Submitted:
18 August 2025
Posted:
27 August 2025
You are already at the latest version
Abstract
Natural wines represent a new trend in winemaking without use of preservatives and starter cultures, revealing unique quality traits of grapes, wine and terroir, but are susceptible to spoilage or undesirable fermentations. This study aims to highlight the diversity and succession of microbiota of natural wines, from the vineyard (grapes) to mature, fermented wines and the effects of different grape varieties, production stages and equipment. Samples of “Limniona”, “Malagouzia” and “Roditis” grape varieties, initial and fermented must, filtered and unfiltered natural Limniona wines were analyzed in order to enumerate key groups of microorganisms and identify beneficial yeasts and bacteria of alcoholic and malolactic fermentation, respectively, as well as potential marker of off-flavors. Although beneficial fermentation microorganisms (especially Saccharomyces yeasts) were scarce in initial grape, where other contaminants or wild yeasts were present, gradually, as fermentation progresses, there was a prevalence of Saccharomyces cerevisiae strains of increased diversity in matured wine, as well as several lactic acid bacteria (LAB) of malolactic fermentation, mostly Lactobacillus and Oenococcus, and other bacteria from environmental sources, irrelevant to alcoholic/malolactic fermentation or spoilage, like Burkholderia. The type of vessel affected the type of LAB that prevail, with an abundance of Oenococcus in clay vessels, versus Lactobacillus species in stainless steel vessels. Notably, some Lactobacillus species like L. parafarraginis can be linked to off-flavors if they represent a high percentage of the wine microbiota. These findings highlight the importance of understanding, monitoring and controling microbial succession during production stages, in order to prevent sensory faults and ensure stable quality of natural wines.

Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Analysis
2.2.1. Microbiological Analysis
2.2.2. Bacterial Isolation from Selective Media
2.3. MALDI-TOF MS Analysis
2.4. NGS-DNA Extraction and Microbial Community Profiling
2.5. Statistical Analysis
3. Results
3.1. Microbial Populations
3.2. MALDI-TOF MS Analysis
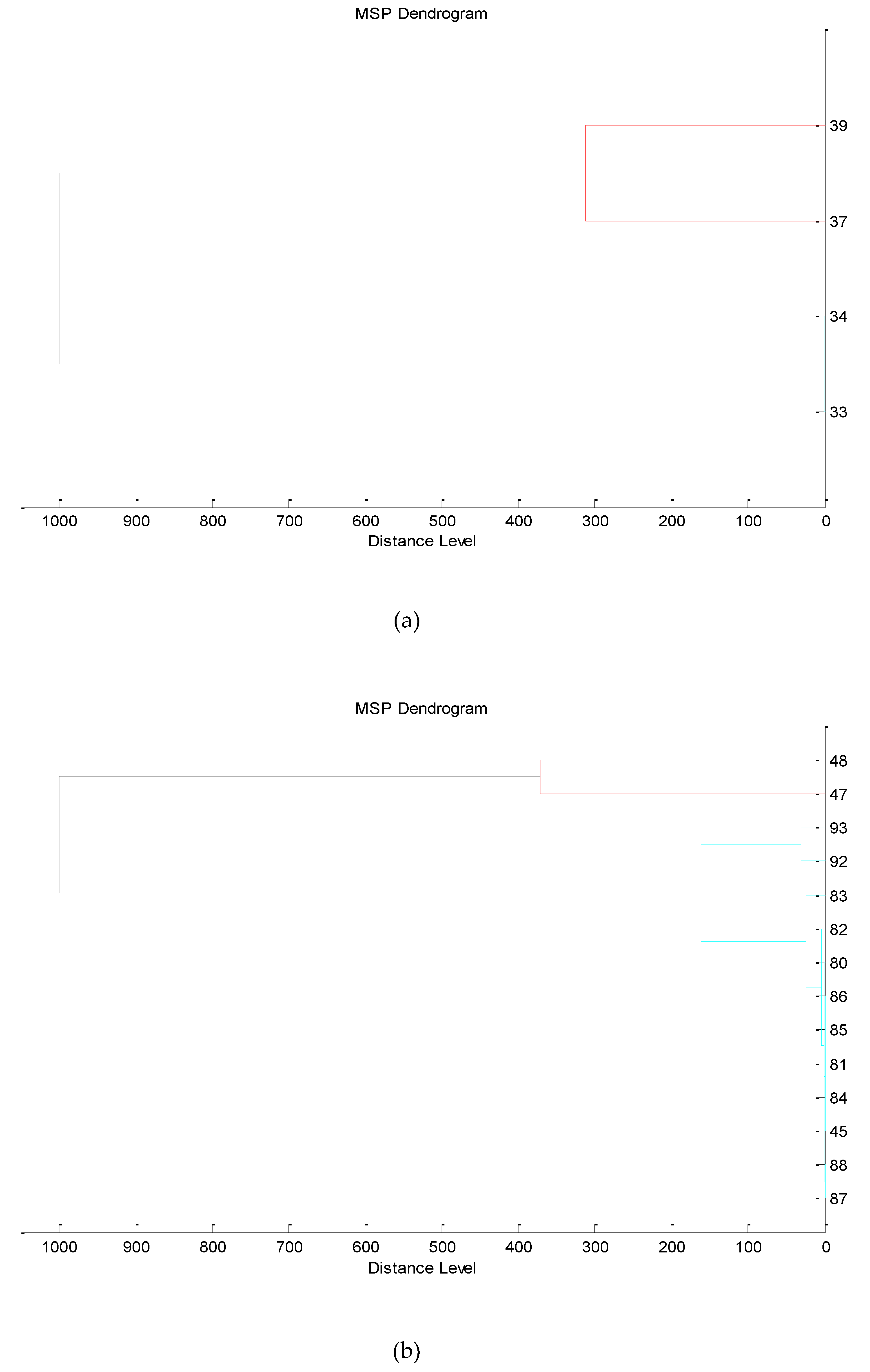
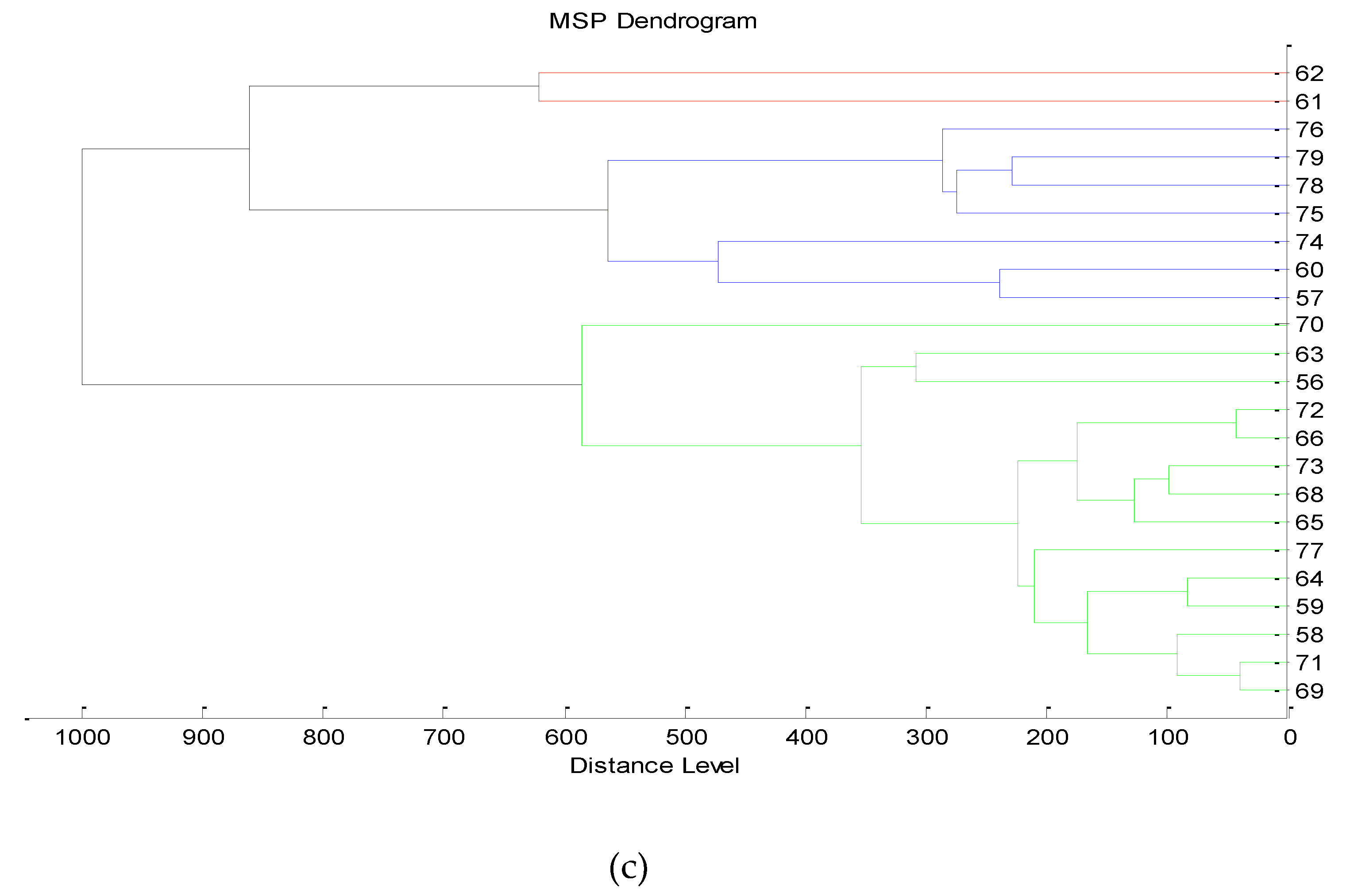
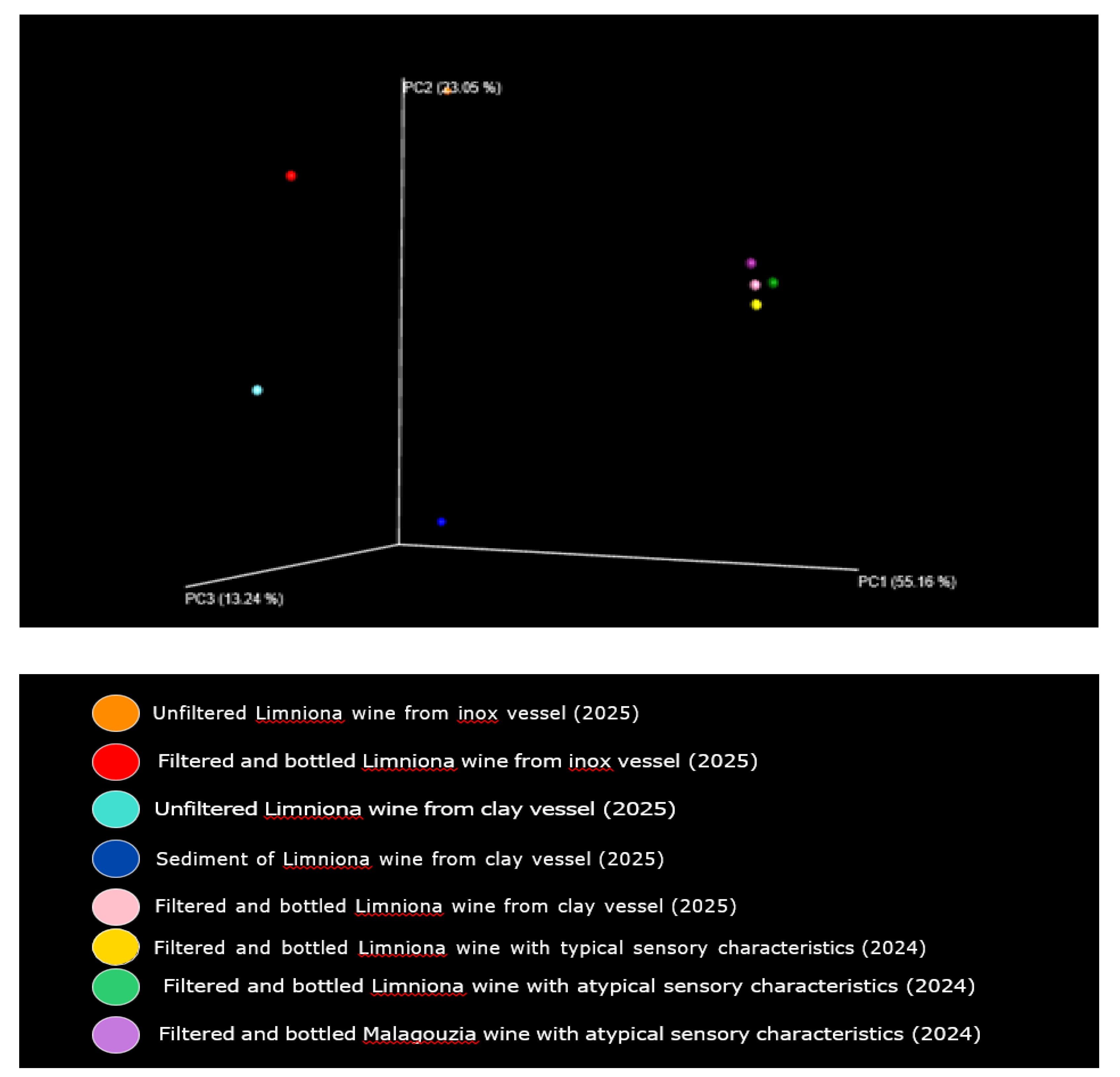
3.3. Next-Generation Sequencing
| Unfiltered wine from clay vessel | Filtered and bottled wine from clay vessel | Sediment from clay vessel | ||||||
|---|---|---|---|---|---|---|---|---|
| Genus | Species | % of valid reads | Genus | Species | % of valid reads | Genus | Species | % of valid reads |
