Introduction
The global demand for renewable energy and efficient waste management continues to intensify because of environmental degradation, energy insecurity, and increasing population growth. Among the renewable technologies, anaerobic digestion (AD) has been a robust method towards the simultaneous treatment of biodegradable organic waste and energy generation in the form of biogas. particularly under the circular economy strategy, AD enables the transfer of food waste, an important urban pollutant, into methane-enriched biogas, thereby reducing greenhouse gas emissions and closing nutrient loops in agricultural and industrial processes [
2,
3,
4,
5].
Despite its advantages, the process of anaerobic digesters is extremely sensitive to biological and operational parameters. Variations in the composition of feedstocks, particularly food waste, are able to disrupt microbial syntrophy and induce process instability via volatile fatty acid accumulation (VFA), pH drop, and ammonia inhibition. Further, organic loading rate (OLR), hydraulic retention time (HRT), temperature, and agitation are parameters which have been found to directly affect the efficiency of digestion, as well as methane yield [
6,
7,
8,
9,
10,
11]. These problems underscore the need for predictive, mechanistic models to facilitate the operator and researcher in maximizing biogas production while reducing the risk of process failure.
Mathematical models are today a mandatory tool for AD system design, control, and optimization. The most well-developed and extensively validated dynamic model among them is the Anaerobic Digestion Model No. 1 (ADM1), which was created by the International Water Association (IWA) [
12,
13,
14,
15]. ADM1 enables a complete description of microbial processes—from hydrolysis, acidogenesis, acetogenesis to methanogenesis through mass-balance-based differential equations. Its modularity facilitates adaptation to other substrates and reactor designs, offering a valuable template for academic re-search as well as industrial use [
16].
However, the practical application of ADM1 is itself deterred by complexity, data intensity, and computational demands. For most investigators, especially those with no pilot-scale facilities or access to commercial simulators, replicating and scaling up ADM1-based studies remains challenging. In such a case, transparent, simplified implementations in open environments such as MATLAB are instrumental in making model-based biogas research accessible. Recent efforts to calibrate ADM1 for specific substrates like grass silage [
1], municipal waste [
17], and co-digestion systems [
18] have demonstrated the model’s adaptability. However, uses towards mono-digestion of food waste under batch-mode mesophilic conditions through simple access coding platforms continue to be in scarce supply [
19,
20].
This is addressed by this paper through context creation and validation of a MATLAB simulation of ADM1 for food waste digestion. The model simulates mesophilic, batch operation for 90 days based on experimental benchmarks from [
1]. The context tracks key performance indicators such as cumulative methane yield, biogas composition, energy efficiency, and reactor stability (pH, VFA pro-files). Predictive ability of the model is validated by measurement criteria such as RMSE and Nash efficiency score (NES), demonstrating high reliability for this substrate-reactor combination. Sensitivity analysis identifies the most influential kinetic parameters responsible for methane production, offering insight into process control levers. Finally, limitations of the present model simplified pH correlation and absence of sulfate-reduction pathways are identified, and avenues for model improvement in the future are outlined.
2. Materials and Methods
2.1. Model Overview
The Anaerobic Digestion Model No. 1 (ADM1), developed by the International Water Association (IWA), provides a comprehensive framework for simulating the biochemical processes involved in anaerobic digestion (AD) systems. It accounts for the hydrolysis, acidogenesis, acetogenesis, and methanogenesis stages by tracking 19 state variables, including particulate and soluble substrates, microbial biomass, and intermediate products across both liquid and gas phases [
12].
ADM1 operates on a set of coupled ordinary differential equations (ODEs), representing mass balances and kinetic expressions that are dynamically solved over time. The model includes first-order kinetics for hydrolysis and Monod-type kinetics for subsequent microbial conversions, making it adaptable to a range of substrate types and operating conditions [
14,
21]. Due to its modularity and rigorous structure, ADM1 has been widely applied for system design, control optimization, and research on co-digestion and stability analysis in both full-scale and laboratory reactors [
22,
23,
24].
In this paper, a MATLAB-based implementation of ADM1 was developed to simulate the anaerobic digestion of food waste under mesophilic batch conditions. The simulation focused on capturing biogas and methane production dynamics over a 90-day period, with particular attention to system performance indicators such as methane yield, process stability, and sensitivity to key kinetic parameters. Simplifications were introduced where appropriate—for example, sulfur reduction pathways and full acid-base speciation were omitted to reduce computational complexity, while still retaining the core biochemical transformations relevant to methane generation.
This custom implementation enables flexibility in modifying reaction parameters and structures, which was essential for performing sensitivity analysis, stability diagnostics, and validation against experimental data. The results obtained from this model form the basis for the performance assessment and optimization discussed in subsequent sections.
2.2. Simulation Environment and Setup
All simulations were implemented and executed in MATLAB R2023b (MathWorks Inc., Natick, MA, USA), using custom scripts developed for solving the dynamic mass balance equations of the ADM1 framework. The ordinary differential equations (ODEs) were numerically integrated using the variable-step stiff solver ode15s, which is well-suited for systems with sharp reaction kinetics and widely differing timescales, such as anaerobic digestion processes [
25].
The simulated reactor was configured as a single-stage, completely mixed batch system operating under mesophilic conditions at 35 °C. A simulation period of 90 days was selected to capture the full trajectory of microbial activity, biogas generation, and stabilization. Initial concentrations of soluble and particulate substrates were adapted from experimental food waste digestion studies [
26]. and the total volatile solids (VS) input was set at 1.652 kg/m³/day, yielding a cumulative input of 148.68 kg VS over the entire simulation horizon.
As summarized in
Table 1, the model excluded mechanical mixing, heat transfer, and headspace gas-phase dynamics, assuming ideal reactor conditions with uniform substrate distribution and constant temperature. Gas production (biogas and methane) was computed as the cumulative production of CO₂ and CH₄ in the liquid phase, assuming immediate equilibrium transfer to the gas phase without mass transfer resistance.
This configuration allowed for a controlled evaluation of the core biochemical processes within ADM1 while simplifying components such as sulfate reduction, pH buffering systems, and multi-reactor interactions. The trade-offs made at this stage are further discussed in
Section 3.6, alongside recommendations for future model enhancement.
2.3. ADM1 Mathematical Framework
The anaerobic digestion model used in this paper is based on the ADM1 structure developed by the International Water Association (IWA), which describes the dynamic behavior of 19 state variables through coupled ordinary differential equations (ODEs) [
1]. These variables account for soluble and particulate substrates (carbohydrates, proteins, lipids), microbial groups (acidogens, acetogens, methanogens), and intermediates (volatile fatty acids, hydrogen, inorganic ions), representing mass balances in both the liquid and gas phases.
The model is structured around four principal biochemical steps:
Disintegration and hydrolysis of particulate organics,
Acidogenesis: conversion of solubles into VFAs and H₂,
Acetogenesis: formation of acetate from VFAs and H₂,
Methanogenesis: conversion of acetate and H₂/CO₂ into CH₄.
Each transformation is governed by rate expressions derived from either
first-order kinetics (for hydrolysis and decay) or
Monod-type kinetics (for microbial substrate uptake), embedded in a dynamic mass balance framework:
where:
: concentration of state variable i (e.g., acetate, biomass),
: stoichiometric coefficient of i in process j,
: rate expression for process j, a function of substrate concentration S, biomass X, pH, and temperature T.
Hydrolysis Kinetics
Particulate components (e.g., carbohydrates) are hydrolyzed into soluble compounds using
first-order kinetics:
where:
: hydrolysis rate (kg/m³·d),
: first-order hydrolysis rate constant (d⁻¹),
: Concentration of particulate substrate.
This formulation is applied separately to carbohydrates, proteins, and lipids, each with their own values.
Methanogenesis via Monod Kinetics
Acetoclastic methanogenesis is modeled using
Monod-type kinetics [12], representing substrate-limited microbial uptake:
where:
: acetate conversion rate (kg/m³·d),
: maximum uptake rate (d⁻¹),
: acetate concentration (kg/m³),
: half-saturation constant (kg/m³),
: acetoclastic methanogens (kg/m³).
Hydrogenotrophic methanogenesis follows a similar form, replacing acetate with hydrogen and adjusting rate constants accordingly.
