1. Introduction
Shrimp aquaculture emerged as an international enterprise during the latter part of the 20th century, delivering nutrition- and protein-dense seafood to meet growing global market demand. With the boom of shrimp farming throughout much of Asia and beyond came a formidable foe: White Spot Syndrome Virus (WSSV), now one of the most virulent pathogens of shrimp in aquaculture. Since the emergence of WSSV in 1992, the global shrimp sector has suffered an estimated USD 8-15 billion in economic losses from this single disease (Verbruggen, Bickley et al. 2016, Panchal, Kumar et al. 2021). For the Asian shrimp industry a loss of about USD 20 billion due to WSD was possibly its worst experience (Davies 2016). The widespread presence of WSSV in global shrimp farms is a continuing nightmare for farmers. The virus can persist in pond sediments and surrounding areas for over 20 months, with studies detecting its presence in ponds soil for over ten months post-outbreak (Quang, Hoa et al. 2009). Notably, water serves as a critical medium for rapid viral dissemination; research has shown that WSSV DNA can be detected in water within six hours of disease onset in shrimp, with shedding intensifying until the host's death (Cox, De Swaef et al. 2023). Early reports of this disease from shrimp farms in China, Thailand, Vietnam, Indonesia, Ecuador and many countries that are among the largest producers of farmed shrimp in the world
(Figure 1). The speed with which the virus spread also reflected how quickly and across borders live shrimp and possibly contaminated water were traded in the globalized shrimp industry. By the early 2000s, WSSV had spread without control in shrimp aquaculture and epidemics appeared to be underway in almost all major shrimp-producing countries globally (Lotz and Soto 2002). While the devastating impacts of the virus were felt worldwide by shrimp farmers and aquaculture scientists, the virus continued to evolve infecting new species and gradually adapting to different environmental conditions. Over the years, WSSV advanced so rapidly – from farms in Bangladesh to shrimp ponds in Brazil – that the industry called for more sustainable farming practices paired with effective disease management (Hasan, Haque et al. 2020).
2. Integrated Review and Analytical Methods
This review presents a systematic effort in understanding the biology, pathology, and diagnostic methods as well as control measures associated with White Spot Syndrome Virus (WSSV), a globally-important pathogen of shrimp aquaculture. Data were synthesized according to a structured methodology adopted from various primary and secondary sources. A semi-structured search of relevant literature was conducted using the databases of Web of Science, Scopus, PubMed, and Google Scholar, focusing on peer-reviewed articles, government reports, and industry publications. Keywords used in the database search included variations and combinations of "WSSV diagnostics," "shrimp viral diseases," "aquaculture sustainability," and "viral transmission in crustaceans". After reviewing these primary sources, additional literature was identified by examining references cited in the initial studies, as well as through subsequent non-systematic searches on Google Scholar, Web of Science, and ScienceDirect. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines were used to further transparency in the entire selection process. A total of 202 articles were found, and the final number of articles that were considered relevant for analysis was 108 after removal of duplicates and articles not relevant. These articles gave insights on the historical emergence and global distribution of WSSV and the biological mechanisms of its infection. To enhance the scope of the review, spatial and phylogenetic analyses were performed using secondary data. GPS coordinates of WSSV-infected zones were collected from government and non-government databases and visualized using ESRI’s ArcGIS software (version 10.8). Phylogenetic trees, depicting the genetic diversity of WSSV, were constructed using the VICTOR algorithm based on nucleotide sequence similarity data sourced from the NCBI database. Moreover, the review covers the economic and ecological impact assessments of WSSV outbreaks and these were conducted by compiling the global production statistics and analyzing co-infection reports with other pathogens. Such an integrative approach allowed the identification of patterns in WSSV spread and resilience mechanisms in shrimp.