| Oenococcus | ND | 22.72 | Lactobacillus | ND | 29 | Oenococcus | oeni | 30.6 |
| Arcobacter | oeni | 19.48 | Lactobacillus | parafarraginis | 25.1 | Arcobacter | venerupis | 8.78 |
| Arcobacter | ND | 16.5 | Lactobacillus | diolivorans | 13.09 | Arcobacter | defluvii | 7.65 |
| Buttiauxella | parvulus | 6.11 | Burkholderiaceae | ND | 3.14 | Buttiauxella | warmboldiae | 5.21 |
| Arcobacter | ND | 3 | Oenococcus | oeni | 1.37 | Arcobacter | ellisii | 2.78 |
| Acetobacter | ND | 2 | Citrobacter | gillenii | 1.7 | |||
4. Discussion
4.1. Microbial Succession and Community Dynamics During Winemaking: Insights from Selective Media Profiling
4.2. Microbial Diversity Across Winemaking Stages
4.3. Intraspecific Variation in Saccharomyces cerevisiae
4.4. Taxonomic Profiling and Microbial Diversity in Wine Samples via 16S rRNA NGS
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ilieva, F.; Petrov, K.; Veličkovska, S. K.; Gunova, N.; Dimovska, V.; Rocha, J. M. F.; Esatbeyoglu, T. Influence of autochthonous and commercial yeast strains on fermentation and quality of wines produced from Vranec and Cabernet Sauvignon grape varieties from Tikveš wine-growing region, Republic of North Macedonia. Appl. Sci. 2021, 11, 6135. [Google Scholar] [CrossRef]