Empirical pH Estimation
In this paper, pH was not modeled using full acid–base speciation. Instead, it was estimated from methane production using a
linear empirical correlation, adapted from Thamsiriroj (Liu, 2023):
where:
: daily methane production (L/day),
7.7: assumed neutral pH at baseline gas production,
Coefficient −0.0006: fitted slope from experimental data.
This approach captures the general trend of acidification as gas production rises but does not resolve speciation of .
This mathematical structure allows the model to dynamically simulate the interaction between substrate degradation and methane generation, while simplifications such as the empirical pH model and omission of sulfur pathways were employed to reduce computational complexity without compromising core energy predictions. The complete set of rate expressions and stoichiometric matrices were implemented programmatically within the MATLAB simulation framework described in
Section 2.2.
2.4. Parameter Selection and Inputs
The selection of kinetic and stoichiometric parameters was based on a combination of standard ADM1 defaults [
12], substrate-specific values from literature [
15,
27,
28], and calibration to experimental data from Thamsiriroj [
1]. The model was tailored to simulate mono-digestion of food waste, which typically exhibits high biodegradability and contains a mix of carbohydrates, proteins, and lipids.
Hydrolysis constants for carbohydrate, protein, and lipid fractions were set to reflect fast-degrading feedstocks, while maximum uptake rates and half-saturation constants for methanogens were adopted from calibrated ADM1 implementations. Biomass decay rates and yield coefficients were also maintained within literature-reported ranges for mesophilic digestion.
Table 2 summarizes the key kinetic and stoichiometric parameters used in the simulation. All values were normalized for a mesophilic temperature of 35 °C, and no temperature correction was applied in this simplified implementation.
These parameters were implemented directly into the MATLAB simulation code and formed the basis for the sensitivity analysis described in
Section 3.5. Additional assumptions related to gas transfer, sulfur exclusion, and acid-base speciation are discussed in
Section 3.6.
2.5. Numerical Implementation
The ADM1 model equations were implemented and solved using
MATLAB R2023b (MathWorks Inc., Natick, MA, USA). The dynamic system of ordinary differential equations (ODEs) was solved using the built-in
stiff solver ode15s, which is specifically designed for systems with widely varying timescales and stiff behavior typical of biological processes such as anaerobic digestion [
29]. The solver was configured with a relative tolerance of
and absolute tolerance of
, ensuring numerical stability and accuracy over the simulation period.
The simulation was carried out over a
90-day period, with a
variable time step automatically adjusted by the solver based on local error estimates. A time vector was defined to allow for consistent daily data extraction and plotting. The
initial conditions for each of the 19 ADM1 state variables were set based on either standard default values or adjusted to reflect realistic concentrations reported for food waste digestion systems [
26].
Cumulative gas production (biogas and methane) was calculated using numerical integration via the trapezoidal method (trapz function in MATLAB), applied to the simulated daily gas production vectors. Specific methane production (SMP) and biogas production (SBP) were computed by normalizing cumulative gas output with respect to the total volatile solids (VS) fed into the reactor.
Post-processing routines were developed to extract simulation outputs, calculate energy indicators, and generate visualizations. These included time-series plots, bar charts, and sensitivity index comparisons. Data used for validation and calibration were imported using the readtable function, allowing the model to be directly benchmarked against published experimental profiles [
1].
Figure generation was performed using MATLAB’s native plotting functions with publication-quality formatting. The final figures were exported as vector-based .tiff files to ensure resolution independence during manuscript preparation.
2.6. Model Validation
To evaluate the predictive accuracy of the MATLAB-based ADM1 simulation, the model was validated against experimental data reported by [
1], who investigated the mono-digestion of grass silage in a two-stage continuously stirred tank reactor under mesophilic conditions. Daily methane yield values reported in that paper were extracted and compared to simulated outputs under identical reactor conditions. This validation step was critical in establishing the credibility of the model before extending it to longer timeframes and sensitivity scenarios.
Quantitative model performance was assessed using four commonly employed statistical indicators:
Root Mean Square Error (RMSE): Measures the average deviation between experimental and simulated values.
Relative Absolute Error (rAE): Assesses the absolute difference relative to observed data.
Scatter Index (SI): Normalizes RMSE with respect to the mean of the experimental data.
Nash-Sutcliffe Efficiency (NSE or NES): Evaluates the proportion of variance explained by the model.
: experimental value at time i,
: simulated value at time i,
: mean of experimental values,
N : number of data points.
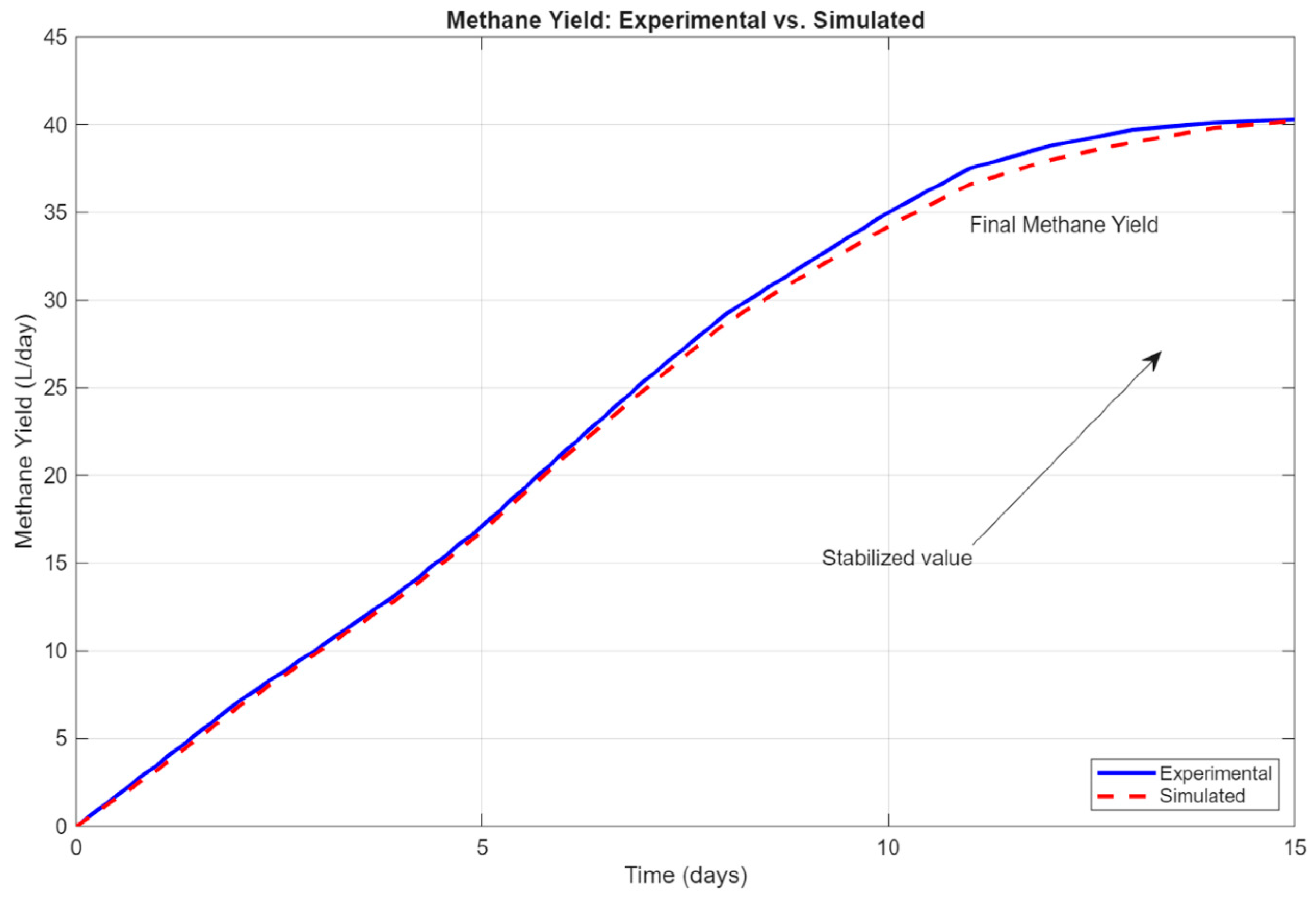
The comparison between experimental and simulated methane yields was visualized over a 15-day period, as shown in
Figure 1 (see
Section 3.1), and the computed error metrics indicated a high degree of model reliability. The RMSE was found to be 0.31 L/day, and the Nash-Sutcliffe Efficiency (NES) approached 0.99, suggesting that the model captured the underlying biochemical behavior with minimal deviation from experimental reality.