3. History of White Spot Syndrome Virus
Shrimp is one of the most valuable species in global aquaculture, prized for its high levels of protein, omega-3 fatty acids, vitamins, and minerals (Sass 2022). The development of modern shrimp farming began in Japan, where Fujinaga pioneered semi-intensive shrimp farming techniques in the mid-20th century (Flegel 1997). His innovations, including advancements in shrimp spawning, larval rearing, and growth techniques, laid the groundwork for the expansion of shrimp farming to other countries such as Taiwan and the United States (Chamberlain 2010). As semi-intensive shrimp farming techniques were adopted globally, inputs such as feed, therapeutic agents, and overstocking were introduced without proper regulation, resulting in outbreaks of various diseases (e.g., white spot syndrome virus (WSSV), Enterocytozoon hepatopenaei (EHP), infectious hypodermal and hematopoietic necrosis (IHHNV), infectious myonecrosis virus (IMNV), yellow head virus (YHV), Taura syndrome virus (TSV), Macrobrachium rosenbergii nodavirus (MrNV), and acute hepatopancreatic necrosis disease (AHPND)) (Jithendran, Navaneeth Krishnan et al. 2021, Patil, Geetha et al. 2021). Among these, WSSV has caused the most devastating financial losses with mortality rates up to 100% within 7-10 days of infection (Talukder, Punom et al. 2021). WSSV was first reported in 1992 in cultured Penaeus japonicas in Taiwan and China (Chou Hy, Wang et al. 1995, Zhan, Wang et al. 1998), quickly spreading to Japan and Korea by 1993 where the disease was reported in farmed Peaeaus japonicas and Penaeus orientalis, respectively (Inouye, Miwa et al. 1994, Park, Lee et al. 1998). The rapidity with which WSSV spread across Asia caused massive devastation to the shrimp aquaculture industry, conservatively estimated at billions of dollars in losses, severely affecting the local economies of those countries. This single disease caused annual losses of over USD 500 million in China during its first prevalence due to reduced shrimp yields (Pereira Dantas Da Rocha Lima 2013). This outbreak seriously affected global supply of shrimp while China was one of the large shrimp producers at the time and continues to maintain its production pace. By 1994, the virus spread throughout Southeast Asia, affecting countries such as Thailand, Malaysia, Indonesia, Vietnam, India and Bangladesh (Flegel 1997, Shankar and Mohan 1998, Wang, Hassan et al. 1999, Sunarto, Widodo et al. 2004). During this period, Thailand was the world's largest shrimp producer, and it was estimated that outbreak caused losses of approximately USD 600 million within one year, crippling the aquaculture industry (Chanratchakool and Phillips 2002). In India, the virus caused annual economic losses exceeding USD 100 million due to a more than 80% reduction in shrimp exports (Kalaimani, Ravisankar et al. 2013). The spread of virus in Vietnam, resulted in annual losses of approximately USD 200 million, seriously damaging the nation's economy (Chanratchakool and Phillips 2002). Indonesia which ranked second globally as a shrimp-producing country by 2001, WSSV induced annual losses ranging between USD 300 million and USD 400 million (Evan and Putri 2021). In Bangladesh, the first major outbreak occurred in semi-intensive shrimp farms in Cox's Bazar in 1994, primarily affecting Penaeus monodon. The outbreak led to widespread devastation, with 90% of shrimp farms impacted, resulting in a 20% decrease in national shrimp production. A subsequent outbreak in 2001, driven by unplanned and uncontrolled expansion of shrimp farming, affected 25% of production (Debnath, Karim et al. 2014). Since 2007, the frequency of outbreaks in Bangladesh has increased, with WSSV remaining the leading cause of production loss (Hasan, Haque et al. 2020). The spread of WSSV was not confined to Asia. By 1995, the virus had reached the United States, likely introduced through frozen shrimp imports. The virus was detected in cultured shrimp in Texas and South Carolina in 1997 and 1998, respectively (Lightner, Redman et al. 1997). Subsequently, in 1999, major WSSV epizootics occurred in Ecuador, Panama, Honduras, Guatemala, Mexico, Cambodia, Nicaragua and South Asian country Philippines, primarily affecting cultured P. monodon (Fe, Karlo et al. 2000, Rodríguez, Bayot et al. 2003, Galavíz-Silva, Molina-Garza et al. 2004, Chamberlain, Lightner et al. 2013). In Ecuador, a major shrimp exporting country in Latin America, losses were estimated at over USD 300 million annually during early 2000s (Stern and Sonnenholzner 2010). WSSV infections were causing losses of more than USD 300 million annually in Mexico during the early years of the outbreak, which prompted widespread adoption of biosecurity measures to reduce the impact of the virus (López-Téllez, Corbalá-Bermejo et al. 2020).