- Sapardani, E.; Melfou, K.; Chatzitheodoridis, F.; Kontogeorgos, A. Technical and scale efficiency of farms producing grapes for wine. KnE Soc. Sci. 2023, 8, 102–118. [Google Scholar] [CrossRef]
- Koufos, G. C.; Mavromatis, T.; Koundouras, S.; Fyllas, N. M.; Theocharis, S.; Jones, G. V. Greek wine quality assessment and relationships with climate: Trends, future projections and uncertainties. Water 2022, 14, 573. [Google Scholar] [CrossRef]
- Wei, R.; Wang, L.; Ding, Y.; Zhang, L.; Gao, F.; Chen, N.; Wang, H. Natural and sustainable wine: A review. Crit. Rev. Food Sci. Nutr. Association des Vins Méthode Nature. Vin Méthode Nature: Charte Vin Méthode Nature. 2020. https://www.vinmethodenature.org/. 2023, 63, 8249–8260. [Google Scholar] [CrossRef]
- Vecchio, R.; Parga-Dans, E.; Alonso González, P.; Annunziata, A. Why consumers drink natural wine? Consumer perception and information about natural wine. Agric. Food Econ. 2021, 9, 22. [Google Scholar] [CrossRef]
- Lappa, I. K.; Kachrimanidou, V.; Pateraki, C.; Koulougliotis, D.; Eriotou, E.; Kopsahelis, N. Indigenous yeasts: Emerging trends and challenges in winemaking. Curr. Opin. Food Sci. 2020, 32, 133–143. [Google Scholar] [CrossRef]
- Eller, M. R.; de Almeida, E. L. M.; Campos, L. B.; Montoya, S. G. Wild wines: The state of the art. In Trending Topics on Fermented Foods; Elsevier: 2024; pp. 341–369.