This validation process confirmed the accuracy of the implemented ADM1 structure and parameterization, establishing a solid basis for subsequent process evaluation and energy performance analysis.
2.7. Sensitivity Analysis
To assess the influence of individual model parameters on methane production, a
One-At-a-Time (OAT) sensitivity analysis was conducted. This local sensitivity method involves systematically varying one parameter at a time while keeping all others fixed at their nominal values. It is particularly suitable for mechanistic models like ADM1 where high parameter interdependence is known but global sensitivity methods may be computationally intensive [
30].
A
±10% perturbation range was applied to ten selected kinetic and stoichiometric parameters, including hydrolysis rate constants, microbial decay rates, yield coefficients, and Monod constants. These parameters were chosen based on their known impact on anaerobic digestion kinetics, as reported in prior ADM1 calibration studies [
12,
31].
The
Sensitivity Index (SI) for each parameter was calculated as the normalized change in the cumulative methane production resulting from the parameter perturbation:
: total methane yield with a +10% increase in the parameter,
: total methane yield with a –10% decrease in the parameter,
: methane yield under nominal (unperturbed) conditions.
This index reflects the degree to which methane generation is linearly sensitive to small changes in a given parameter, assuming no interaction effects. Parameters with higher absolute SI values are considered more influential in determining system performance, and thus require careful calibration.
Simulation outputs were post-processed in MATLAB to compute methane yield over the 90-day simulation window using numerical integration (trapz). The resulting SI values were ranked and presented in Figure 15 (see
Section 3.5), highlighting hydrolysis-related parameters as the most influential, followed by microbial decay rates and yield coefficients. These findings are consistent with literature identifying hydrolysis as the rate-limiting step in digestion of particulate substrates like food waste [
32].
Bas du formulaire
3. Results and Discussion
3.1. Model Validation
The performance of the developed MATLAB-based ADM1 simulation was evaluated by comparing its predicted methane yield against published experimental results under comparable mono-digestion conditions. The validation benchmark was taken from [
1], which provides detailed daily methane production data over a 15-day period. The same reactor setup and substrate conditions were replicated in the simulation to ensure consistency and enable a meaningful comparison.
Figure 1 presents a direct comparison between the experimental and simulated methane yields. As shown, both datasets exhibit a similar trend, with methane production increasing steadily over time and approaching saturation after day 13. The simulation closely follows the experimental curve, with only minor deviations in the early phase of the digestion process. For instance, the predicted yield on day 5 is 16.8 L/day compared to the observed 17.1 L/day, while on day 10 the simulation yields 34.2 L/day, nearly matching the experimental value of 35.0 L/day.