Table 1.
Recent outbreak of WSSV in different countries whole over the world (DISEASES-Cefas , 2022).
Table 1.
Recent outbreak of WSSV in different countries whole over the world (DISEASES-Cefas , 2022).
| Country |
Year |
Country |
Year |
| Mozambique |
2019 |
Ecuador |
2019 |
| China |
2019 |
Bangladesh |
2012 |
| India |
2018 |
Brunei Darussalam |
2013 |
| Indonesia |
2019 |
Colombia |
2005 |
| Japan |
2020 |
El Salvador |
2005 |
| Malaysia |
2009 |
Honduras |
2013 |
| Philippines |
2019 |
Hong Kong |
2013 |
| South Korea |
2019 |
Iran |
2013 |
| Taiwan |
2019 |
Madagascar |
2013 |
| Thailand |
2019 |
Myanmar (Burma) |
2012 |
| Vietnam |
2019 |
Nicaragua |
2013 |
| Costa Rica |
2019 |
Peru |
2013 |
| Mexico |
2019 |
Iran |
2011 |
| Panama |
2019 |
Saudi Arabia |
2012 |
| United States |
2020 |
Venezuela |
2011 |
| Australia |
2020 |
Argentina |
2010 |
| Brazil |
2019 |
|
|
By 2000, the virus had spread to Costa Rica, where it was first detected in Litopenaeus vannamei farms in the Gulf of Nicoya (Peña Navarro, Castro Vásquez et al. 2020). In the same period (between 1995 and 2001), the virus was detected in shrimp farms in several European Union (EU) countries including Greece, Italy, and Spain, and later in Turkey (Stentiford and Lightner 2011). In 2002, France reported its first WSSV outbreak, traced back to wild crustaceans (Rosenberry 2002, Stentiford and Lightner 2011). The Middle East was affected by WSSV, with the first outbreak in L. vannamei reported in the Khuzestan province of Iran in 2001 (Afsharnasab, Kakoolaki et al. 2014). Brazil recorded its first WSSV outbreak in L. vannamei farms in the Laguna province in 2005 (Cavalli, Romano et al. 2011), and by 2008, the virus was detected in Argentina (Martorelli, Overstreet et al. 2010). WSSV was reported in Saudi Arabia in 2010 and off the coast of Iraq in wild penaeids in 2012 (Tang, Navarro et al. 2012, Jassim and Al-Salim 2015). In Africa, the first detection of WSSV occurred at the Aquapesca shrimp farm in Quelimane, Mozambique, in 2011, with a subsequent outbreak in Madagascar in 2012 (Chamberlain, Lightner et al. 2013). Most recently, in November 2016, WSSV was identified in a prawn farm near Brisbane, Queensland, Australia (Knibb, Le et al. 2018).