- Legodi, L. M.; Lekganyane, M. A.; Moganedi, K. L. M. Morula tree: From fruit to wine through spontaneous fermentation and the potential of deriving other value-added products. Processes 2022, 10, 1633. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Stasinou, V.; Tzamourani, A.; Kotseridis, Y.; Dimopoulou, M. Malolactic fermentation—Theoretical advances and practical considerations. Fermentation 2022, 8, 521. [Google Scholar] [CrossRef]
- Mendes Ferreira, A.; Mendes-Faia, A. The role of yeasts and lactic acid bacteria on the metabolism of organic acids during winemaking. Foods 2020, 9, 1231. [Google Scholar] [CrossRef]
- Devi, A.; Anu-Appaiah, K. A.; Lin, T. F. Timing of inoculation of Oenococcus oeni and Lactobacillus plantarum in mixed malo-lactic culture along with compatible native yeast influences the polyphenolic, volatile and sensory profile of the Shiraz wines. LWT 2022, 158, 113130. [Google Scholar] [CrossRef]
- Englezos, V.; Jolly, N. P.; Di Gianvito, P.; Rantsiou, K.; Cocolin, L. Microbial interactions in winemaking: ecological aspects and effect on wine quality. Trends Food Sci. Technol. 2022, 127, 99–113. [Google Scholar] [CrossRef]
- Gonzalez, R.; Morales, P. Truth in wine yeast. Microb. Biotechnol. Article in press. 2022. [Google Scholar] [CrossRef]
- Goode, J. The Science of Wine: From Vine to Glass, 3rd ed.; Mitchell Beazley: London, UK, 2021. [Google Scholar]
- Eller, M. R.; de Almeida, E. L. M.; Campos, L. B.; Montoya, S. G. Wild wines: The state of the art. In Trending Topics on Fermented Foods; Elsevier: 2024; pp. 341–369.
- Wei, R.; Wang, L.; Ding, Y.; Zhang, L.; Gao, F.; Chen, N.; Wang, H. Natural and sustainable wine: a review. Crit. Rev. Food Sci. Nutr. 2023, 63, 8249–8260. [Google Scholar] [CrossRef] [PubMed]
- Maykish, A.; Rex, R.; Sikalidis, A. K. Organic winemaking and its subsets: biodynamic, natural, and clean wine in California. Foods 2021, 10, 1975. [Google Scholar] [CrossRef]
- Hu, S. Natural Wine: The Conundrum; 2020. https://www.researchgate. 2020. Available online: https://www.researchgate.net/publication/347470167_Natural_Wine_The_Conundrum.
