Submitted:
15 August 2025
Posted:
18 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
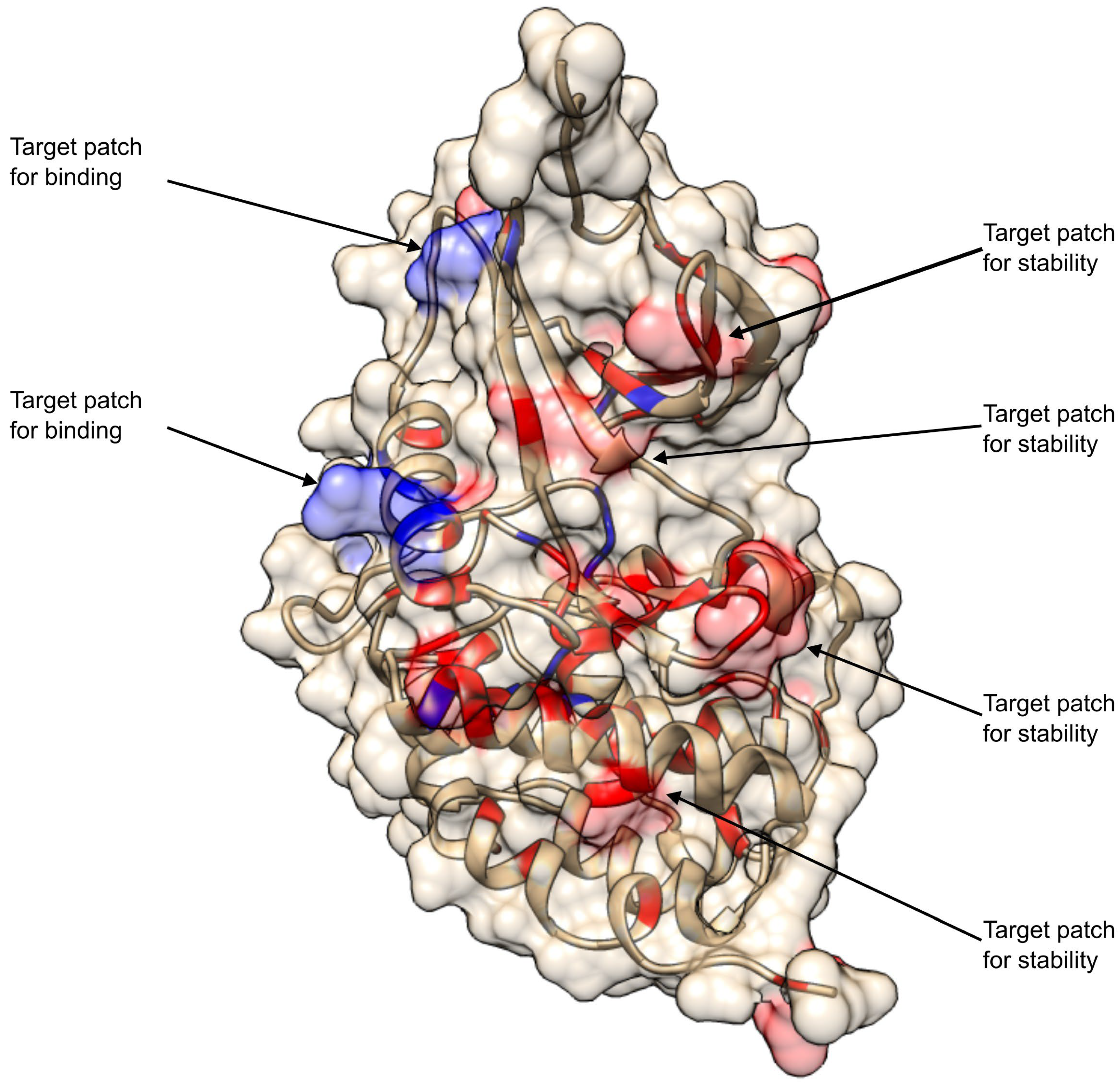
2.1. CDKL5 Variants, Structure and Binding Partners
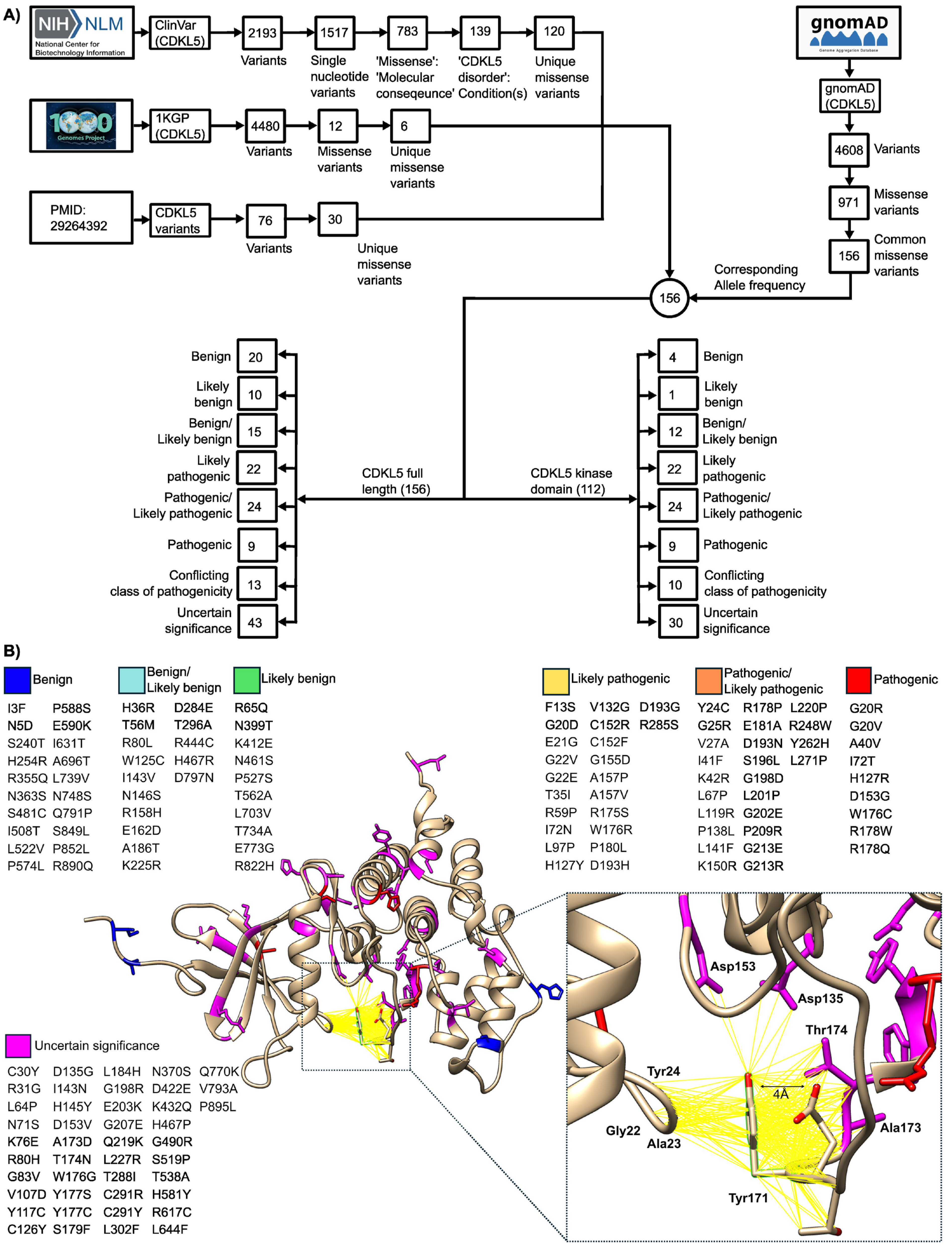
2.1.1. Curation and Structural Mapping of Missense CDKL5 Variants Associated with CDD
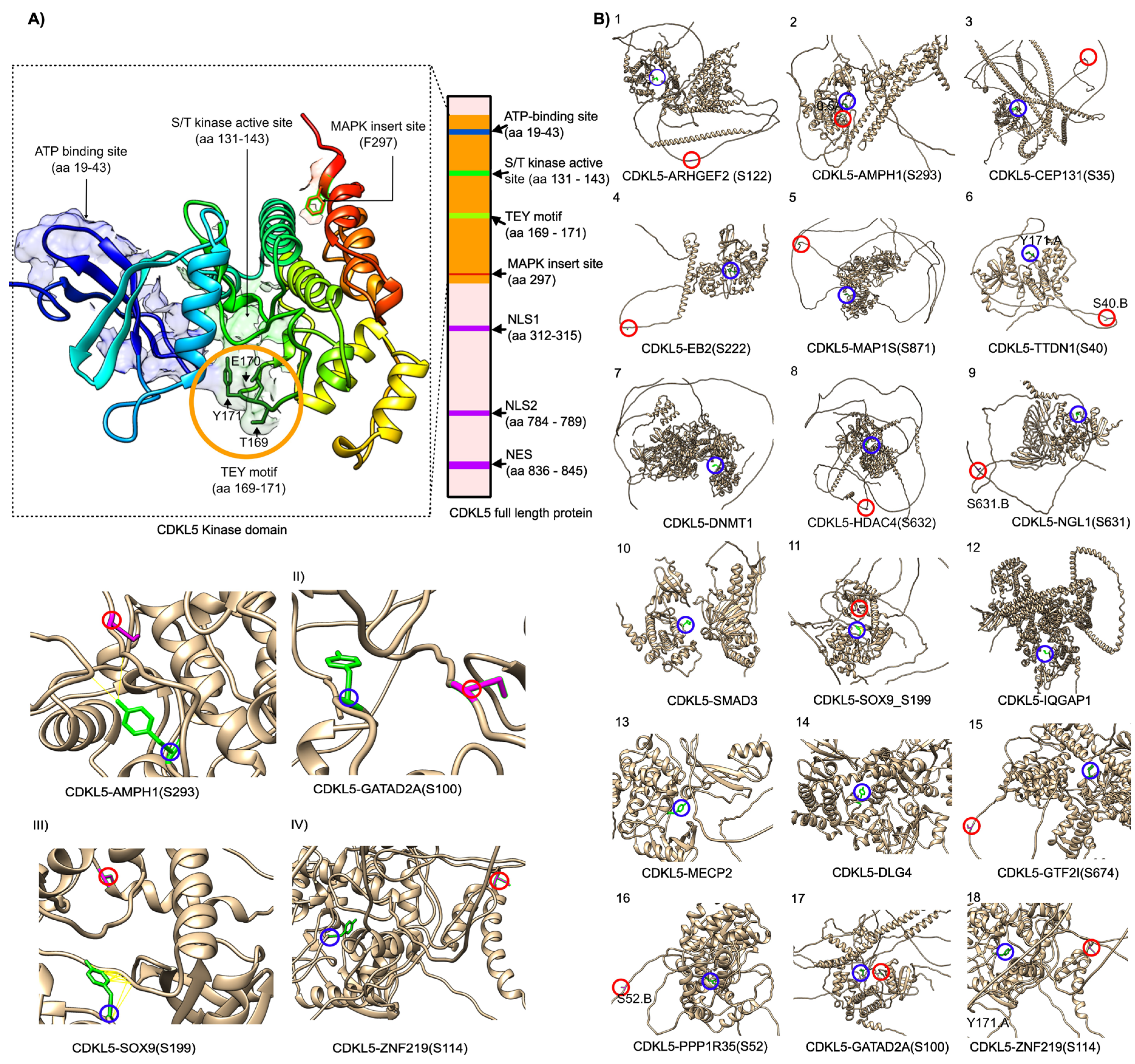
2.1.2. CDKL5 partners
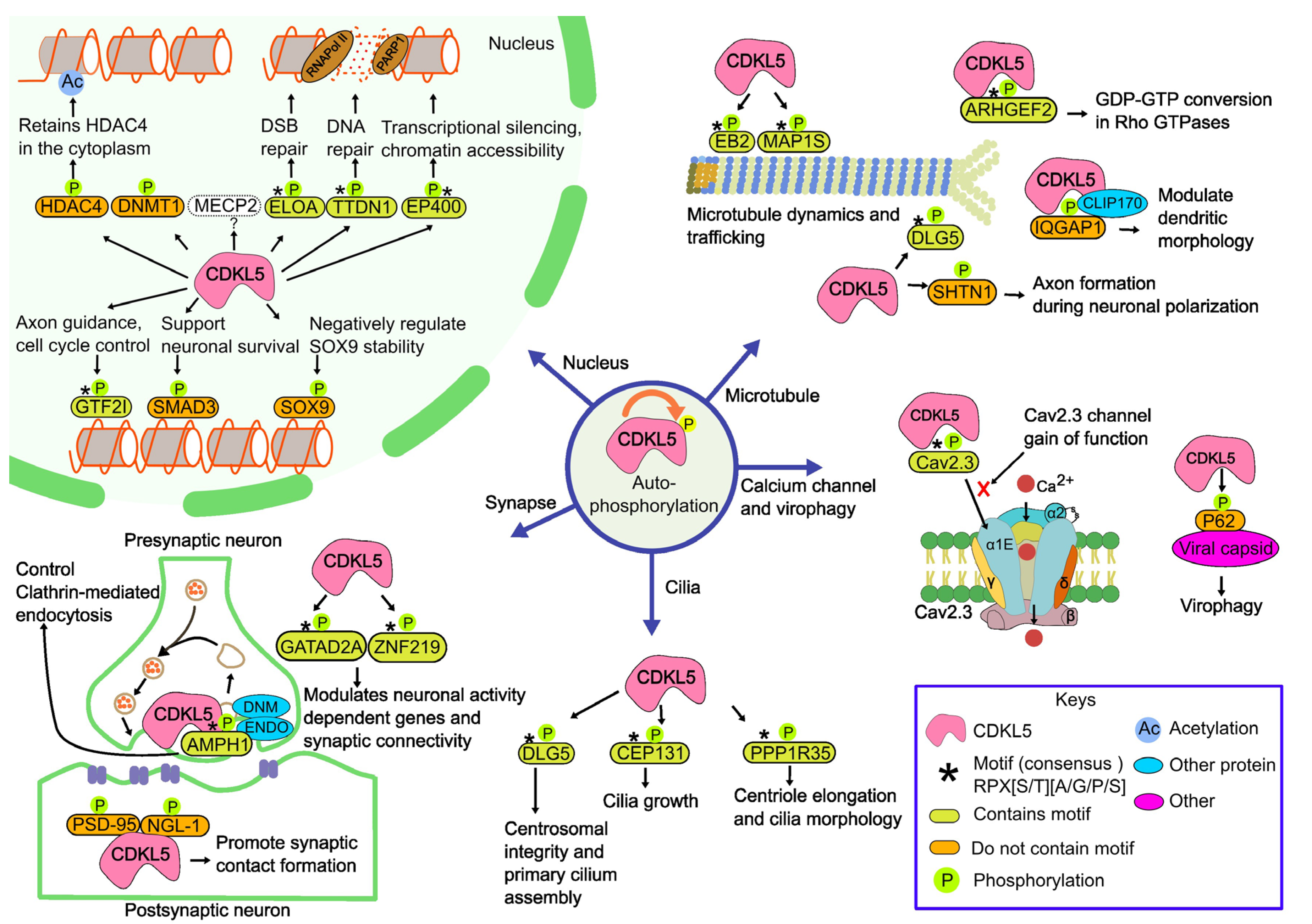
| SL | UniProt | Gene | pSite |
Consensus Motif (RPX[S/T][A/G/P/S]) |
Protein | Source |
| 1 | Q92974 | ARHGEF2 | S122 | TIRERPSsAIYPS | Rho guanine nucleotide exchange factor 2 | [31] |
| 2 | P49418 | AMPH1 | S293 | PAPARPRsPSQTR | Amphiphysin1 | [31] |
| 3 | Q9UPN4 | CEP131 | S35 | PVSRRPGsAATTK | Centrosomal protein of 131 kDa | [31] |
| 4 | Q8TDM6 | DLG5 | S1115 | QKRRRPKsAPSFR | Disks large homolog 5 | [31] |
| 5 | Q14241 | ELOA | S311 | EENRRPPsGDNAR | Elongin A | [31] |
| 6 | Q96L91 | EP400 | S729 | SPVNRPSsATNKA | EE1A-binding protein p400 | [31] |
| 7 | Q66K74 | MAP1S | S871, S900 | KAPARPSsASATP, DRASRPLsARSEP | Microtubule-associated protein 1S | [31] |
| 8 | Q15555 | EB2/MAPRE2 | S222 | STPSRPSsAKRAS | Microtubule-associated protein RP/EB family member 2 | [31] |
| 9 | Q8TAP9 | TTDN1 | S40 | GGGPRPPsPRDGY | TTD non-photosensitive 1 protein | [31] |
| 10 | P26358 | DNMT1 | N/A | DNA methyltransferase 1 | [31] | |
| 11 | P56524 | HDAC4 | S632 | RPLSRAQsSPASAtF | Histone deacetylase 4 | [31] |
| 12 | Q9HCJ2 | NGL-1/KIAA1580/LRRC4C | S631 | PLLIRMNsKDNVQET | Netrin-G ligand-1 | [31] |
| 13 | P84022 | SMAD3 | N/A | N/A | Mothers against decapentaplegic homolog 3 | [31] |
| 14 | P48436 | SOX9 | S199 | ATEQTHIsPNAIFKA | Transcription factor SOX-9 | [31] |
| 15 | P46940 | IQGAP1 | N/A | N/A | IQ Motif Containing GTPase Activating Protein 1 | [31] |
| 16 | P51608 | MeCP2 | N/A | N/A | Methyl-CpG binding protein 2 | [31] |
| 17 | P78352 | PSD95/DLG4 | N/A | N/A | Postsynaptic density protein 95 | [31] |
| 18 | A0MZ66 | SHTN1/SHOT1 | N/A | N/A | Shootin1 | [31] |
| 19 | P78347 | GTF2I | S674 | QSPKRPRsPGSNS | General transcription factor II-I | [42] |
| 20 | Q8TAP8 | PPP1R35 | S52 | SLSPRPDsPQPRH | Protein phosphatase 1 regulatory subunit 35 | [42] |
| 21 | Q86YP4 | GATAD2A | S100 | KSERRPPsPDVIV | GATA zinc finger domain containing 2A | [42] |
| 22 | Q9P2Y4 | ZNF219 | S114 | HQPERPRsPAARL | Zinc finger protein 219 | [42] |
| 23 | Q15878 | CACNA1E/Cav2.3 | S14 | AVVARPGsGDGD | Voltage-dependent R-type calcium channel subunit alpha-1E (Cav2.3) | [60] |
| 24 | Q13501 | SQSTM1/p62 | T269/S272 | RSRLTPVsPESS, GGKRSRLtPVSP | Sequestosome-1(p62) | [61] |
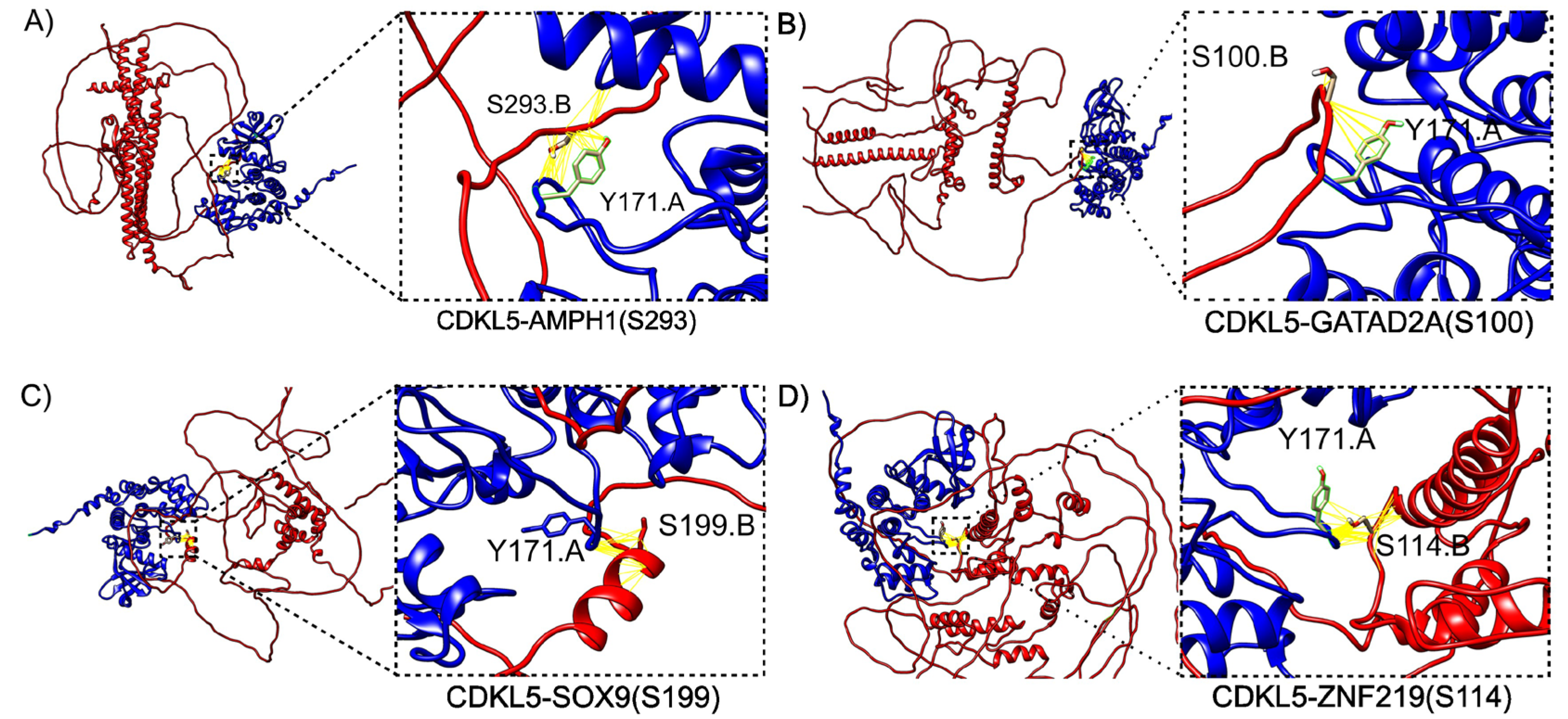
2.2. Homology Modeling of the CDKL5 Kinase Domain and CDKL5-Target Complex Prediction Using ColabFold and CDKL5-target protien-protein docking using HADDOCK3
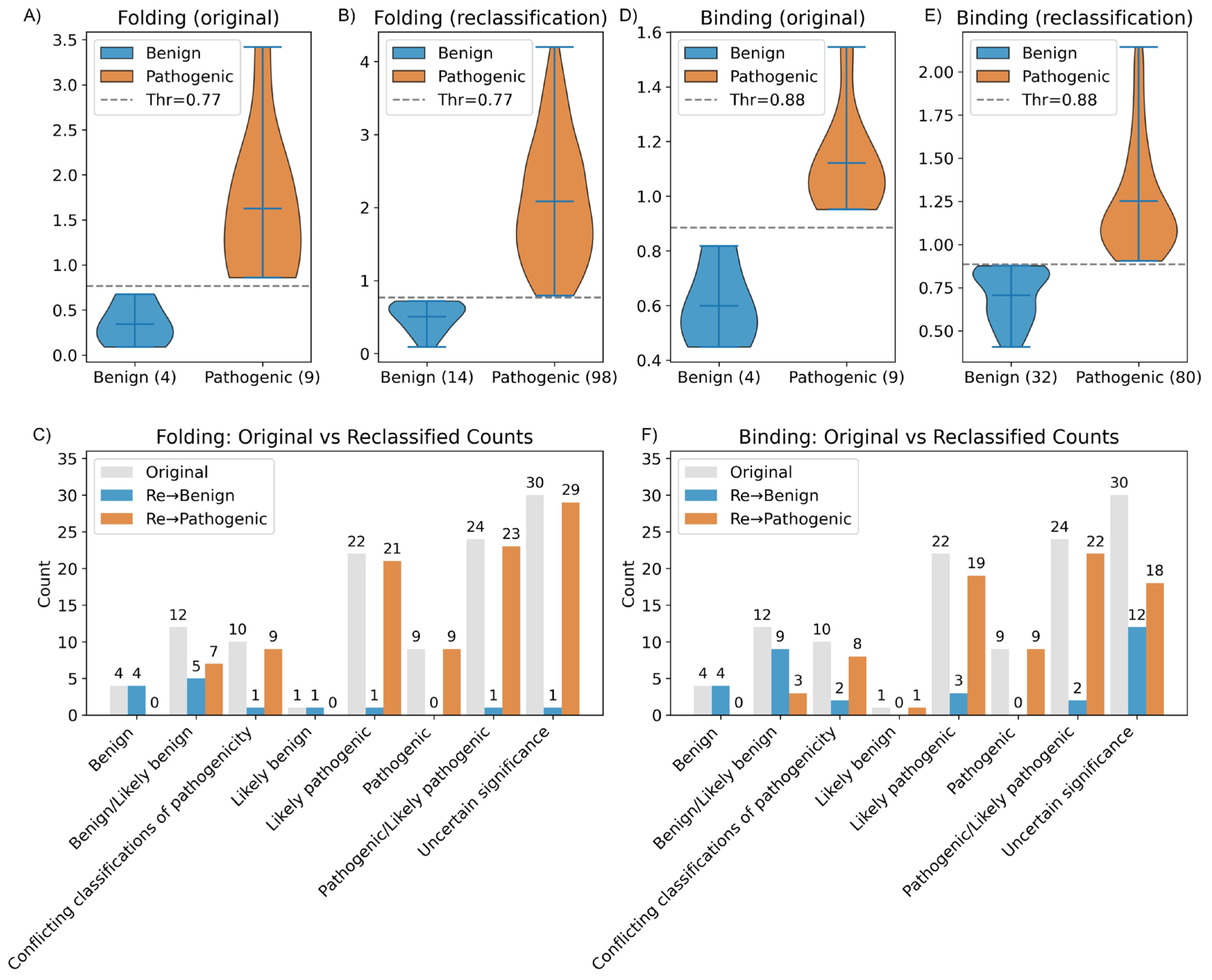
2.3. Folding, Docking ΔΔGfolding and ΔΔGbinding Analysis and Variant Reclassification
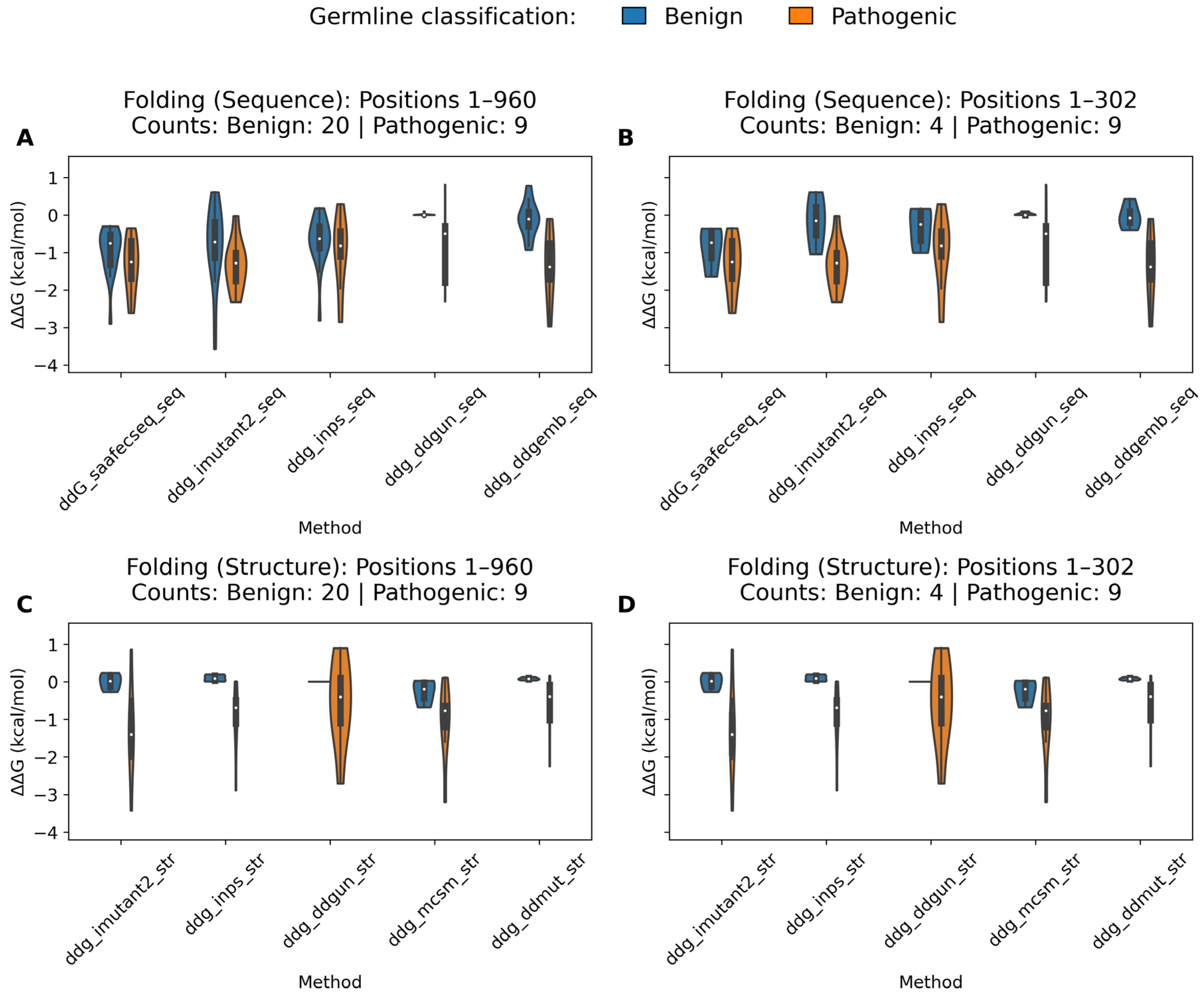
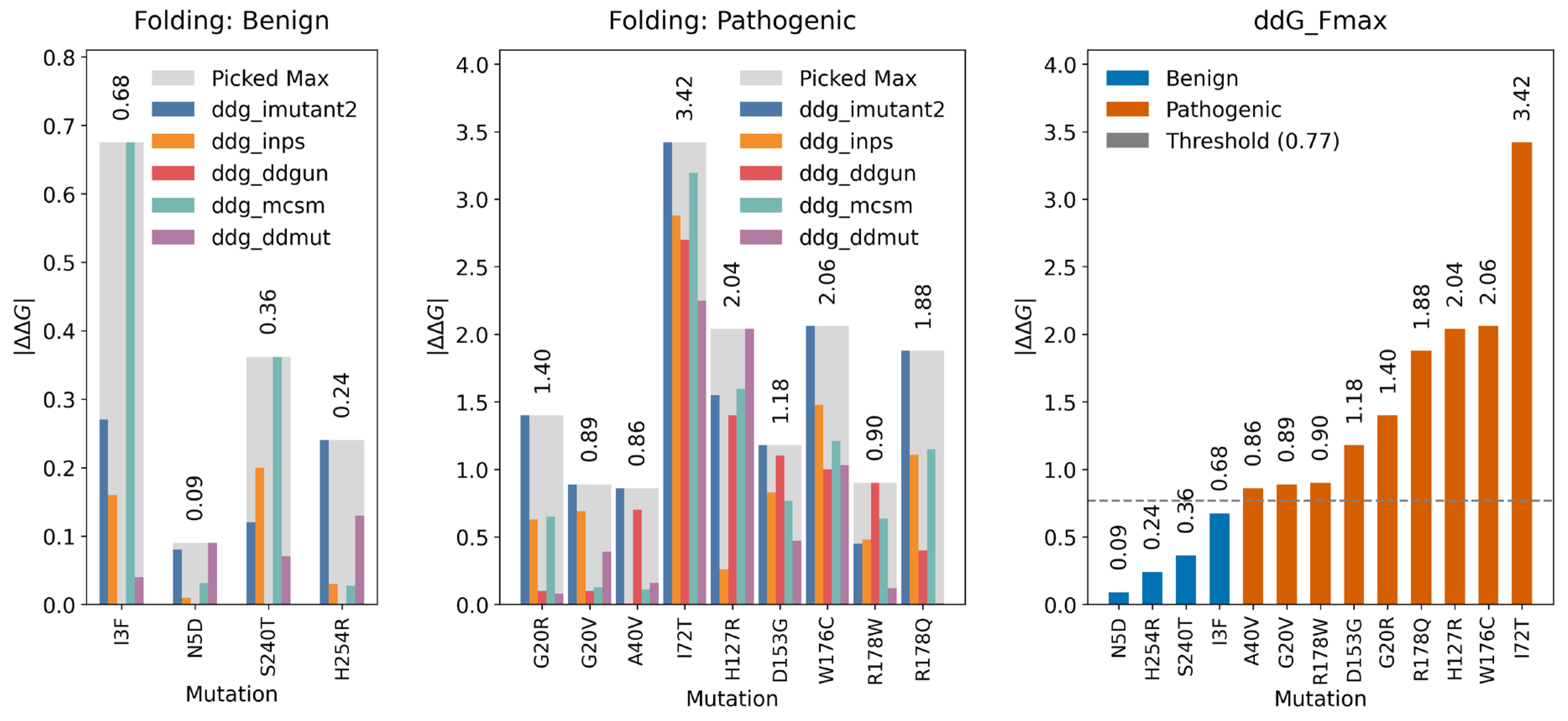
2.3.1. Folding Free Energy Change (ΔΔGfolding)
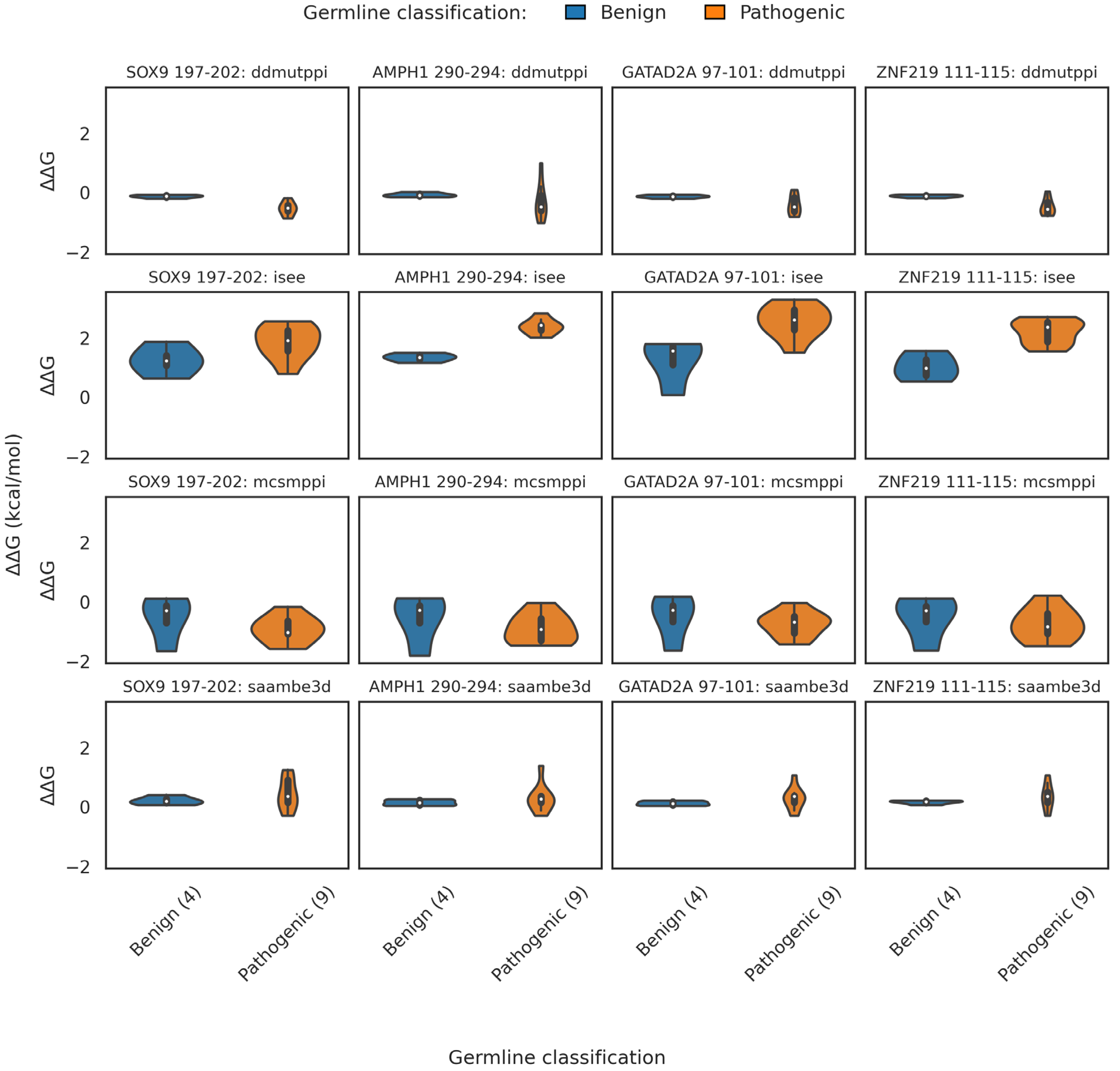
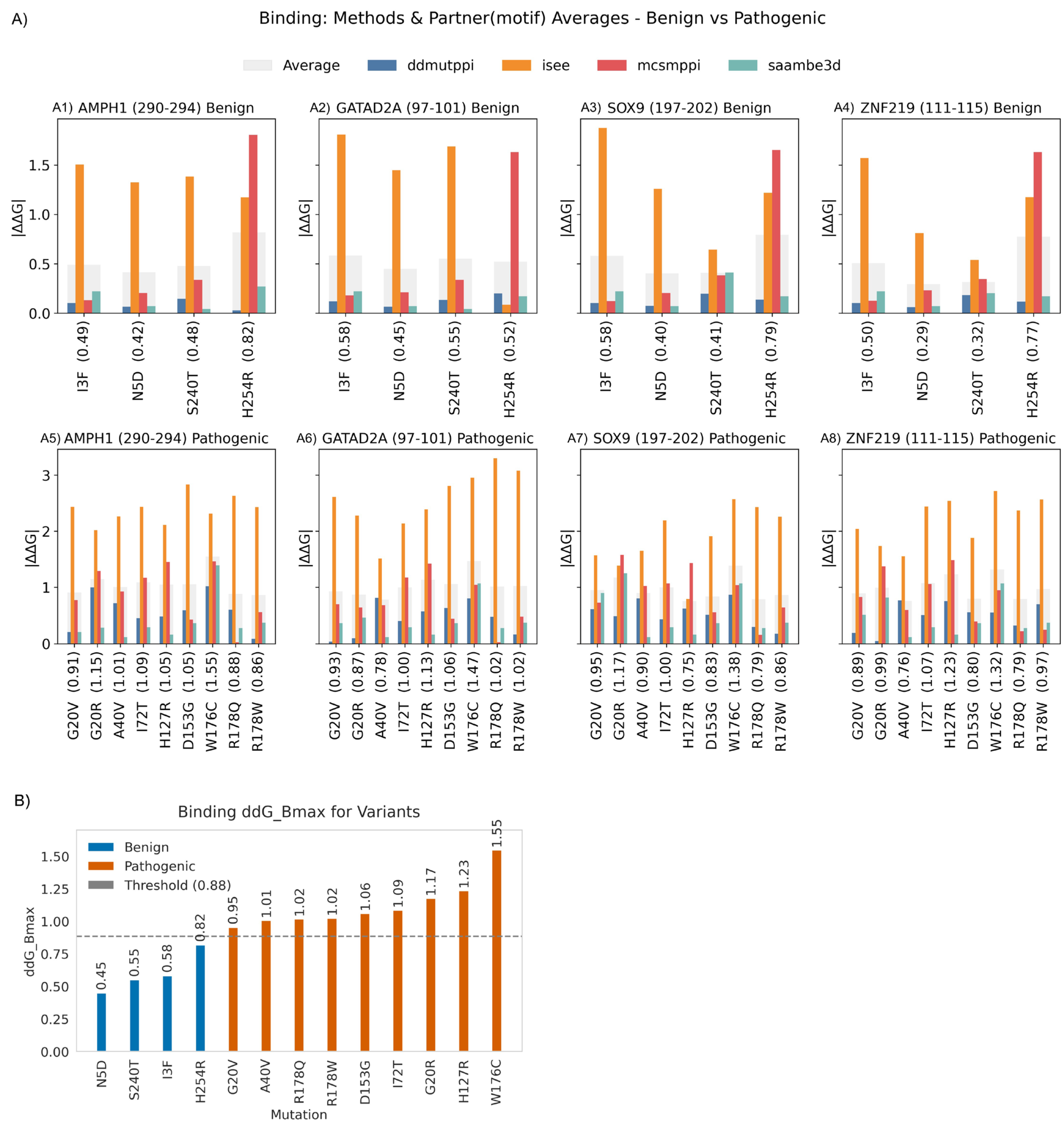
2.3.2. Binding Free Energy