4. Biology of WSSV
4.1. Taxonomy, Evolution and Protein Homology of WSSV with Other Taxa
WSSV was officially named in 2005 after multiple reclassifications (Fauquet, Mayo et al. 2005). It was earlier described under various names in literature, including hypodermal and hematopoietic necrosis baculovirus (HHNBV) (Miao, Tong et al. 2000), rod-shaped nuclear virus of P. japonicus (RV-PJ) (Miao, Tong et al. 2000), Chinese baculovirus (CBV) (Nadala, Tapay et al. 1997), systemic ectodermal and mesodermal baculovirus (SEMBV) (Sahul Hameed, Anilkumar et al. 1998), penaeid rod-shaped DNA virus (PRDV) (Inouye, Yamano et al. 1996), and white spot baculovirus (WSBV) (Miao, Tong et al. 2000). Initially what is now WSSV was considered a non-occluded Baculovirus due to its cylindrical morphological characteristics and histological injuries observed at the onset of the virus (Wongteerasupaya, Vickers et al. 1995). However, it was found to differ genetically and ultrastructurally from them. It was later reclassified as the only member of the genus Whispovirus, in the family Nimaviridae by the International Committee of Taxonomy of Viruses on the basis of its thread-like polar extension – the distinguishing morphological feature of the family (Wang, Hirono et al. 2019).
WSSV taxonomy thus reflects not only its unique structure but also represents a distant phylogenetic relation to other large dsDNA viruses, which include members of families Baculoviridae, Ascoviridae, Asfarviridae, phycodnaviridae, and Iridoviridae (Wang, Hirono et al. 2019). WSSV presents a unique genomic organization, and shares a relatively small subset of conserved genes with the earlier aforementioned viral families, indicating a distant evolutionary relationship (
for details see supplementary Table 1). Thus, large-dsDNA viruses are characterized by comparative phylogenetic studies with genetic conservatism, particularly in genes involved in DNA replication and repair. These observations suggest that WSSV and other virus families may have diverged from other virus families and evolved over time into distinct genomic features. Evidence of this evolutionary linkage is further supported through detailed protein homology analysis, which reflects notable sequence alignments between WSSV proteins and those of other dsDNA viruses.
Table 2 of this article shows homologous relationships of WSSV proteins to those of several viral families. For example, wsv459 of WSSV shares full identity with a hypothetical protein from PBCV-1 (Phycodnaviridae), with an E-value of 3e-04. This strong conservation among those viral families suggests that this protein may have a very important role in the virus life cycle. On the other hand, wsv360 and wsv143 are homologous to proteins of Asfarviridae and Ascoviridae, showing identity values of 86% and 96%, with E-values of 0.008 and 0.022, respectively. The low values of the E-parameter indicate that the observed homologies are statistically significant and are not due to chance alignments of sequences. Probably the most significant information deduced from this is that conserved proteins, such as ribonucleotide reductase, exist across viral families, indicating shared molecular mechanisms crucial for viral replication (Sakowski, Munsell et al. 2014). Apart from ribonucleotide reductase, other WSSV proteins homologous to those from various viral families like Poxviridae, Mimiviridae, and Baculoviridae are listed in
Table 2. For instance, wsv486 shares 90% identity with the variola B22R protein from FWPV (Poxviridae) with an E-value of 0.041, suggesting functional relatedness between these proteins. These conserved proteins may play roles in vital viral functions like DNA replication, immune evasion, and virion assembly (Sánchez-Paz 2010). The existence of such proteins in different viral families could be the result of evolutionary convergence in which homologous genes have been retained across different lineages due to similar functional imperatives (Wang, Li et al. 2020).
The presence of conserved proteins between WSSV and other large dsDNA viruses bears very strong implications for the understanding the evolutionary history of WSSV. WSSV also shares several essential genes with viruses infecting different hosts, such as plants and vertebrates, would implies that these are maintained through evolutionary pressures due to their functionality. This information enhances our understanding of how WSSV may have adapted to its crustacean hosts and developed its pathogenic capabilities. Moreover, the conservation of viral proteins across families has practical applications in the development of anti-viral strategies.