- Kwofie, M. K.; Bukari, N.; Adeboye, O. Probiotics potential of yeast and lactic acid bacteria fermented foods and the impact of processing: a review of indigenous and continental food products. Adv. Microbiol. 2020, 10, 771–793. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A. T.; Dahunsi, S. O.; Olayanju, A. Synergistic microbial interactions between lactic acid bacteria and yeasts during production of Nigerian indigenous fermented foods and beverages. Food Control 2020, 110, 106963. [Google Scholar] [CrossRef]
- Maicas, S. The role of yeasts in fermentation processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- Franco, W.; Benavides, S.; Valencia, P.; Ramírez, C.; Urtubia, A. Native yeasts and lactic acid bacteria isolated from spontaneous fermentation of seven grape cultivars from the Maule region (Chile). Foods 2021, 10, 1737. [Google Scholar] [CrossRef]
- Torres-Guardado, R.; Esteve-Zarzoso, B.; Reguant, C.; Bordons, A. Microbial interactions in alcoholic beverages. Int. Microbiol. 2022, 25, 1–15. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Bauer, F.F. Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 2015, 6, 1358. [Google Scholar] [CrossRef] [PubMed]
- Kántor, A.; Mareček, J.; Ivanišová, E.; Terentjeva, M.; Kačániová, M. Microorganisms of grape berries. Proc. Latv. Acad. Sci. 2017, 71, 502–507. [Google Scholar] [CrossRef]
- Renouf, V.; Claisse, O.; Lonvaud-Funel, A. Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 2007, 75, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Martini, A.; Ciani, M.; Scorzetti, G.; Scorzetti, G. Direct enumeration and isolation of wine yeasts from grape surfaces. Am. J. Enol. Vitic. 1996, 47, 435–440. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 2016, 7, e00631–e16. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F. Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 2015, 1, 1–6. [Google Scholar] [CrossRef]
- Hierro, N.; González, Á.; Mas, A.; Guillamón, J.M. Diversity and evolution of non-Saccharomyces yeast populations during alcoholic fermentation: Effect of grape ripeness and cold maceration. FEMS Yeast Res. 2006, 6, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. The microorganisms of winemaking—Isolation, enumeration and identification. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Chur, Switzerland, 1993; pp. 1–25. [Google Scholar]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From vineyard soil to wine fermentation: Microbiome approximations to explain the 'terroir' concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef] [PubMed]
- Kántor, A.; Kačániová, M. Diversity of bacteria and yeasts on the surface of table grapes. Sci. Pap. Anim. Sci. Biotechnol. 2015, 48, 149–154. [Google Scholar]
- Renouf, V.; Claisse, O.; Lonvaud-Funel, A.L. Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Aust. J. Grape Wine Res. 2005, 11, 316–327. [Google Scholar] [CrossRef]
- Ghanbarzadeh, B.; Sampaio, J.P.; Arzanlou, M. Biodiversity of epiphytic yeasts on post-harvest table grapes in markets of Tabriz, Iran. Mycol. Iran. 2019, 6, 101–111. [Google Scholar]
- Koulougliotis, D.; Eriotou, E. Isolation and identification of endogenous yeast strains in grapes and must solids of Mavrodafni Kefalonias and antioxidant activity of the produced red wine. Ferment. Technol. 2016, 5, 1. [Google Scholar] [CrossRef]
- Drumonde-Neves, J.; Čadež, N.; Reyes-Domínguez, Y.; Gallmetzer, A.; Schuller, D.; Lima, T.; Franco-Duarte, R. Clavispora santaluciae sp. nov., a novel ascomycetous yeast species isolated from grapes. Int. J. Syst. Evol. Microbiol. 2020, 70, 6307–6312. [Google Scholar] [CrossRef]
- Kurtzman, C. P.; Fell, J. W. The yeasts, a taxonomic study, Ed.; Elsevier: New York, NY, USA, 1998. [Google Scholar]
- Cabañes, F. animals. In Pathogenic Yeasts; Springer: Berlin/Heidelberg, Germany, 2009; pp. 253–279. [Google Scholar]
- Campaniello, D.; Sinigaglia, M. Wine spoiling phenomena. In The Microbiological Quality of Food; Woodhead Publishing: Cambridge, UK, 2017; pp. 237–255. [Google Scholar]
- Benavent-Gil, Y.; Berbegal, C.; Lucio, O.; Pardo, I.; Ferrer, S. A new fear in wine: Isolation of Staphylococcus epidermidis histamine producer. Food Control 2016, 62, 142–149. [Google Scholar] [CrossRef]
- Nisiotou, A. A.; Rantsiou, K.; Iliopoulos, V.; Cocolin, L.; Nychas, G. J. E. Bacterial species associated with sound and Botrytis-infected grapes from a Greek vineyard. Int. J. Food Microbiol. 2011, 145, 432–436. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed]