2.3.3. Folding Threshold
2.3.4. Binding Threshold
2.3.5. Variants Reclassification Based on ΔΔGfolding and ΔΔGbinding Thresholds
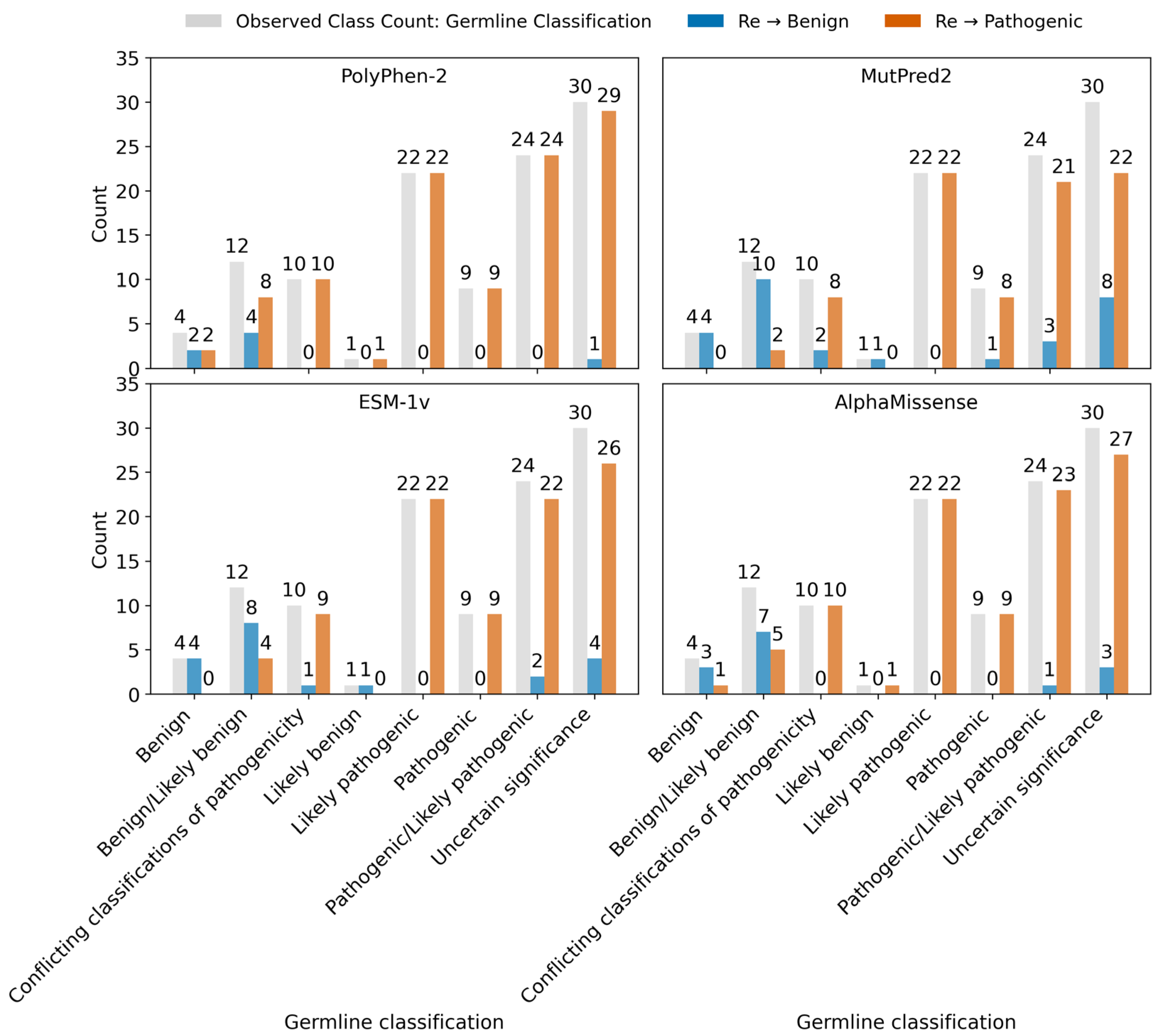
2.3.6. Variants Reclassification Based on Pathogenicity Score
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. CDKL5 Structure Preparation and Prediction of Complex with it’s binding partners
4.3. Folding Free Energy Calculations
4.4. HADDOCK3 Protein-Protein Docking
4.5. Binding Free Energy Calculations
4.6. CDKL5 Variant Reclassification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Abbreviations
| 1KGP | The 1000 Genomes Project |
| AMPH1 | Amphiphysin1 |
| ARHGEF2 | Rho guanine nucleotide exchange factor 2 |
| CACNA1E/Cav2.3 | Voltage-dependent R-type calcium channel subunit alpha-1E (Cav2.3) |
| CDD | CDKL5 Deficiency Disorder |
| CDKL5 | Cyclin-Dependent Kinase-Like 5 |
| CEP131 | Centrosomal protein of 131 kDa |
| CVI | Cortical/cerebral visual impairment |
| DLG5 | Disks large homolog 5 |
| DNMT1 | DNA methyltransferase 1 |
| EB2/MAPRE2 | Microtubule-associated protein RP/EB family member 2 |
| ELOA | Elongin A |
| EP400 | EE1A-binding protein p400 |
| GATAD2A | GATA zinc finger domain containing 2A |
| GRCh38 | The Genome Reference Consortium Human Build 38 |
| GTF2I | General transcription factor II-I |
| HDAC4 | Histone deacetylase 4 |
| IGSR | The International Genome Sample Resource |
| IQGAP1 | IQ Motif Containing GTPase Activating Protein 1 |
| MAP1S | Microtubule-associated protein 1S |
| MeCP2 | Methyl-CpG binding protein 2 |
| NGL-1/KIAA1580/LRRC4C | Netrin-G ligand-1 |
| PPP1R35 | Protein phosphatase 1 regulatory subunit 35 |
| PSD95/DLG4 | Postsynaptic density protein 95 |
| SHTN1/SHOT1 | Shootin1 |
| SMAD3 | Mothers against decapentaplegic homolog 3 |
| SOX9 | Transcription factor SOX-9 |
| SQSTM1/p62 | Sequestosome-1(p62) |
| STK9 | Serine threonine kinase 9 |
| TTDN1 | TTD non-photosensitive 1 protein |
| VDW | Van der Walls |
| XCI | X-chromosome inactivation |
| ZNF219 | Zinc finger protein 219 |
| ΔΔGbinding | Binding free energy change |
| ΔΔGfolding | Folding free energy changes |
| ΔΔGBmax | Maximum binding free energy change: For each variant, the maximum complex-averaged |ΔΔGbinding| across selected CDKL5-target protein-protein interactions. |
| ΔΔGFmax | Maximum folding free energy change: For each variant, the highest ΔΔGfolding value across several computational methods. |
References
- Olson, H.E.; Demarest, S.T.; Pestana-Knight, E.M.; Swanson, L.C.; Iqbal, S.; Lal, D.; Leonard, H.; Cross, J.H.; Devinsky, O.; Benke, T.A. Cyclin-Dependent Kinase-Like 5 Deficiency Disorder: Clinical Review. Pediatr Neurol 2019, 97, 18–25. [Google Scholar] [CrossRef]
- Akiba, T.; Shimada, S.; Imai, K.; Takahashi, S. A case of CDKL5 deficiency disorder with a novel intragenic multi-exonic duplication. Hum Genome Var 2024, 11, 1–3. [Google Scholar] [CrossRef]
- Jakimiec, M.; Paprocka, J.; Śmigiel, R. CDKL5 Deficiency Disorder-A Complex Epileptic Encephalopathy. Brain Sci 2020, 10, 107. [Google Scholar] [CrossRef]
- Leonard, H.; Downs, J.; Benke, T.A.; Swanson, L.; Olson, H.; Demarest, S. CDKL5 deficiency disorder: clinical features, diagnosis, and management. Lancet Neurol 2022, 21, 563–576. [Google Scholar] [CrossRef]