4.2. Global Genetic Distribution of WSSV (Genome)
The VICTOR program has produced the following neighbour-joining phylogenetic tree (
Figure 2), which illustrates evolutionary relationships between WSSV isolates from a broad geographical range. The tree is rooted by midpoint rooting and displays genetic diversity among the WSSV isolates based on nucleotide sequence similarity. This tree contains high (~100%) bootstrap values for most of the branches, providing very high confidence in clustering, particularly between the more closely related isolates. The isolates from China (NC 075105.1) and Bangladesh (PP134839.1, PP134840.1, PP134841.1) were grouped in one clade, which was strongly supported by a bootstrap value of 100, suggesting an extremely recent common ancestor or a closely related evolutionary origin. Indian isolates, EU327500, EU327499, and Thai, KX501222.1, KX501223.1, were phylogenetically tight, indicating regional phylogeographic patterns. This is further reflected in the presence of distant isolates, such as those from Mexico (MG432477.1) and Germany (KF981443.1), outside of primary clusters, indicating significant genetic divergence that might relate to geographical and environmental differences influencing WSSV evolution. Isolates from Saudi Arabia (KF976716.1) and Brazil (HQ130032.1) occupy an intermediate position, with a likely migration or trade-related virus spread. Another distinction involves the groupings from South Korea (GQ328029.1) and Australia (MF161441.1), which, further downstream, split into two lineages diverging from the other Asian core isolates. The branch lengths within the tree themselves are indicative of the mutation rates across the given isolates; some have longer branches, such as Germany and Brazil, indicating higher rates of evolution or separate mutational events.
4.3. Transmission Dynamics of WSSV
The WSSV is the most virulent pathogen affecting global shrimp aquaculture, and unraveling its dynamics of transmission is crucial for effective mitigation strategies. WSSV is a highly contagious, lethal, double-stranded DNA virus of the Nimaviridae family that often causes large scale mortalities (Oakey, Smith et al. 2019). It has vertical and horizontal routes of transmission, both contributing to the rapid spread of the disease in different shrimp farms and even in nature. Horizontal transmission is the more common mode of transmission; this includes direct contamination through waterborne contact, infected shrimp, and organic materials such as feces and molts (Tuyen, Verreth et al. 2014, Kim, Kim et al. 2023). Waterborne transmission is most important because infected shrimp release viral particles into the water through gill shedding, from body surfaces, or during decomposition, hence producing a heavily contaminated milieu. It has been observed that even an extremely low level of virus-contaminated water may mediate the spread of WSSV, and viral shedding can be detectable within hours of infection onset (Kim, Kim et al. 2023). This environmental transmission is a serious concern in densely populated shrimp farming systems where high stocking densities enhance the risk of infection. In infected shrimp, WSSV advances with rapid and lethal progression.
Significantly, it has been found that compared to the important transmission route of cannibalism or ingestion of infected tissues, waterborne exposure poses a greater infection risk in high-density farming (Pradeep, Rai et al. 2012, Verbruggen, Bickley et al. 2016). Clearly, this makes the design of biosecurity protocols very relevant; it implies that control over water quality and reduction of waterborne exposures should be emphasized above preventing cannibalism. On the other hand, vertical transmission becomes evident when the virus-carrying broodstock is used to transmit the virus to progeny through spawning (Vijayan, Anand et al. 2024). This has been considered a very injurious route of infection in hatcheries since asymptomatic carriers can spread the virus unknowingly to those populations.