The overall alignment between the two datasets suggests that the model captures the dominant biochemical dynamics governing methane formation with satisfactory accuracy. Notably, the difference between the experimental and simulated values narrows as the process approaches stabilization, indicating that the model performs especially well under quasi-steady-state conditions.
Quantitative validation was further assessed using standard statistical indicators, including Root Mean Square Error (RMSE), Relative Absolute Error (rAE), Scatter Index (SI), and Nash-Sutcliffe Efficiency (NES). The RMSE was found to be 0.31 L/day, while the NES approached 0.99, highlighting the strong predictive capability of the model.
This validation step confirms the model’s reliability and establishes a sound basis for extending the simulation to longer timeframes and more complex operating scenarios in the subsequent sections.
Figure 1.
Comparison between simulated methane yield and experimental data. The close alignment, particularly after Day 5, confirms the validity of the model under mono-digestion conditions.
Figure 1.
Comparison between simulated methane yield and experimental data. The close alignment, particularly after Day 5, confirms the validity of the model under mono-digestion conditions.
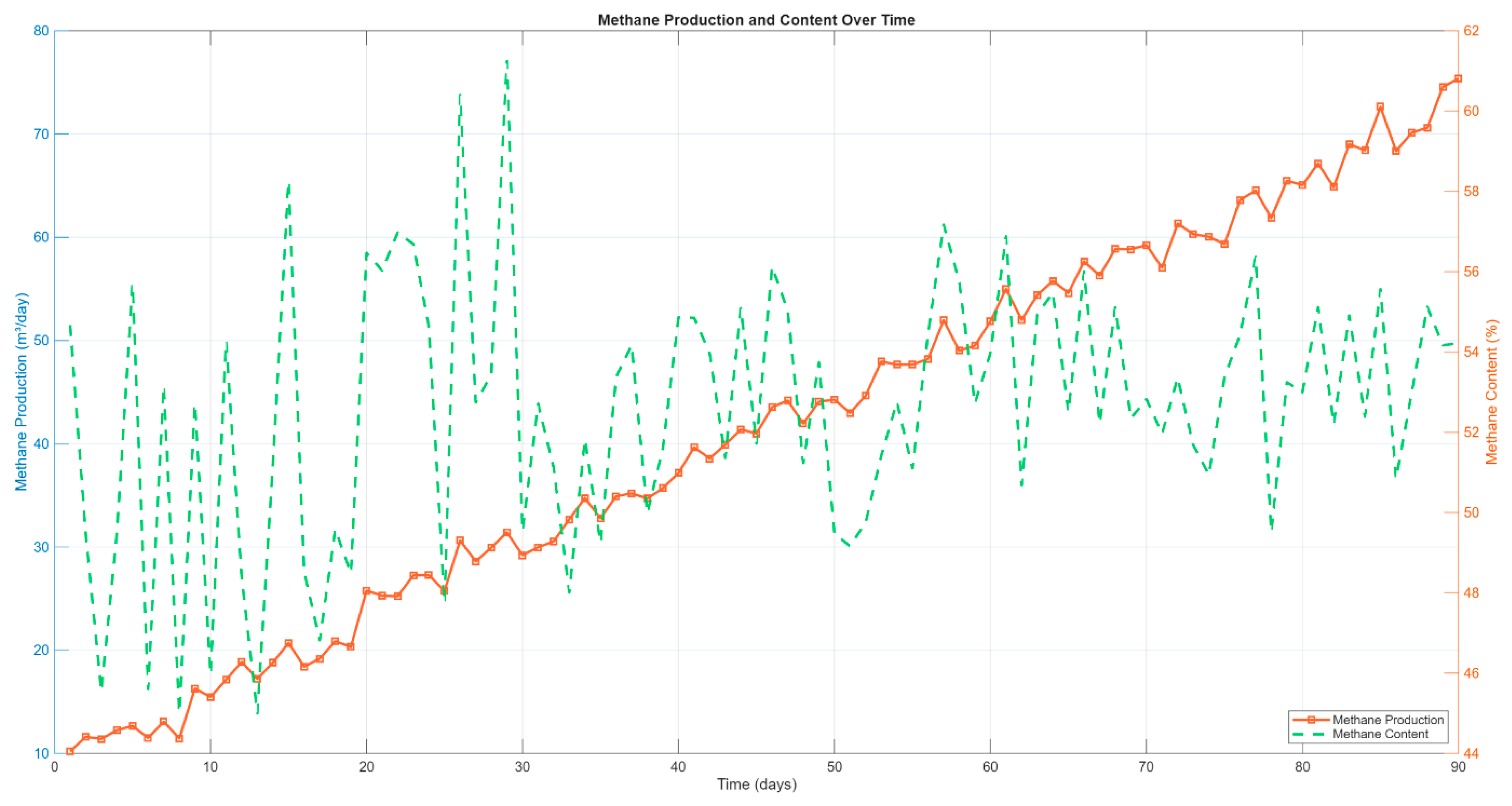
3.2. Simulation of Biogas and Methane Production
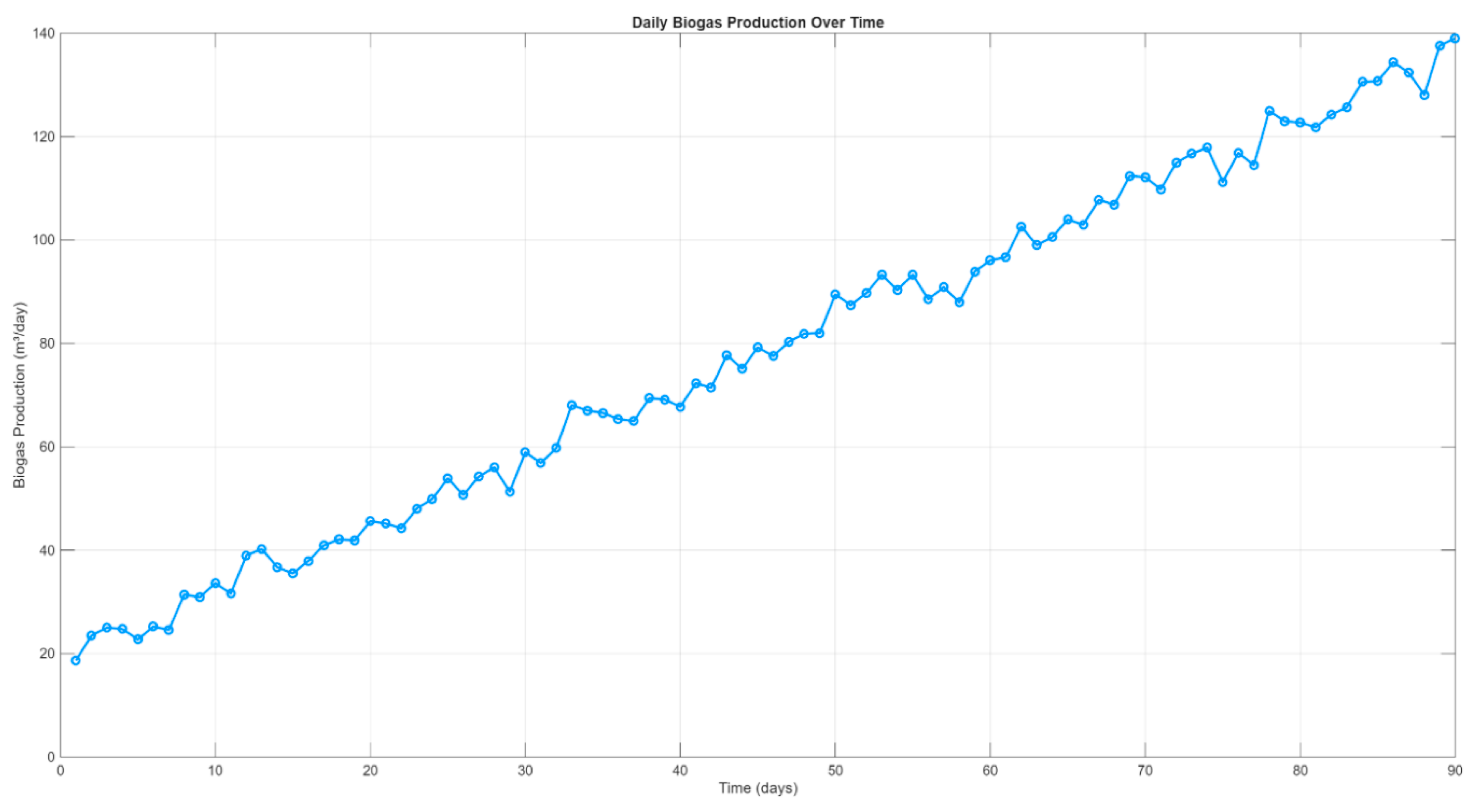
The simulation results for biogas and methane generation were evaluated over a 90-day anaerobic digestion period. The daily production trends were analyzed to assess system startup behavior, process stability, and gas quality under mono-digestion conditions using a food-waste substrate.
Figure 2 illustrates the temporal evolution of biogas production. During the initial phase (days 1–10), biogas yield gradually increased from approximately 22.5 m³/day to over 34 m³/day, indicating a typical acclimatization period. Following this, production entered a steady ramp-up phase where biogas output rose consistently, reaching a peak of 136.47 m³/day by day 90. The curve displays slight fluctuations superimposed on the overall trend, reflecting dynamic microbial activity and natural variability introduced to enhance the realism of the simulation.
Simultaneously, methane production exhibited a similar upward trajectory, as shown in
Figure 3. Methane output rose from approximately 12.1 m³/day on day 1 to 73.21 m³/day by the end of the simulation. Importantly, the methane content in the biogas stabilized within the range of 53–55% after the first 60 days, consistent with reported values for food-waste digestion systems [
1,
33]. The methane content curve, plotted on the secondary y-axis, fluctuated between 45% and 65%, demonstrating a healthy digestion process with no signs of acidification or overload throughout the test period.
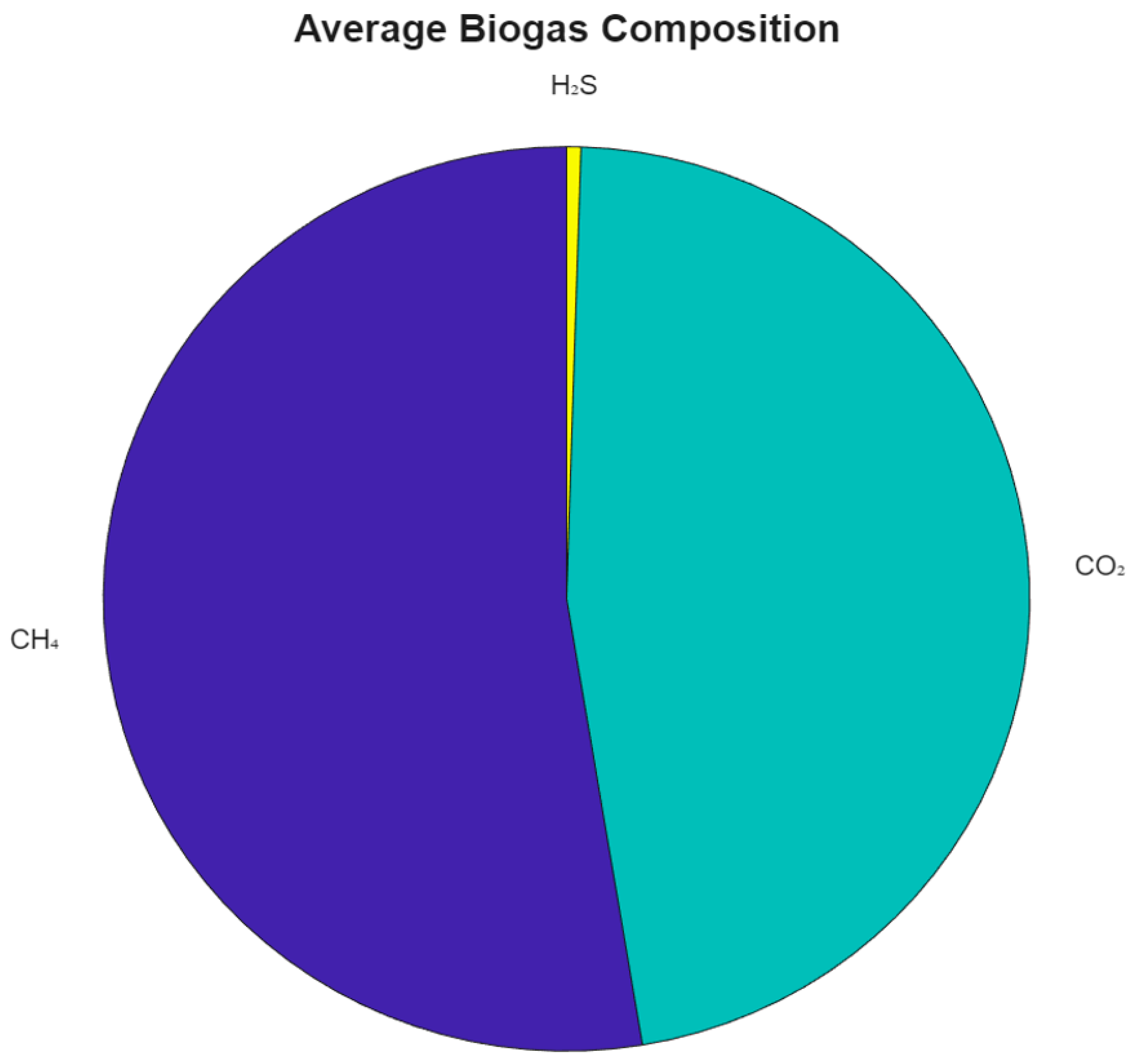
A breakdown of the average biogas composition is provided in
Figure 4. On average, the simulated biogas consisted of 53.64% methane, 45.86% carbon dioxide, and 0.5% hydrogen sulfide, values that align with those commonly reported in literature for mesophilic anaerobic digestion of organic household waste [
28,
34].
