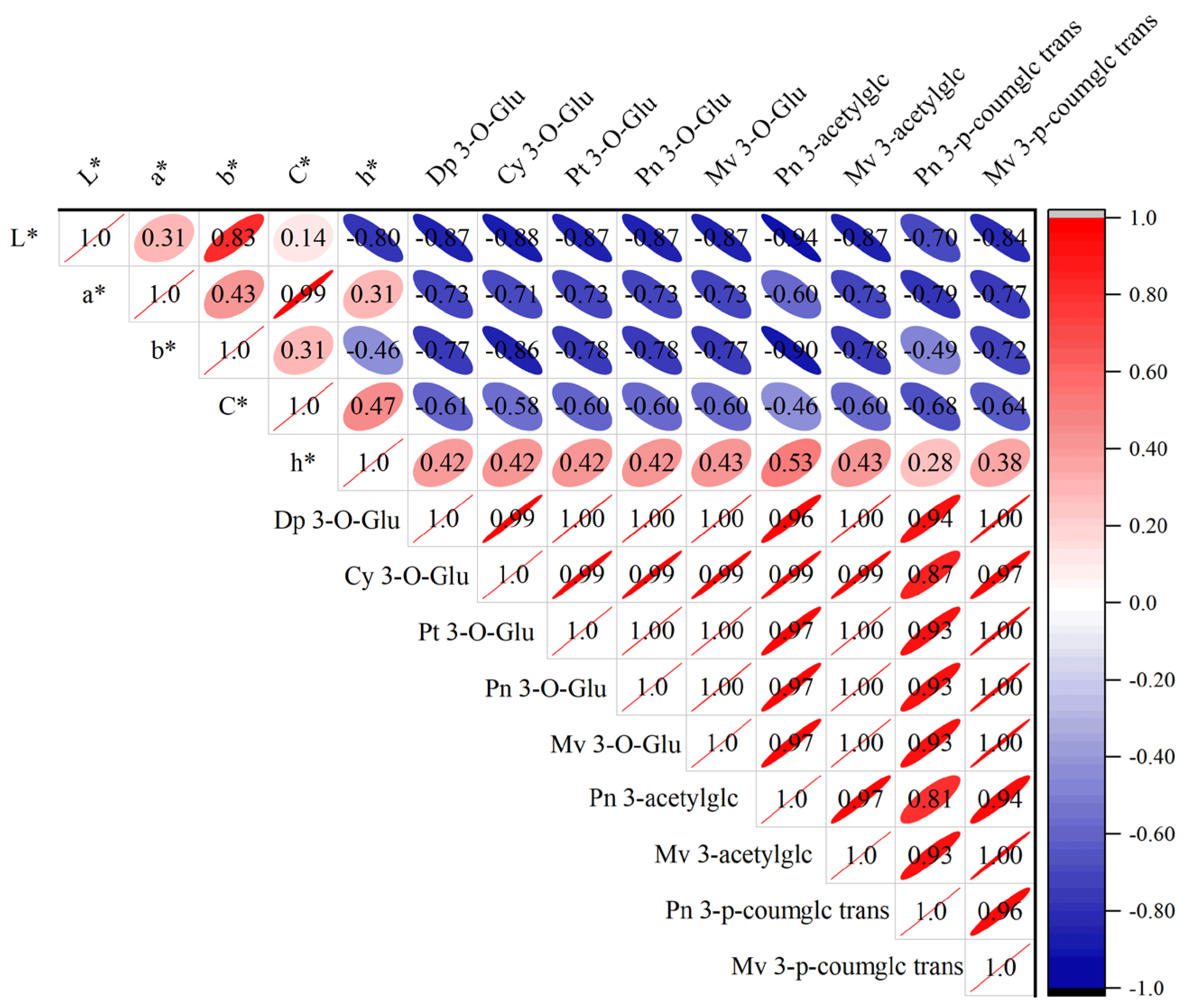
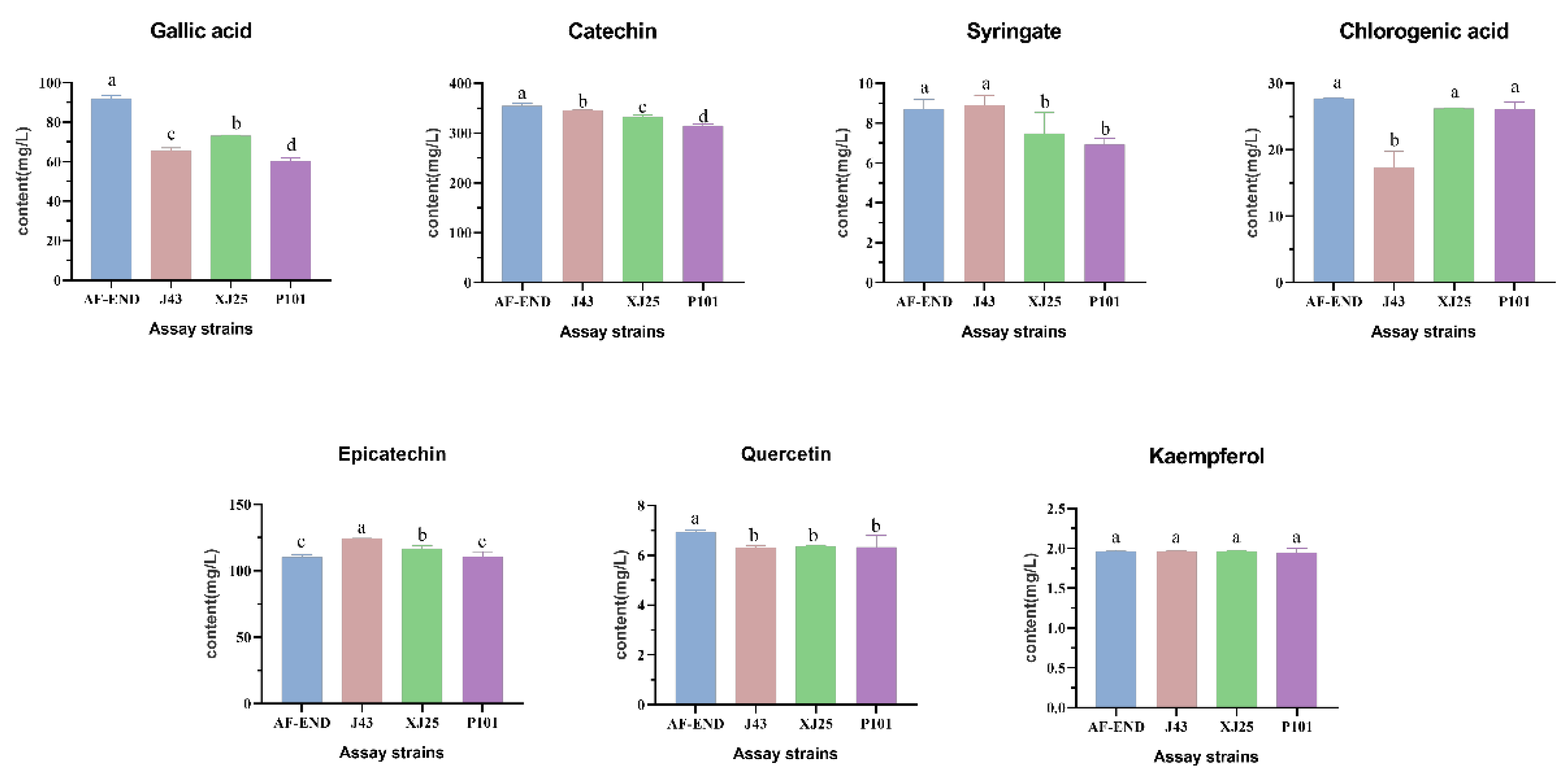
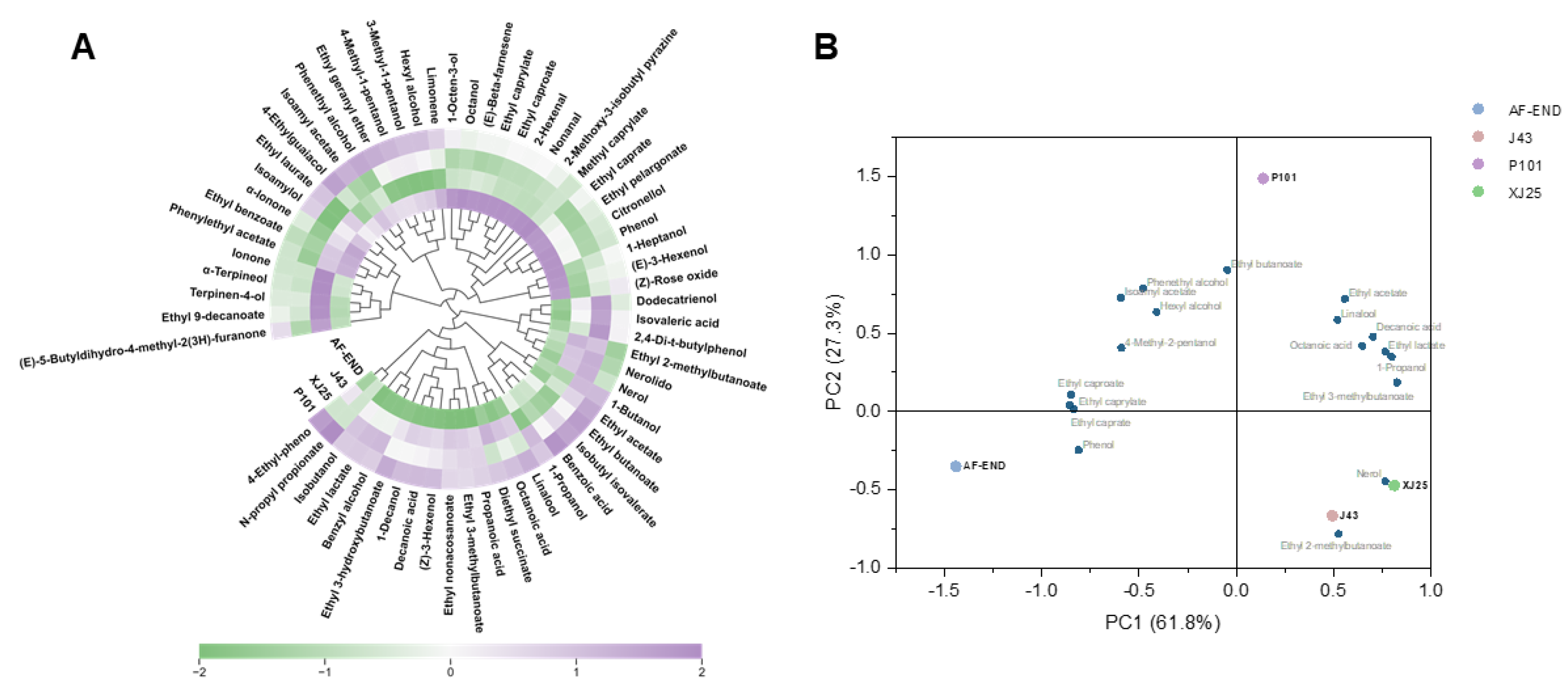
| Strain | pH | Total Acid g/L |
Glucose g/L |
Fructose g/L |
Glycerin g/L |
Alcohol (%) (v/v) |
Total Phenol mg/L |
|---|---|---|---|---|---|---|---|
| AF-END | 3.61±0.00c | 7.04±0.15a | 1.39±0.01c | 7.72±0.10a | 4.85±0.14a | 14.26±0.08a | 3,504.96±186.20a |