- Daniels, C.; Greene, C.; Smith, L.; Pestana-Knight, E.; Demarest, S.; Zhang, B.; Benke, T.A.; Poduri, A.; Olson, H. CDKL5 deficiency disorder and other infantile-onset genetic epilepsies. Dev Med Child Neurol 2024, 66, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Siri, B.; Varesio, C.; Freri, E.; Darra, F.; Gana, S.; Mei, D.; Porta, F.; Fontana, E.; Galati, G.; Solazzi, R.; et al. CDKL5 deficiency disorder in males: Five new variants and review of the literature. Eur J Paediatr Neurol 2021, 33, 9–20. [Google Scholar] [CrossRef]
- Dell’Isola, G.B.; Fattorusso, A.; Pisani, F.; Mastrangelo, M.; Cordelli, D.M.; Pavone, P.; Parisi, P.; Ferretti, A.; Operto, F.F.; Elia, M.; et al. CDKL5 deficiency-related neurodevelopmental disorders: a multi-center cohort study in Italy. J Neurol 2024, 271, 5368–5377. [Google Scholar] [CrossRef]
- Lombardo, A.; Sinibaldi, L.; Genovese, S.; Catino, G.; Mei, V.; Pompili, D.; Sallicandro, E.; Falasca, R.; Liambo, M.T.; Faggiano, M.V.; et al. A Case of CDKL5 Deficiency Due to an X Chromosome Pericentric Inversion: Delineation of Structural Rearrangements as an Overlooked Recurrent Pathological Mechanism. Int J Mol Sci 2024, 25, 6912. [Google Scholar] [CrossRef]
- Evans, J.C.; Archer, H.L.; Colley, J.P.; Ravn, K.; Nielsen, J.B.; Kerr, A.; Williams, E.; Christodoulou, J.; Gécz, J.; Jardine, P.E.; et al. Early onset seizures and Rett-like features associated with mutations in CDKL5. Eur J Hum Genet 2005, 13, 1113–1120. [Google Scholar] [CrossRef]
- Martinez, D.; Jiang, E.; Zhou, Z. Overcoming genetic and cellular complexity to study the pathophysiology of X-linked intellectual disabilities. J Neurodev Disord 2024, 16, 5. [Google Scholar] [CrossRef]
- Adhikari, A.; Buchanan, F.K.B.; Fenton, T.A.; Cameron, D.L.; Halmai, J.A.N.M.; Copping, N.A.; Fink, K.D.; Silverman, J.L. Touchscreen cognitive deficits, hyperexcitability and hyperactivity in males and females using two models of Cdkl5 deficiency. Hum Mol Genet 2022, 31, 3032–3050. [Google Scholar] [CrossRef]
- Galvani, G.; Mottolese, N.; Gennaccaro, L.; Loi, M.; Medici, G.; Tassinari, M.; Fuchs, C.; Ciani, E.; Trazzi, S. Inhibition of microglia overactivation restores neuronal survival in a mouse model of CDKL5 deficiency disorder. J Neuroinflammation 2021, 18, 155. [Google Scholar] [CrossRef]
- Benke, T.A.; Demarest, S.; Angione, K.; Downs, J.; Leonard, H.; Saldaris, J.; Marsh, E.D.; Olson, H.; Haviland, I. CDKL5 Deficiency Disorder. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle: Seattle (WA), 1993. [Google Scholar]
- Olson, H.E.; Daniels, C.I.; Haviland, I.; Swanson, L.C.; Greene, C.A.; Denny, A.M.M.; Demarest, S.T.; Pestana-Knight, E.; Zhang, X.; Moosa, A.N.; et al. Current neurologic treatment and emerging therapies in CDKL5 deficiency disorder. Journal of Neurodevelopmental Disorders 2021, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Dell’Isola, G.B.; Perinelli, M.G.; Frulli, A.; D’Onofrio, G.; Fattorusso, A.; Siciliano, M.; Ferrara, P.; Striano, P.; Verrotti, A. Exploring neurodevelopment in CDKL5 deficiency disorder: Current insights and future directions. Epilepsy Behav 2025, 171, 110504. [Google Scholar] [CrossRef] [PubMed]