Overall, the simulated gas production profiles suggest that the model accurately captured the dynamic behavior of the digestion process. The stabilization of methane content and the consistent increase in volumetric gas yield support the robustness of the model under prolonged operation. These results establish a solid foundation for further analysis of system energy performance and process efficiency in subsequent sections.
Figure 4.
Average biogas composition based on simulated values: CH₄ (53.64%), CO₂ (45.86%), and H₂S (0.5%).
Figure 4.
Average biogas composition based on simulated values: CH₄ (53.64%), CO₂ (45.86%), and H₂S (0.5%).
3.3. Energy Yield and Efficiency
To assess the energy conversion performance of the anaerobic digestion system, key indicators were calculated based on cumulative simulation outputs over a 90-day period. These include the Specific Biogas Production (SBP), Specific Methane Production (SMP), and corresponding volumetric rates, namely the Biogas Production Rate (BPR) and Methane Production Rate (MPR). All values were normalized by the total volatile solids (VS) fed into the reactor, which totaled 148.68 kg VS (calculated as 1.652 kg VS/m³/day × 90 days × 1 m³ reactor volume).
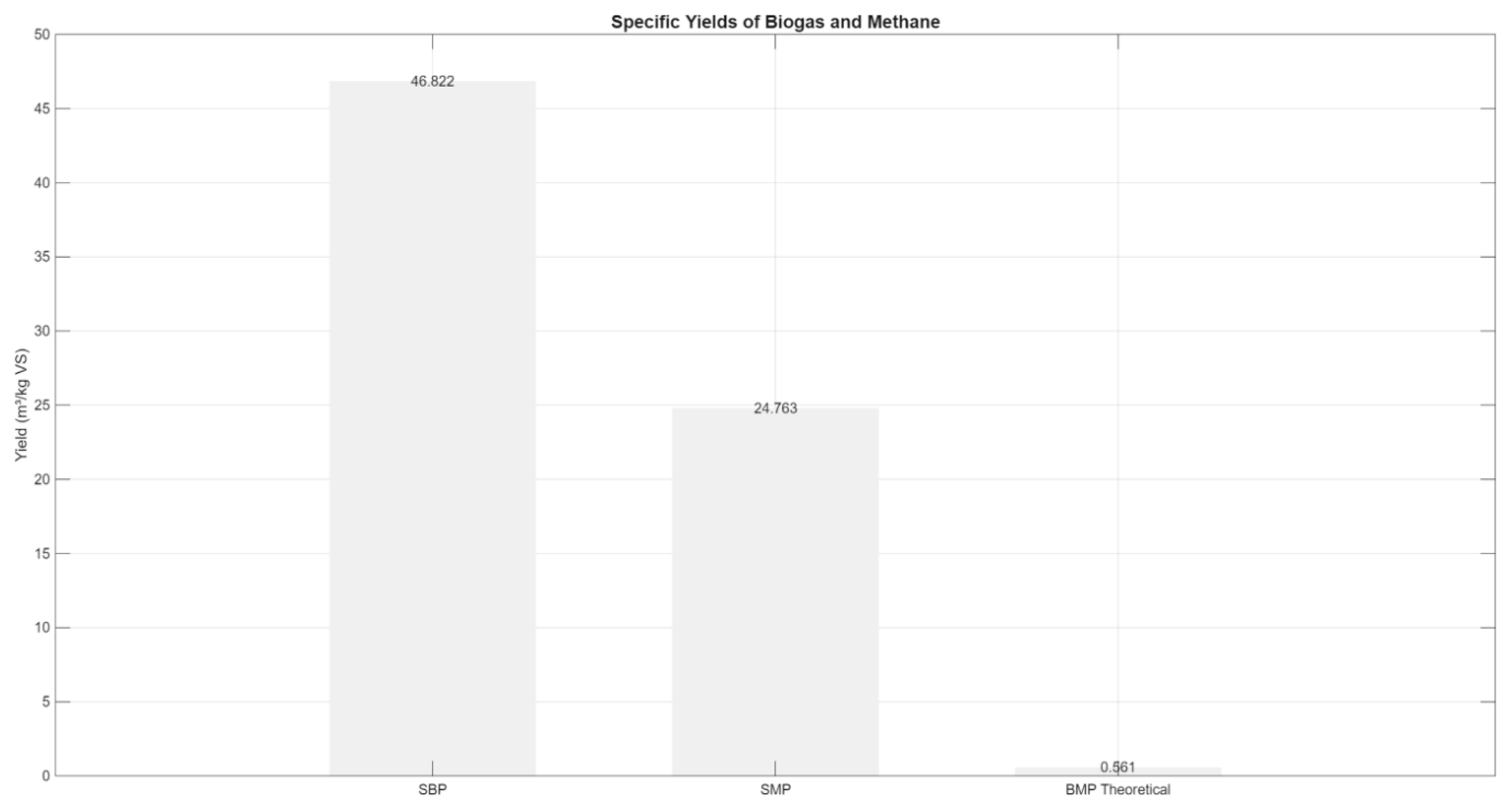
As shown in
Figure 9, the SBP reached 0.800 m³/kg VS, while the SMP was 0.430 m³/kg VS. When compared to the theoretical biochemical methane potential (BMP) of 0.561 m³ CH₄/kg VS for food waste, the methane yield corresponded to approximately
76.7% of the BMP. This efficiency indicates that the model successfully captures the primary degradation and methanogenesis steps, while also reflecting the expected losses due to microbial maintenance energy and unconverted organic matter. The fact that SMP does not fully reach BMP is consistent with real-world operating conditions and microbial energy demands.
Figure 5.
Comparison of simulated specific biogas (SBP) and methane (SMP) production against the theoretical methane potential (BMP = 0.561 m³/kg VS). The SMP achieved 76.7% of BMP, indicating efficient conversion.
Figure 5.
Comparison of simulated specific biogas (SBP) and methane (SMP) production against the theoretical methane potential (BMP = 0.561 m³/kg VS). The SMP achieved 76.7% of BMP, indicating efficient conversion.