| J43 | 3.76±0.00b | 5.90±0.17b | 1.37±0.03c | 6.24±0.17b | 4.34±0.10b | 13.68±0.38b | 3,047.19±74.38b |
| XJ25 | 3.79±0.00a | 5.30±0.34c | 1.54±0.02a | 5.93±0.06b | 4.42±0.03b | 14.02±0.06ab | 3,411.33±57.70a |
| P101 | 3.79±0.00a | 5.06±0.29c | 1.46±0.04b | 6.06±0.30b | 4.41±0.02b | 13.99±0.10ab | 3,264.50±168.12ab |
| Strain | L* | a* | b* | C*ab | h*ab | △E*ab |
|---|---|---|---|---|---|---|
| AF-END | 16.86±0.21c | 45.25±0.35a | 20.53±0.26a | 49.68±0.42a | 0.43±0.00a | - |
| J43 | 18.26±0.67bc | 46.54±1.03a | 21.58±1.32a | 51.04±1.25a | 0.43±0.00a | 2.17±0.25b |
| XJ25 | 18.13±0.62bc | 46.41±0.86a | 20.85±0.63a | 50.88±1.04a | 0.42±0.00a | 1.75±0.22c |
| P101 | 19.20±1.22a | 45.57±3.51a | 21.50±0.60a | 49.75±4.12a | 0.41±0.01a | 2.55±0.40a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