- Melikishvili, G.; Sharkov, A.; Gachechiladze, T.; Tomenko, T.; Pivovarova, A.; Volkov, I.; Andrade, M.-T.; Castellanos, A.; Bienvenu, T.; Dulac, O.; et al. Epileptic spasms with terror during sleep in CDKL5 encephalopathy. Sleep Adv 2022, 3, zpac010. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Helbig, I.; Jansen, C.; Bast, T.; Guerrini, R.; Jähn, J.; Muhle, H.; Auvin, S.; Korenke, G.C.; Philip, S.; et al. Retrospective evaluation of low long-term efficacy of antiepileptic drugs and ketogenic diet in 39 patients with CDKL5-related epilepsy. Eur J Paediatr Neurol 2016, 20, 147–151. [Google Scholar] [CrossRef]
- Klein, K.M.; Yendle, S.C.; Harvey, A.S.; Antony, J.H.; Wallace, G.; Bienvenu, T.; Scheffer, I.E. A distinctive seizure type in patients with CDKL5 mutations: Hypermotor-tonic-spasms sequence. Neurology 2011, 76, 1436–1438. [Google Scholar] [CrossRef]
- Massey, S.; Quigley, A.; Rochfort, S.; Christodoulou, J.; Van Bergen, N.J. Cannabinoids and Genetic Epilepsy Models: A Review with Focus on CDKL5 Deficiency Disorder. Int J Mol Sci 2024, 25, 10768. [Google Scholar] [CrossRef]
- Sun, X.; Wang, T. Research progress on the pathogenesis of CDKL5 pathogenic variants and related encephalopathy. Eur J Pediatr 2023, 182, 3049–3056. [Google Scholar] [CrossRef]
- Silvestre, M.; Dempster, K.; Mihaylov, S.R.; Claxton, S.; Ultanir, S.K. Cell type-specific expression, regulation and compensation of CDKL5 activity in mouse brain. Mol Psychiatry 2024, 29, 1844–1856. [Google Scholar] [CrossRef]
- Saldaris, J.M.; Jacoby, P.; Marsh, E.D.; Suter, B.; Leonard, H.; Olson, H.E.; Rajaraman, R.; Pestana-Knight, E.; Weisenberg, J.; Price, D.; et al. Adapting a measure of gross motor skills for individuals with CDKL5 deficiency disorder: A psychometric study. Epilepsy Res 2024, 200, 107287. [Google Scholar] [CrossRef]
- Wong, K.; Junaid, M.; Demarest, S.; Saldaris, J.; Benke, T.A.; Marsh, E.D.; Downs, J.; Leonard, H. Factors influencing the attainment of major motor milestones in CDKL5 deficiency disorder. Eur J Hum Genet 2023, 31, 169–178. [Google Scholar] [CrossRef]
- Brock, D.; Fidell, A.; Thomas, J.; Juarez-Colunga, E.; Benke, T.A.; Demarest, S. Cerebral Visual Impairment in CDKL5 Deficiency Disorder Correlates With Developmental Achievement. J Child Neurol 2021, 36, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Quintiliani, M.; Ricci, D.; Petrianni, M.; Leone, S.; Orazi, L.; Amore, F.; Gambardella, M.L.; Contaldo, I.; Veredice, C.; Perulli, M.; et al. Cortical Visual Impairment in CDKL5 Deficiency Disorder. Front Neurol 2021, 12, 805745. [Google Scholar] [CrossRef] [PubMed]
- Peikes, T.; Hartley, J.N.; Mhanni, A.A.; Greenberg, C.R.; Appendino, J.P. Reflex Seizures in a Patient with CDKL5 Deficiency Disorder. Can J Neurol Sci 2019, 46, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Downs, J.; Jacoby, P.; Saldaris, J.; Leonard, H.; Benke, T.; Marsh, E.; Demarest, S. Negative impact of insomnia and daytime sleepiness on quality of life in individuals with the cyclin-dependent kinase-like 5 deficiency disorder. J Sleep Res 2022, 31, e13600. [Google Scholar] [CrossRef]