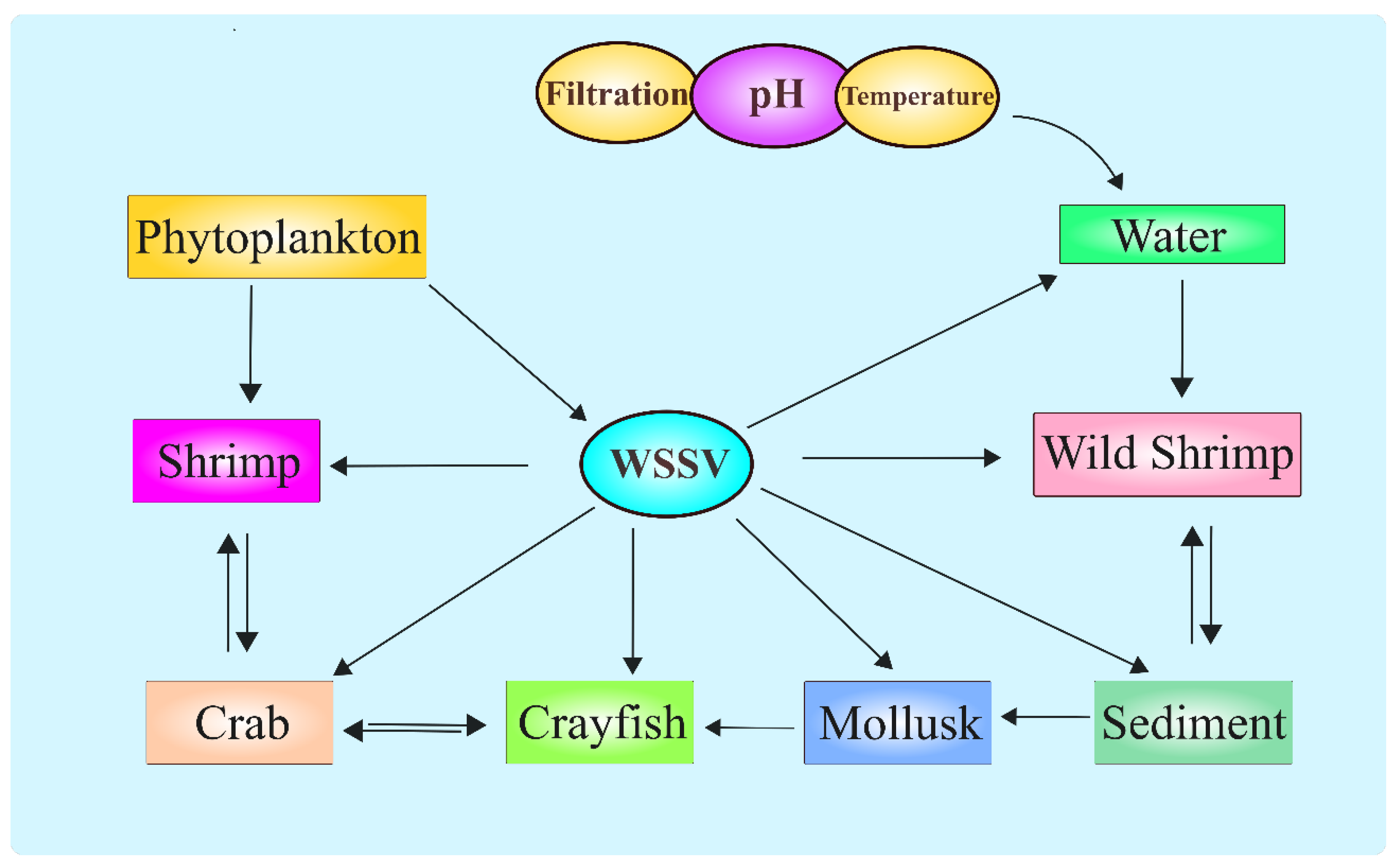
WSSV does not exclusively affect shrimp, but can affect a wide variety of crustacean and non-crustacean species as carriers and vectors both in aquaculture and the wild (Peng, Lo et al. 1998, Otta, Shubha et al. 1999, Hossain, Chakraborty et al. 2001, Joseph, James et al. 2015). For instance, WSSV was isolated from crabs (Pratapa, Kumar et al. 2023), crayfish (Lee, Kim et al. 2021), and other decapods (Wang, Lo et al. 1998) (
Figure 3), broadening the circle of potential inter-species infection and increasing the degree of difficulty in containment measures. These findings further stress the generalist nature of WSSV, able to thrive under varying conditions of brackish and freshwater systems. Such adaptability carries further control complications, especially in open systems where farmed and wild populations interact. Probably one of the main stumbling blocks in interpreting the transmission routes of WSSV is the variation in impact due to different environmental factors like temperature, salinity, and pH (Van Thuong, Van Tuan et al. 2016). Studies have demonstrated that the rate of virus replication as well as the speed of diffusion are higher at elevated temperatures, while variation in salinity can lead to differences in susceptibility among shrimp to the virus (Millard, Ellis et al. 2021). For example, in tropical regions, where temperatures are consistently high, WSSV outbreaks are more serious with rapid disease development and higher mortalities. Moreover, the virus appears to be stable within a broad range of salinity levels, which allows it to infect shrimp in both marine and freshwater aquaculture.