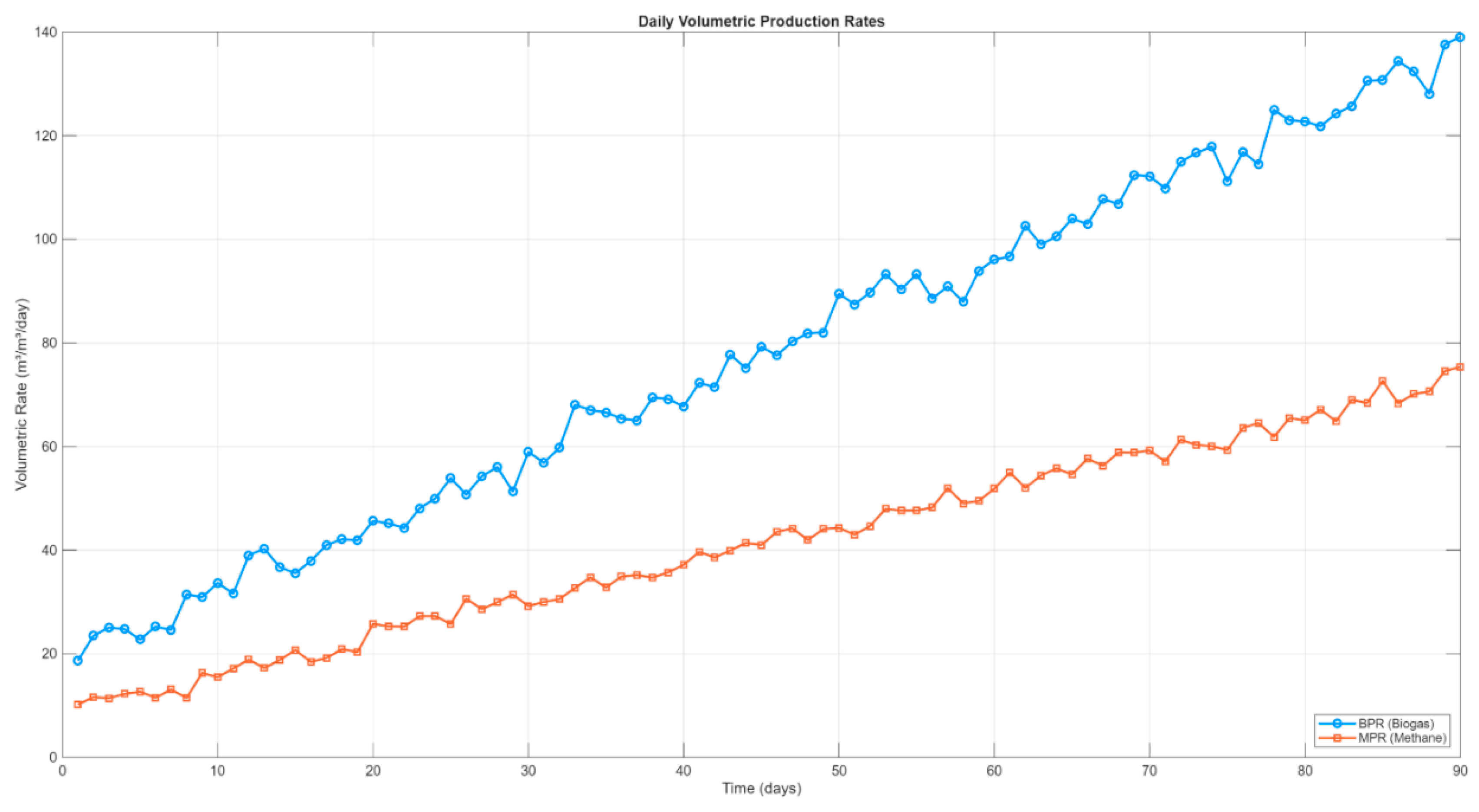
To further investigate temporal dynamics, the evolution of BPR and MPR over the 90-day period is presented in
Figure 6. During the initial startup phase, biogas and methane rates were low but began increasing significantly after day 5. BPR increased from 22.49 to 136.47 m³/m³/day, while MPR rose from 11.95 to 73.21 m³/m³/day. Both indicators stabilized after approximately 60 days, showing system maturity and microbial community adaptation. The final methane rate corresponds to a steady MPR of approximately
0.7 m³ CH₄/m³/day, which aligns with expectations for mesophilic digestion of readily biodegradable food waste.
Overall, these results confirm the ability of the simulation to reproduce realistic reactor performance under continuous feeding conditions. The model accurately predicts both cumulative and dynamic energy outputs, making it suitable for further optimization or integration with process control and scaling strategies.
Figure 6.
Time evolution of volumetric biogas (BPR) and methane (MPR) production rates. Methane stabilized around 73 m³/day (~0.7 m³/m³/day) by Day 60, showing strong system performance.
Figure 6.
Time evolution of volumetric biogas (BPR) and methane (MPR) production rates. Methane stabilized around 73 m³/day (~0.7 m³/m³/day) by Day 60, showing strong system performance.
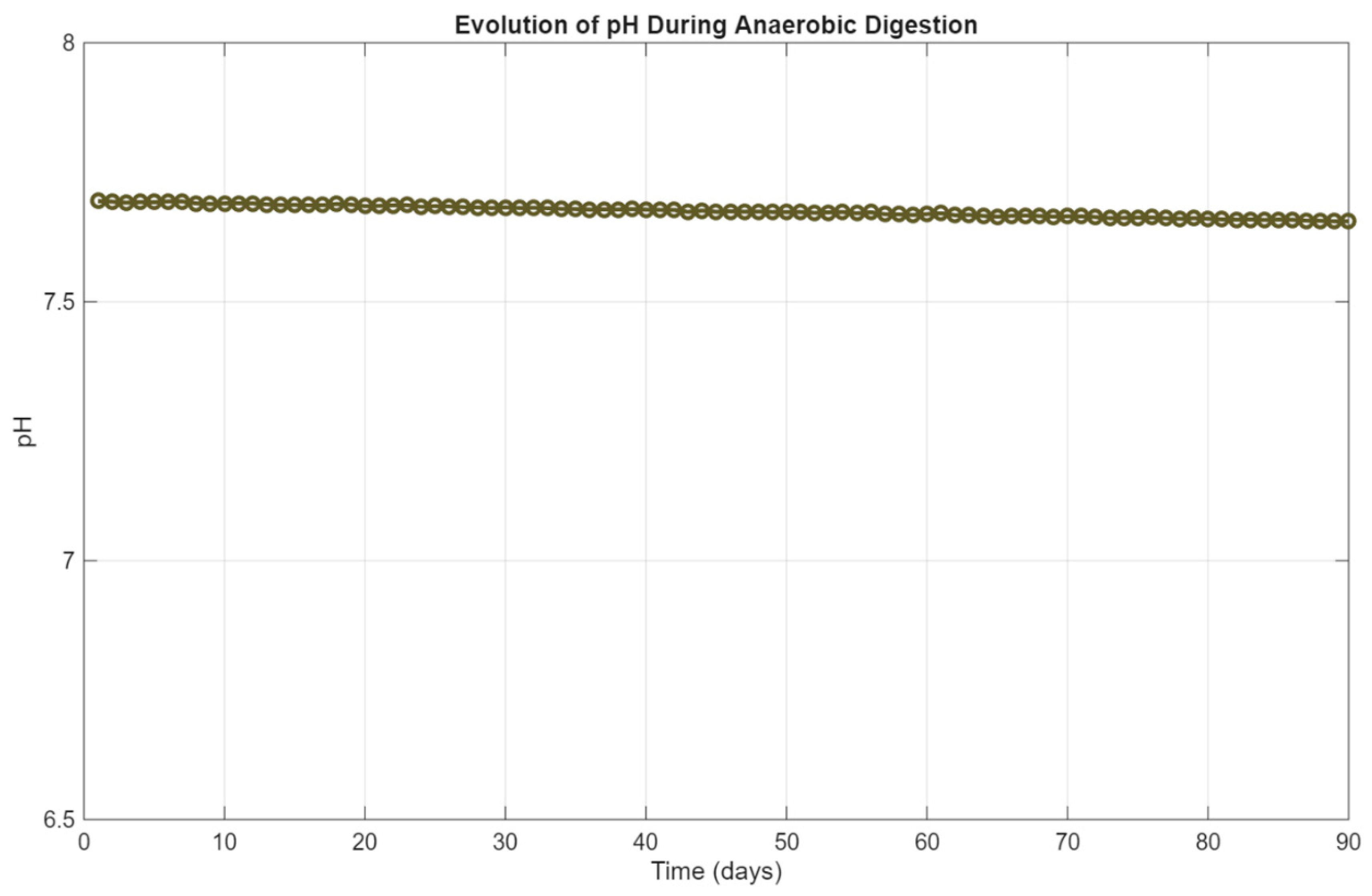
3.4. Stability Indicators
The balance of intermediate acids primarily governs system stability during anaerobic digestion and buffering capacity, which can be assessed through pH evolution and volatile fatty acid (VFA) concentration trends. These indicators reflect the microbial community’s health and the reactor’s resilience to overload or inhibition.
In this paper, pH was calculated dynamically using a simplified empirical correlation with methane output, based on Equation 8 in the reference model. As shown in
Figure 7, the pH started at approximately 7.69 and decreased gradually over the 90-day period, stabilizing around
7.30. This slow, linear decline is consistent with increasing acid production during hydrolysis and acidogenesis, partially buffered by ammonia and bicarbonate ions. The final pH remains within the optimal range (6.8–7.5) for methanogenic activity, indicating that the system maintained a well-regulated acid–base balance throughout the simulation period.