- Amin, S.; Monaghan, M.; Aledo-Serrano, A.; Bahi-Buisson, N.; Chin, R.F.; Clarke, A.J.; Cross, J.H.; Demarest, S.; Devinsky, O.; Downs, J.; et al. International Consensus Recommendations for the Assessment and Management of Individuals With CDKL5 Deficiency Disorder. Front Neurol 2022, 13, 874695. [Google Scholar] [CrossRef]
- La Montanara, P.; Hervera, A.; Baltussen, L.L.; Hutson, T.H.; Palmisano, I.; De Virgiliis, F.; Kong, G.; Chadwick, J.; Gao, Y.; Bartus, K.; et al. Cyclin-dependent-like kinase 5 is required for pain signaling in human sensory neurons and mouse models. Sci Transl Med 2020, 12, eaax4846. [Google Scholar] [CrossRef]
- Specchio, N.; Trivisano, M.; Lenge, M.; Ferretti, A.; Mei, D.; Parrini, E.; Napolitano, A.; Rossi-Espagnet, C.; Talenti, G.; Longo, D.; et al. CDKL5 deficiency disorder: progressive brain atrophy may be part of the syndrome. Cereb Cortex 2023, 33, 9709–9717. [Google Scholar] [CrossRef]
- Van Bergen, N.J.; Massey, S.; Quigley, A.; Rollo, B.; Harris, A.R.; Kapsa, R.M.I.; Christodoulou, J. CDKL5 deficiency disorder: molecular insights and mechanisms of pathogenicity to fast-track therapeutic development. Biochem Soc Trans 2022, 50, 1207–1224. [Google Scholar] [CrossRef]
- Chowdhury, I.; Dashi, G.; Keskitalo, S. CMGC Kinases in Health and Cancer. Cancers (Basel) 2023, 15, 3838. [Google Scholar] [CrossRef]
- Quadalti, C.; Sannia, M.; Humphreys, N.E.; Baldassarro, V.A.; Gurgone, A.; Ascolani, M.; Zanella, L.; Giardino, L.; Gross, C.T.; Croci, S.; et al. A new knockin mouse carrying the E364X patient mutation for CDKL5 deficiency disorder: neurological, behavioral and molecular profiling. Heliyon 2024, 10, e40165. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, F.F.; Myers, C.T.; Cossette, P.; Lemay, P.; Spiegelman, D.; Laporte, A.D.; Nassif, C.; Diallo, O.; Monlong, J.; Cadieux-Dion, M.; et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am J Hum Genet 2017, 101, 664–685. [Google Scholar] [CrossRef] [PubMed]
- Benke, T.A.; Demarest, S.; Angione, K.; Downs, J.; Leonard, H.; Saldaris, J.; Marsh, E.D.; Olson, H.; Haviland, I. CDKL5 Deficiency Disorder. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle: Seattle (WA), 1993. [Google Scholar]
- Katayama, S.; Sueyoshi, N.; Inazu, T.; Kameshita, I. Cyclin-Dependent Kinase-Like 5 (CDKL5): Possible Cellular Signalling Targets and Involvement in CDKL5 Deficiency Disorder. Neural Plast 2020, 2020, 6970190. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, L.; Salvatoni, L.; Giudici, L.; Bertani, I.; Kilstrup-Nielsen, C.; Broccoli, V.; Landsberger, N. CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J Biol Chem 2008, 283, 30101–30111. [Google Scholar] [CrossRef]
- Muñoz, I.M.; Morgan, M.E.; Peltier, J.; Weiland, F.; Gregorczyk, M.; Brown, F.C.; Macartney, T.; Toth, R.; Trost, M.; Rouse, J. Phosphoproteomic screening identifies physiological substrates of the CDKL5 kinase. EMBO J 2018, 37, e99559. [Google Scholar] [CrossRef]