However, despite advances in understanding WSSV transmission dynamics, substantial knowledge gaps still remain, particularly relating to the role of non-crustacean species and environmental reservoirs in the perpetuation of the virus. Further, the presence of wild species that could be carriers or reservoirs of WSSV complicates efforts to establish WSSV-free zones in aquaculture. Additionally, the persistence of the virus in the environment, even in the absence of host species, raises concerns about the long-term sustainability of shrimp farming in such locations (Quang, Hoa et al. 2009). One proposed solution has been compartmentalization farming, a concept where shrimp are reared in biosecure units, inhibiting the spread of pathogens at both farm and external environment levels (Tidbury, Ryder et al. 2020). These techniques have achieved limited success, and the high cost of retrofitting has precluded widespread adoption. Future research should focus on elucidating the precise mechanisms of WSSV transmission in mixed-species environments, as well as developing novel strategies for disease prevention and control. With such diverse routes of virus transmission, further investigation into the host range, whether through natural or experimental infection, together with interaction of host proteins with the virus during replication and dissemination, will clarify the complex epidemiology of WSSV.
4.4. Host Species Reported to Be Naturally or Experimentally with WSSV
A wide range of host including economically important different shrimp species and organisms from both fresh and marine environment have been found to be infected with viral pathogens. For WSSV, hosts in a wide array of shrimp species have been detected from
Penaeus monodon (Wang, Hassan et al. 1999),
Penaeus vannamei (Jang, Qiao et al. 2014),
Marsupenaeus japonicus (Zhang, Koiwai et al. 2018), and
Penaeus chinensis (Yang, Zhang et al. 2008). In addition to these economically valuable penaeid species, WSSV has been isolated from crabs (
Scylla olivacea,
Neohelice granulate) (Pratapa, Kumar et al. 2023), copepods (Chang, Chen et al. 2011), lobsters (Rajendran, Vijayan et al. 1999), crayfish i.e.,
Procambarus clarkii (Huang, He et al. 2022), and freshwater species such as
Macrobrachium rosenbergii (Peng, Lo et al. 1998). Furthermore, it was confirmed that WSSV primarily afflicts decapod crustaceans, but recent studies demonstrate that its host range is further expanding. A study by Desrina
et al. emphasized that WSSV infects species from more than 50 families, including non-crustacean hosts such as mollusks, though crustaceans remain the primary hosts (Desrina, Prayitno et al. 2022). In experimental conditions, the transmission and replication of WSSV have been confirmed in non-target species like
Metapenaeus ensis (Chang, Peng et al. 2004),
Exopalaemon orientalis, and
Calappa lophos (Pradeep, Rai et al. 2012), where non-target species might act as reservoirs or vectors (for more details see
Table 3). The wide host range of WSSV, comprising different species of shrimps and other crustaceans, partly due to the ability of WSSV to bind a wide variety of host proteins that are conserved or functionally similar across hosts.