- Maulani, S.; Hosseini, S. M.; Elikaie, A.; Mirnurollahi, S. M. Isolated microorganisms from Iranian grapes and its derivatives. Iran. J. Microbiol. 2012, 4, 25–32. [Google Scholar] [PubMed]
- Lorenzini, M.; Zapparoli, G. Epiphytic bacteria from withered grapes and their antagonistic effects on grape-rotting fungi. Int. J. Food Microbiol. 2020, 319, 108505. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, S.; Wang, D.; Xue, J.; Guo, D.; Song, X.; Luan, C. Complete genome sequence of Bacillus amyloliquefaciens B15 isolated from grape skin, a strain of strong inhibitory activity against fungi. J. Biotechnol. 2016, 228, 28–29. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Kang, X.; Ma, X.; Ran, L.; Zhen, Z. New strain of Bacillus amyloliquefaciens G1 as a potential downy mildew biocontrol agent for grape. Agronomy 2024, 14, 1532. [Google Scholar] [CrossRef]
- Guo, J.; Ma, G.; Zhu, H.; Fu, J. Isolation and identification of a Bacillus amyloliquefaciens strain against grape downy mildew and optimization for its liquid fermentation medium. In Advances in Applied Biotechnology; Springer: Heidelberg, Germany, 2015; pp. 409–418. [Google Scholar]
- Le Jeune, C.; Erny, C.; Demuyter, C.; Lollier, M. Evolution of the population of Saccharomyces cerevisiae from grape to wine in a spontaneous fermentation. Food Microbiol. 2006, 23, 709–716. [Google Scholar] [CrossRef]
- Martini, A. Origin and domestication of the wine yeast Saccharomyces cerevisiae. J. Wine Res. 1993, 4, 165–176. [Google Scholar] [CrossRef]
- Cappello, M. S.; Bleve, G.; Grieco, F.; Dellaglio, F.; Zacheo, G. Characterization of S. cerevisiae strains isolated from must of grapes grown in an experimental vineyard. J. Appl. Microbiol. 2004, 97, 1274–1280. [Google Scholar] [CrossRef]
- de Ponzzes-Gomes, C. M.; Cadete, R. M.; Soares, M. A.; Barbosa Júnior, A. M.; Rocha, K. K.; de Souza Graça, A.; Rosa, C. A. Isolation of indigenous S. cerevisiae strains from fermented must of wine grapes in a tropical semiarid region of Brazil. Braz. J. Microbiol. 2025, in press.
- Romano, P.; Capece, A.; Jespersen, L. Taxonomic and ecological diversity of food and beverage yeasts. In Yeasts in Food and Beverages, 2nd ed.; Querol, A., Fleet, G. H., Eds.; Springer: Berlin, Germany, 2006; pp. 13–53. [Google Scholar]
- Liu, P. T.; Lu, L.; Duan, C. Q.; Yan, G. L. The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of Cabernet Sauvignon wines by spontaneous fermentation. LWT–Food Sci. Technol. 2016, 71, 356–363. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F. Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 2015, 1, 1–6. [Google Scholar] [CrossRef]
- Fleet, G. H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Egas, C.; Gomes, A. C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef]
- Bartowsky, E. J.; Borneman, A. R. Genomic variations of Oenococcus oeni strains and the potential to impact malolactic fermentation and aroma compounds in wine. Appl. Microbiol. Biotechnol. 2011, 92, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L. M. T.; Endo, A. Taxonomic status of lactic acid bacteria in wine and key characteristics to differentiate species. S. Afr. J. Enol. Vitic. 2009, 30, 72–90. [Google Scholar]
- Lonvaud-Funel, A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie van Leeuwenhoek 1999, 76, 1–4. [Google Scholar] [CrossRef]
- Hall, M.E. , and Wilcox, W.F. Identification and frequencies of endophytic microbes within healthy grape berries. Am. J. Enol. Viticul., 2019, 70, 212–219. [Google Scholar] [CrossRef]
- Esmaeel, Q. , Jacquard, C., Sanchez, L., Clément, C., & Ait Barka, E. The mode of action of plant associated Burkholderia against grey mould disease in grapevine revealed through traits and genomic analyses. Sci. Rep. 2020, 10, 19393. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. (Eds.) Handbook of Enology, Volume 1: The Microbiology of Wine and Vinifications, 2nd ed.; Wiley: Chichester, UK, 2006. [Google Scholar]
- Capozzi, V.; Garofalo, C.; Chiriatti, M. A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2011, 166, 675–683. [Google Scholar] [CrossRef]
- Parish, M. E.; Fleet, G. H. Wine. In Food Microbiology: Fundamentals and Frontiers; Doyle, M. P.; Klaenhammer, T. R.; Deibel, R. H., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 915–947.