- Baltussen, L.L.; Negraes, P.D.; Silvestre, M.; Claxton, S.; Moeskops, M.; Christodoulou, E.; Flynn, H.R.; Snijders, A.P.; Muotri, A.R.; Ultanir, S.K. Chemical genetic identification of CDKL5 substrates reveals its role in neuronal microtubule dynamics. EMBO J 2018, 37, e99763. [Google Scholar] [CrossRef]
- Eyers, P.A. A new consensus for evaluating CDKL5/STK9-dependent signalling mechanisms. EMBO J 2018, 37, e100848. [Google Scholar] [CrossRef]
- Li, C.; Liu, Y.; Luo, S.; Yang, M.; Li, L.; Sun, L. A review of CDKL: An underestimated protein kinase family. International Journal of Biological Macromolecules 2024, 277, 133604. [Google Scholar] [CrossRef]
- Massey, S.; Ang, C.-S.; Davidson, N.M.; Quigley, A.; Rollo, B.; Harris, A.R.; Kapsa, R.M.I.; Christodoulou, J.; Van Bergen, N.J. Novel CDKL5 targets identified in human iPSC-derived neurons. Cell Mol Life Sci 2024, 81, 347. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014, 42, D980–985. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium; Auton, A. ; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Hector, R.D.; Kalscheuer, V.M.; Hennig, F.; Leonard, H.; Downs, J.; Clarke, A.; Benke, T.A.; Armstrong, J.; Pineda, M.; Bailey, M.E.S.; et al. CDKL5 variants: Improving our understanding of a rare neurologic disorder. Neurol Genet 2017, 3, e200. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2024, 625, 92–100. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol 2016, 17, 122. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fong, C.S.; Ozaki, K.; Tsou, M.-F.B. PPP1R35 ensures centriole homeostasis by promoting centriole-to-centrosome conversion. Mol Biol Cell 2018, 29, 2801–2808. [Google Scholar] [CrossRef]
- Archambault, D.; Cheong, A.; Iverson, E.; Tremblay, K.D.; Mager, J. Protein phosphatase 1 regulatory subunit 35 is required for ciliogenesis, notochord morphogenesis, and cell-cycle progression during murine development. Dev Biol 2020, 465, 1–10. [Google Scholar] [CrossRef]
- Panda, P.; Kovacs, L.; Dzhindzhev, N.; Fatalska, A.; Persico, V.; Geymonat, M.; Riparbelli, M.G.; Callaini, G.; Glover, D.M. Tissue specific requirement of Drosophila Rcd4 for centriole duplication and ciliogenesis. J Cell Biol 2020, 219, e201912154. [Google Scholar] [CrossRef]
- Lepanto, P.; Badano, J.L.; Zolessi, F.R. Neuron’s little helper: The role of primary cilia in neurogenesis. Neurogenesis (Austin) 2016, 3, e1253363. [Google Scholar] [CrossRef]
- Adams, J.W.; Vinokur, A.; de Souza, J.S.; Austria, C.; Guerra, B.S.; Herai, R.H.; Wahlin, K.J.; Muotri, A.R. Loss of GTF2I promotes neuronal apoptosis and synaptic reduction in human cellular models of neurodevelopment. Cell Rep 2024, 43, 113867. [Google Scholar] [CrossRef]
- López-Tobón, A.; Shyti, R.; Villa, C.E.; Cheroni, C.; Fuentes-Bravo, P.; Trattaro, S.; Caporale, N.; Troglio, F.; Tenderini, E.; Mihailovich, M.; et al. GTF2I dosage regulates neuronal differentiation and social behavior in 7q11.23 neurodevelopmental disorders. Sci Adv 2023, 9, eadh2726. [Google Scholar] [CrossRef]
- Barak, B.; Feng, G. Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat Neurosci 2016, 19, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.-O.; Makeyev, A.V.; Ruddle, F.H.; Bayarsaihan, D. Identification of the TFII-I family target genes in the vertebrate genome. Proc Natl Acad Sci U S A 2008, 105, 9006–9010. [Google Scholar] [CrossRef] [PubMed]