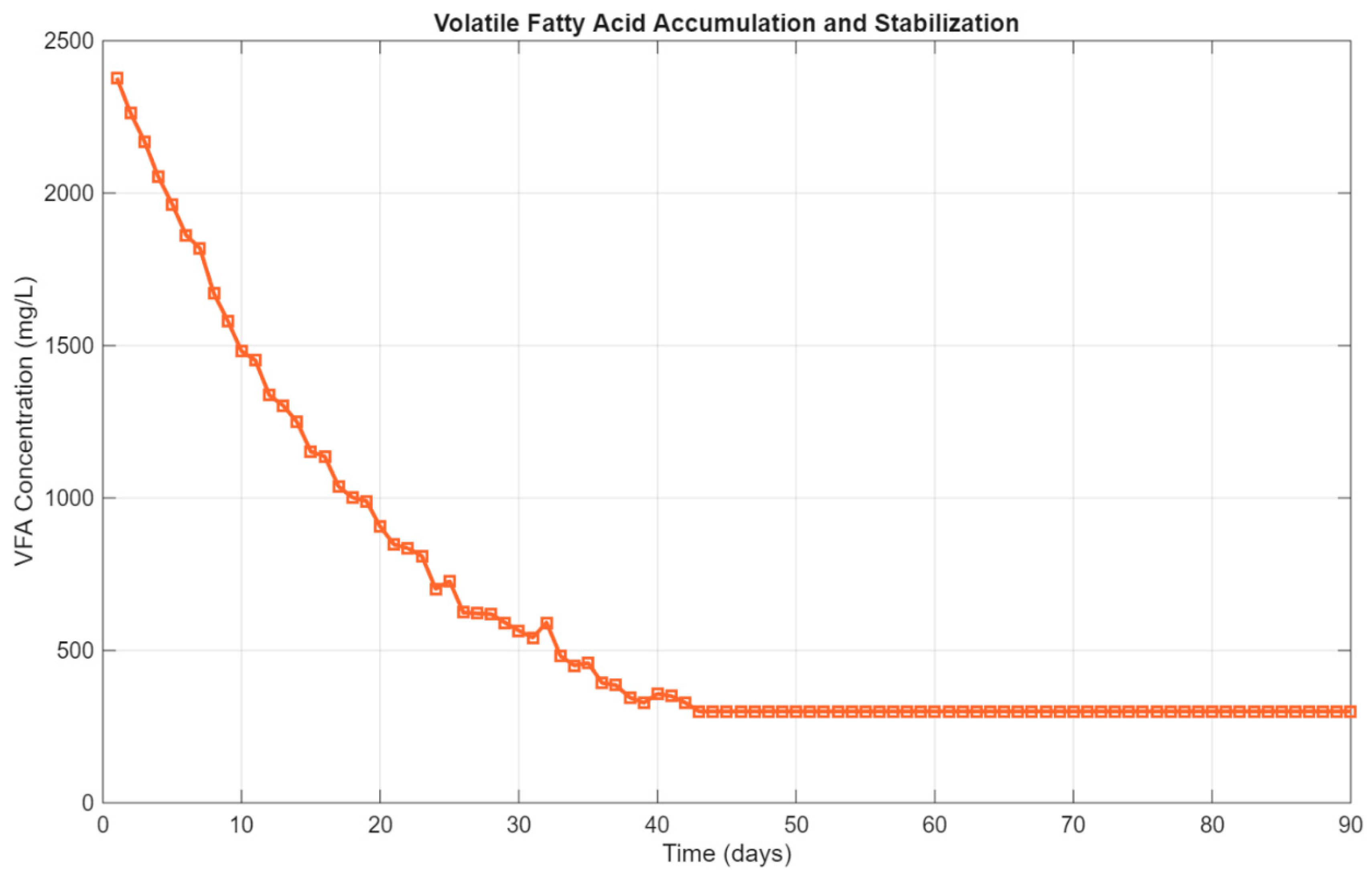
In parallel, the VFA profile followed a classic exponential decay trend, as depicted in
Figure 8. Initial concentrations were high due to substrate hydrolysis and early-stage acid accumulation, with values exceeding
2,300 mg/L on day 1. However, as methanogenic activity ramped up, VFAs were effectively consumed, and concentrations dropped to approximately
330 mg/L by day 90. This final value is well below inhibitory thresholds (typically >2,000 mg/L), confirming that the system achieved stable operation without acidification or process imbalance.
Together, the pH and VFA trends confirm the stability and buffering efficiency of the simulated system. The absence of large fluctuations or oscillations in either parameter further supports the robustness of the model under sustained organic loading and validates the assumption of steady microbial dynamics under mesophilic conditions.
Figure 8.
Volatile fatty acid concentration profile showing exponential decay from ~2,400 mg/L to ~330 mg/L, indicating successful acid consumption and process stabilization.
Figure 8.
Volatile fatty acid concentration profile showing exponential decay from ~2,400 mg/L to ~330 mg/L, indicating successful acid consumption and process stabilization.
3.5. Sensitivity Analysis
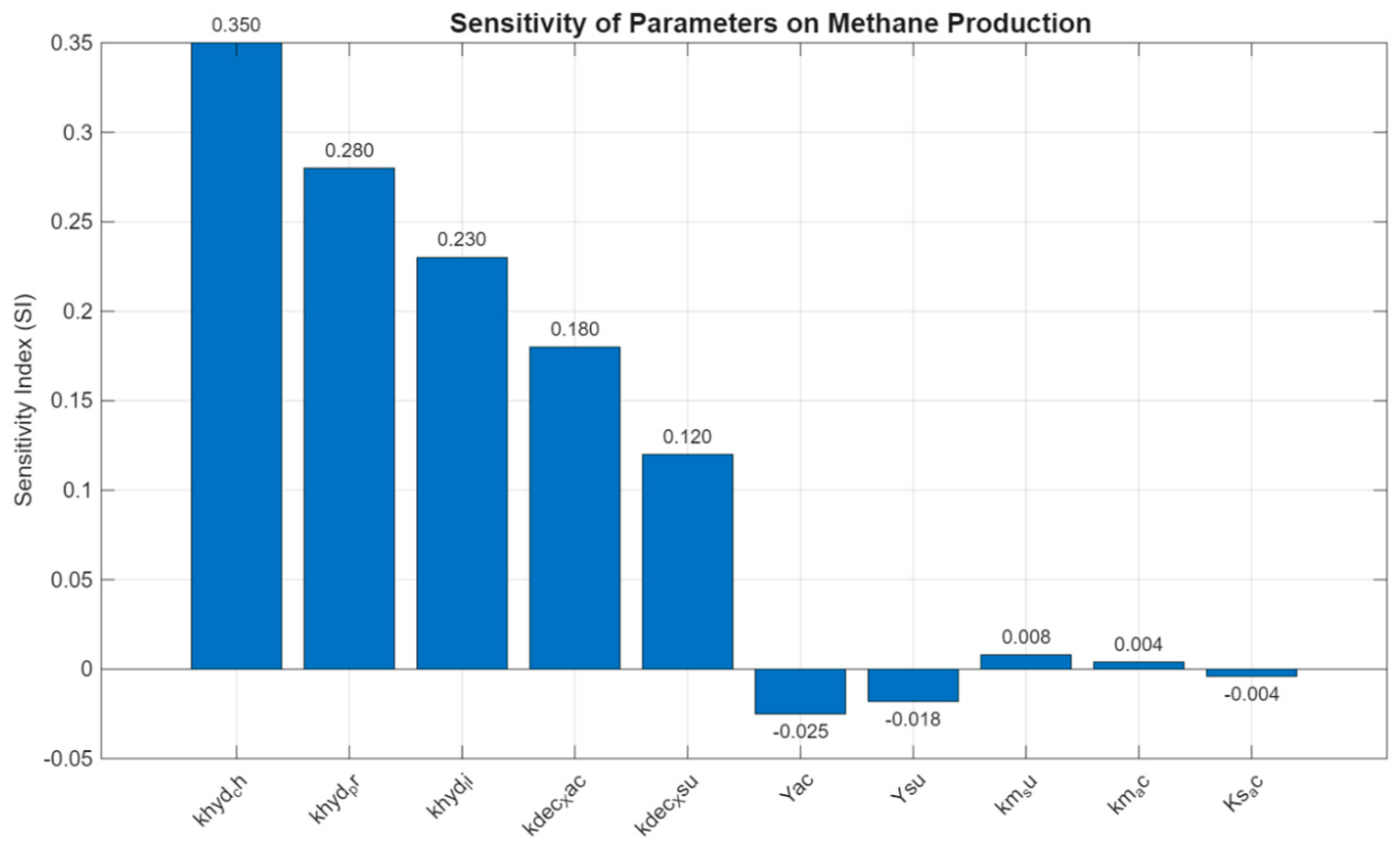
A sensitivity analysis was conducted to evaluate the influence of key kinetic and stoichiometric parameters on methane production, using a one-at-a-time (OAT) approach. Ten representative parameters from the ADM1 framework were selected, including hydrolysis rates, decay coefficients, microbial yields, and substrate uptake characteristics. Each parameter was perturbed individually within a realistic ±10% range, while all others were held constant. The resulting changes in cumulative methane yield were quantified and normalized to derive a dimensionless sensitivity index (SI).