- Bae, S.; Fleet, G. H.; Heard, G. M. Lactic acid bacteria associated with wine grapes from several Australian vineyards. J. Appl. Microbiol. 2006, 100, 712–727. [Google Scholar] [CrossRef] [PubMed]
| Prematured grape varieties and regions | Microbial groups/Substrate | |||
|---|---|---|---|---|
| Lactococci (M17 agar) | Lactobacilli (MRS agar) | Yeasts (PDA agar) | Yeasts (WORT agar | |
| Roditis Mourko | 1.00±0.29 | <DL | 1±0.00 | 1.16±0.27 |
| Malagouzia Raches | <DL | <DL | <DL | <DL |
| Malagouzia Damaki | <DL | <DL | <DL | <DL |
| Limniona Zisi | 1.39±0.36 | <DL | 1.44±0.42 | <DL |
| Limniona Polia | 1.10±0.74 | 1.33±0.28 | 1.50±0.87 | 1.83±1.44 |
| Sampling stages | Microbial groups | |||
|---|---|---|---|---|
| Lactococci (M17 agar) | Lactobacilli (MRS agar) | Yeasts (PDA) | Yeasts (WORT agar) | |
| Mature grapes (Limniona) | 4.62±0.02 | 4.59±0.05 | 5.35±0.06 | 5.25±0.07 |
| Initial must (Limniona) | 4.24±0.06 | 4.25±0.07 | 4.28±0.09 | 4.28±0.07 |
| Fermenting must (Limniona) | 7.49±0.09 | 7.68±0.27 | 7.50±0.04 | 7.65±0.39 |
| Wine sample (Limniona) | 5.07±0.13 | 4.95±0.02 | 5.11±0.13 | 4.40±0.67 |
| Sample | Identification | Number of isolates | Range of identification score values |
|---|---|---|---|
| Mature grapes (Limniona) | Candida lusitaniae | 6 | 1.850-2.084 (2 isolates < 2.000) |
| Bacillus amyloliquefaciens_ssp_plantarum | 3 | 1.834-1.99 | |
| Staphylococcus epidermidis | 2 | 1.934-2.077 (1 isolate < 2.000) |
|
| Candida krusei | 2 | 2.043-2.079 | |
| Initial must (Limniona) | Saccharomyces cerevisiae | 4 | 1.780-1.936 (4 isolates < 2.000) |
| Serratia marcescens | 2 | 2.131-2.298 | |
| Klebsiella aerogenes | 1 | 2.204 | |
| Fermenting must (Limniona) | Saccharomyces cerevisiae | 14 | 1.754-2.011 (11 isolates < 2.000) |
| Serratia marcescens | 1 | 2.273 | |
| Klebsiella aerogenes | 1 | 2.046 | |
| Unfiltered wine (Limniona) | Saccharomyces cerevisiae | 23 | 1.793-2.004 (21 isolates < 2.000) |
| Unfiltered wine from inox vessel | Filtered and bottled wine from inox vessel | ||||
|---|---|---|---|---|---|
| Genus | Species | % of valid reads | Genus | Species | % of valid reads |
| Acetobacter | ND | 39 | Burkholderia | ND | 34 |
| Burkholderia | ND | 23.73 | Acetobacter | ND | 17 |
| Rhizobium | ND | 5.30 | Rhizobium | ND | 6.8 |
| Pseudoalteromonas | ND | 3.92 | Oenococcus | oeni | 5.53 |
| Bacillus | ND | 1.91 | |||
| Lactobacillus | diolivorans | 1.5 | |||
| Bottled “Limniona” red wine with typical sensory characteristics | Bottled “Limniona” red wine with atypical sensory characteristics | Bottled “Malagouzia” white wine with atypical sensory characteristics | ||||||
|---|---|---|---|---|---|---|---|---|
| Genus | Species | % of valid reads | Genus | Species | % of valid reads | Genus | Species | % of valid reads |
| Lactobacillus | ND | 39 | Lactobacillus | ND | 30.6 | Lactobacillus | parafarraginis | 63.16 |
| Lactobacillus | parafarraginis | 19.19 | Lactobacillus | parafarraginis | 28.16 | Acetobacter | ND | 11 |
| Lactobacillus | diolivorans | 14.11 | Lactobacillus | diolivorans | 10.17 | Lactobacillus | ND | 2 |
| Oenococcus | oeni | 1.97 | Burkholderia | ND | 2.37 | Oenococcus | oeni | 2.07 |
| Burkholderia | ND | 1.68 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