- Hakre, S.; Tussie-Luna, M.I.; Ashworth, T.; Novina, C.D.; Settleman, J.; Sharp, P.A.; Roy, A.L. Opposing functions of TFII-I spliced isoforms in growth factor-induced gene expression. Mol Cell 2006, 24, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Ungaro, F.; Hambrock, M.; Rademacher, N.; Stefanelli, G.; Brambilla, D.; Sessa, A.; Magagnotti, C.; Bachi, A.; Giarda, E.; et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 2012, 14, 911–923. [Google Scholar] [CrossRef]
- Yan, M.; Guo, X.; Xu, C. Revealing the complex role of CDKL5 in developmental epilepsy through a calcium channel related vision. Acta Epileptol 2024, 6, 15. [Google Scholar] [CrossRef]
- Sampedro-Castañeda, M.; Baltussen, L.L.; Lopes, A.T.; Qiu, Y.; Sirvio, L.; Mihaylov, S.R.; Claxton, S.; Richardson, J.C.; Lignani, G.; Ultanir, S.K. Epilepsy-linked kinase CDKL5 phosphorylates voltage-gated calcium channel Cav2.3, altering inactivation kinetics and neuronal excitability. Nat Commun 2023, 14, 7830. [Google Scholar] [CrossRef]
- Thinwa, J.W.; Zou, Z.; Parks, E.; Sebti, S.; Hui, K.; Wei, Y.; Goodarzi, M.; Singh, V.; Urquhart, G.; Jewell, J.L.; et al. CDKL5 regulates p62-mediated selective autophagy and confers protection against neurotropic viruses. J Clin Invest 2024, 134, e168544. [Google Scholar] [CrossRef]
- Canning, P.; Park, K.; Gonçalves, J.; Li, C.; Howard, C.J.; Sharpe, T.D.; Holt, L.J.; Pelletier, L.; Bullock, A.N.; Leroux, M.R. CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function. Cell Rep 2018, 22, 885–894. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: making protein folding accessible to all. Nat Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pandey, P.; Alexov, E. Most Monogenic Disorders Are Caused by Mutations Altering Protein Folding Free Energy. Int J Mol Sci 2024, 25, 1963. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Ghimire, S.; Wu, B.; Alexov, E. On the linkage of thermodynamics and pathogenicity. Curr Opin Struct Biol 2023, 80, 102572. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Panday, S.K.; Rimal, P.; Ancona, N.; Alexov, E. Predicting the Effect of Single Mutations on Protein Stability and Binding with Respect to Types of Mutations. Int J Mol Sci 2023, 24, 12073. [Google Scholar] [CrossRef] [PubMed]
- Poudel, P.; Shapovalov, I.; Panday, S.K.; Nouri, K.; Davies, P.L.; Greer, P.A.; Alexov, E. In Silico Screening for Small Molecules to Alter Calpain Proteolysis through Modulating Conformation Changes Induced by Heterodimerization. J Chem Inf Model 2025, 65, 5528–5543. [Google Scholar] [CrossRef]
- Zhang, Z.; Witham, S.; Petukh, M.; Moroy, G.; Miteva, M.; Ikeguchi, Y.; Alexov, E. A rational free energy-based approach to understanding and targeting disease-causing missense mutations. J Am Med Inform Assoc 2013, 20, 643–651. [Google Scholar] [CrossRef]