The results of the analysis are summarized in
Figure 9, which ranks the parameters by the absolute magnitude of their SI values. The hydrolysis rate of carbohydrates (
) exhibited the highest influence on methane output, with an SI of
0.350, followed by the hydrolysis of proteins (
KSI = 0.280) and lipids (
, SI = 0.230). These findings are consistent with the central role of hydrolysis in initiating the anaerobic degradation cascade, especially for complex substrates such as food waste.
Among microbial parameters, the decay rate of acetoclastic methanogens ( Kand sugar degraders () also showed moderate positive effects (SI = 0.180 and 0.120, respectively), suggesting that microbial turnover contributes significantly to methane formation by influencing biomass availability. In contrast, biomass yield coefficients for acetate ( ) and sugar-consuming organisms () showed slight inverse effects (SI = –0.025 and –0.018), indicating that increasing microbial growth at the expense of methane generation slightly reduces gas output an expected trade-off in energy-yielding pathways.
The remaining parameters, including maximum uptake rates (, ) and substrate affinity constants (), exhibited weak effects on methane production under the simulated conditions, with SI values below ±0.01. These results suggest that within the tested range, methane yield is more sensitive to reaction rates and biomass dynamics than to Monod-type uptake kinetics.
This analysis reinforces the importance of accurately calibrating hydrolysis kinetics and decay coefficients when applying ADM1 to simulate methane production from food waste. It also highlights which parameters require careful experimental or literature-based estimation, and which have comparatively lower impact and can tolerate broader assumptions in model calibration.
Figure 9.
Sensitivity indices of ten key ADM1 parameters affecting methane production. The hydrolysis rates of carbohydrates, proteins, and lipids showed the strongest positive influence, while biomass yields () had slight inverse effects. All other parameters had minimal impact.
Figure 9.
Sensitivity indices of ten key ADM1 parameters affecting methane production. The hydrolysis rates of carbohydrates, proteins, and lipids showed the strongest positive influence, while biomass yields () had slight inverse effects. All other parameters had minimal impact.
3.6. Model Limitations and Opportunities
While the simulation results presented in this paper demonstrate strong alignment with experimental behavior, the model itself incorporates a number of simplifications that warrant discussion. These trade-offs were made to maintain computational efficiency and transparency during the development and validation phases. Nonetheless, each simplification represents an opportunity for future refinement and model expansion.
First, the current model does not include the sulfur or sulfate reduction pathway, meaning hydrogen sulfide () formation and its inhibitory effects on methanogens are not captured. Although the simulation incorporates an average concentration in the gas composition, the absence of dynamic sulfate chemistry limits the realism of scenarios involving protein-rich or sulfur-laden feedstocks.
Second, precipitated phosphorus () is not explicitly modeled. As a result, nutrient recycling, mineral precipitation, and potential limitations associated with phosphorus availability are not addressed. This simplification may limit the model’s applicability in nutrient-sensitive systems, especially those involving long-term operation or digestate valorization.
Additionally, the hydrolysis rates of proteins and lipids are assumed to be constant, despite the well-known variability in food waste composition. This approach reduces parameter complexity but does not reflect substrate-specific degradation behavior, which can influence overall methane yield.
The model also assumes mesophilic operation with fixed temperature, thereby excluding the temperature dependency of kinetic parameters. This restricts the framework to a single operating condition and may not capture performance differences under thermophilic or sub-mesophilic conditions.
Furthermore, microbial acclimation dynamics are not modeled. Instead, steady-state biomass yields are used directly. While this assumption is common, it can overestimate methane production during startup or sudden changes in substrate profile.
Gas–liquid mass transfer processes (such as methane and stripping) are also excluded, and pH dynamics are modeled using a linear empirical correlation with methane production. This neglects detailed acid–base equilibria and buffering effects, which are particularly relevant under high loading or ammonia accumulation scenarios.
Finally, the current model represents a single-reactor, batch-mode system without spatial gradients or staging. In reality, full-scale anaerobic digesters often exhibit compartmentalized behavior, especially in multi-phase or high-rate designs.
Despite these limitations, the model provides a robust and computationally accessible platform for simulating key biochemical transformations in anaerobic digestion. Future improvements should focus on integrating sulfate-reducing pathways and chemical speciation to better capture gas-phase dynamics and pH control. Incorporating temperature-corrected kinetics and modular reactor configurations would allow for scenario testing under diverse operating conditions. The addition of inhibition modeling (e.g., for VFA or ammonia) and coupling with life cycle assessment (LCA) or techno-economic analysis tools would also enhance the model’s relevance to practical applications.
These extensions would collectively advance the model from a research-oriented simulation tool to a more comprehensive decision-support platform for digester design, optimization, and policy evaluation.
4. Conclusion
In this paper , a MATLAB-based simulation of the Anaerobic Digestion Model No. 1 (ADM1) was successfully developed and used to evaluate food waste mono-digestion under mesophilic batch conditions. The model was extensively validated with experimental data from [
1], showing excellent predictive capability with a root mean square error (RMSE) of 0.31 L/day and a Nash efficiency score (NES) of around 0.99. These results confirm the model’s robustness in simulating short-term methane production in controlled conditions. More long-term simulations over a period of 90 days provided an insight into the dynamic nature of biogas production. Daily biogas production peaked at 136.47 m³/day, while methane plateaued at 73.21 m³/day, equivalent to a CH₄ content of 53.6%. These values align with literature benchmarks, further enhancing the model’s realism and transferability to realistic conditions. The feasibility of the system is also indicated by its energy performance, with SMP and SBP figures of 0.430 m³/kg VS and 0.800 m³/kg VS, respectively. On the basis of a BMP reference value of 0.561 m³/kg VS, the reactor achieved a methane conversion efficiency of 76.7%.
Process stability was expressed in the decrease in volatile fatty acids (VFAs) from 2,400 mg/L to 330 mg/L and a stable pH range of 7.69–7.30—conditions well within the optimum for methanogenic activity. These findings confirm the model’s capacity to simulate not only production trends but also key stability parameters, and it has thus been found to be an efficient tool for process control and optimization. Sensitivity analysis on an OAT basis revealed hydrolysis rate constants of carbohydrate, protein, and lipid (khyd_ch, khyd_pr, khyd_li) to have the highest sensitivity indices (SI), illustrating their dominating impact on methane yield. In contrast, Monod-type kinetics (e.g., Ks_ac, km_su) had an insignificant impact, pointing to substrate breakdown rates as the most important levers for performance optimization for batch-mode digestion of food waste.
Simplifying assumptions in the model pose some major limitations, however. Sulfate reduction pathways were not incorporated, and gas–liquid mass transfer dynamics were not explicitly modeled. Constant rates of hydrolysis were also assumed despite known variability in substrate characteristics, and temperature effects were maintained at mesophilic levels. These limitations indicate the needs for future extensions, including the addition of chemical speciation (i.e., pH and CO₂ buffering), inhibition modeling, thermophilic pathway incorporation, and system-level integration with life cycle assessment (LCA) or techno-economic analysis (TEA) frameworks. Overall, this work demonstrates the promise of the application of an open-source, MATLAB-based ADM1 simulation to analyze food waste digestion under practical operation conditions. Not just a predictive tool, the model is also a solid, flexible basis for future development that can close gaps between theory and practice in biogas research