- Poudel, P.; Miteva, M.A.; Alexov, E. Strategies for in Silico Drug Discovery to Modulate Macromolecular Interactions Altered by Mutations. FBL 2025, 30, 26339. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res 2020, 48, D941–D947. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Savojardo, C.; Manfredi, M.; Martelli, P.L.; Casadio, R. DDGemb: predicting protein stability change upon single- and multi-point variations with embeddings and deep learning. Bioinformatics 2024, 41, btaf019. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef]
- Kumar, M.D.S. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Research 2006, 34, D204–D206. [Google Scholar] [CrossRef] [PubMed]
- Stourac, J.; Dubrava, J.; Musil, M.; Horackova, J.; Damborsky, J.; Mazurenko, S.; Bednar, D. FireProtDB: database of manually curated protein stability data. Nucleic Acids Res 2021, 49, D319–D324. [Google Scholar] [CrossRef]
- Dehouck, Y.; Kwasigroch, J.M.; Gilis, D.; Rooman, M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics 2011, 12, 151. [Google Scholar] [CrossRef]
- Zhou, Y.; Pan, Q.; Pires, D.E.V.; Rodrigues, C.H.M.; Ascher, D.B. DDMut: predicting effects of mutations on protein stability using deep learning. Nucleic Acids Res 2023, 51, W122–W128. [Google Scholar] [CrossRef]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci 2021, 30, 60–69. [Google Scholar] [CrossRef]
- Li, G.; Panday, S.K.; Alexov, E. SAAFEC-SEQ: A Sequence-Based Method for Predicting the Effect of Single Point Mutations on Protein Thermodynamic Stability. Int J Mol Sci 2021, 22, 606. [Google Scholar] [CrossRef]
- Montanucci, L.; Capriotti, E.; Frank, Y.; Ben-Tal, N.; Fariselli, P. DDGun: an untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinformatics 2019, 20, 335. [Google Scholar] [CrossRef]
- Eddy, S.R. Where did the BLOSUM62 alignment score matrix come from? Nat Biotechnol 2004, 22, 1035–1036. [Google Scholar] [CrossRef]
- Sasidharan Nair, P.; Vihinen, M. VariBench: a benchmark database for variations. Hum Mutat 2013, 34, 42–49. [Google Scholar] [CrossRef]
- Savojardo, C.; Fariselli, P.; Martelli, P.L.; Casadio, R. INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics 2016, 32, 2542–2544. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 2005, 33, W306–310. [Google Scholar] [CrossRef]
- Giulini, M.; Reys, V.; Teixeira, J.M.C.; Jiménez-García, B.; V Honorato, R.; Kravchenko, A.; Xu, X.; Versini, R.; Engel, A.; Verhoeven, S.; et al. HADDOCK3: A Modular and Versatile Platform for Integrative Modeling of Biomolecular Complexes. J Chem Inf Model 2025, 65, 7315–7324. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pahari, S.; Li, G.; Murthy, A.K.; Liang, S.; Fragoza, R.; Yu, H.; Alexov, E. SAAMBE-3D: Predicting Effect of Mutations on Protein-Protein Interactions. Int J Mol Sci 2020, 21, 2563. [Google Scholar] [CrossRef]
- Geng, C.; Vangone, A.; Folkers, G.E.; Xue, L.C.; Bonvin, A.M.J.J. iSEE: Interface structure, evolution, and energy-based machine learning predictor of binding affinity changes upon mutations. Proteins 2019, 87, 110–119. [Google Scholar] [CrossRef]
- Gribskov, M.; McLachlan, A.D.; Eisenberg, D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A 1987, 84, 4355–4358. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet, 7. [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat Commun 2020, 11, 5918. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