4.5. WSSV Virion Proteins
The WSSV virion is an extremely infective particle and thus very important in the process of disease transmission (Sánchez-Paz 2010). Structurally, this virion is a rod-shaped enveloped, non-occluded particle composed of macromolecules arranged to protect and convey the viral genome to effect infection in host organisms. The width of the WSSV virion ranges from 70 to 170 nm, while the length falls between 210 and 420 nm. The virion is composed of three layers: the tegument layer, the envelope, and the nucleocapsid (300 by 70 nm and enveloped by a layer of capsids) (Wang, Hirono et al. 2019). Each of these layers plays a role in the integrity and infectivity of the virus. WSSV has at least 58 structural proteins, of which localization data is available for 48 (Li, Lin et al. 2007). Among these, 33 are envelope proteins, nine are nucleocapsid proteins, and five are tegument proteins (Wang, Hirono et al. 2019). Of these envelope proteins, the major ones are VP28 and VP26 proteins, making up about 60% of the envelope proteins. The envelope proteins play an important part in WSSV's infectivity by means of binding. VP28 is a well-known protein that handles cell surface recognition as a receptor for the virus to attach to the host cell membrane (van Hulten, Witteveldt et al. 2001, Tsai, Wang et al. 2004, Yi, Wang et al. 2004). This mechanism of binding is critical for successful infection in shrimp because it allows the virus to pass into the cytoplasm of the host cell. Thus, VP28 may be considered a prime target for antiviral treatment because inhibiting this protein is normally adequate to block the virus from attaching to or entering host cells.
Besides VP28, other envelope proteins VP31, VP33, VP36A, VP110, VP136A and VP664 contain cell attachment motifs that may facilitate the initial stage of viral infection (Tsai, Wang et al. 2004, Leu, Tsai et al. 2005, Xie, Xu et al. 2006). These motifs allow the virus to attach to the host cell surface, making them an important feature for the development of therapeutic strategies. Among them, VP664 is one of the most abundant and largest proteins, comprising 6,077 amino acids, and plays a very important role in viral replication. Interfering with the functioning of VP664 might disturb replication of the virus and potentially offer another avenue for pharmaceutical intervention. A more detailed breakdown of the structural proteins (VP28, VP39B, VP31A, VP41B, VP51A, VP51B, VP68, VP124, VP150, VP187, VP281, and VP292) found in the WSSV envelope (Li, Lin et al. 2007) and other proteins (VP190, VP466, VP15, VP51, and VP76) derived from a collagen-like protein in the nucleocapsid of WSSV (Li, Chen et al. 2004) are listed in Table 2 according to their respective locations in the virion: envelope, tegument, or nucleocapsid. For instance, proteins such as VP124, VP187, and VP466, their kDa and ORF values reflect their capability to enhance the WSSV virion’s invasiveness into host cells; thus, further understanding of their structure and function could eventually facilitate targeted treatments (Xie, Xu et al. 2006).
Information integrated in Table 2 on WSSV proteins provides further potential targets that could be used in drug development. For instance, proteins such as VP26 and VP28 have roles in maintaining the structural integrity of the virion and are also involved in a series of steps which result in infection (Valdez, Yepiz-Plascencia et al. 2014, Taengchaiyaphum, Nakayama et al. 2017). Small molecules or peptides could be designed to interfere with the structural roles of these proteins, preventing proper virion assembly or entry into host cells (Chang, Liu et al. 2008). These can be targeted therapeutically by devising means through which the functions of these proteins are disrupted to inhibit the virus from spreading. Another idea that might be significant is that proteins with glutathione S-transferase fusion, like ORF151-VP466, can be targets that allow improvements in the host immune response, or inhibit viral processes (Ha, Soo-Jung et al. 2008). Indeed, these key identifications allow the possibility of developing vaccines or antiviral medications targeting the virus life cycle at points intended to interfere with infecting and replicating within shrimp, thereby reducing aquaculture losses attributed to WSSV.
4.6. Molecular Mechanisms of WSSV Life Cycle: Host Protein Contributions
The molecular underpinning of the life cycle mechanisms involves an elaborate interplay between viral components and host cell machinery. Interactions of host proteins with viral proteins at many steps in the infection process are indeed critical to the successful replication and spread of the virus through the host organism. These proteins facilitate not only the entry of the virus into the host cell but also contribute to intracellular trafficking, viral replication, assembly, and egress. Here, we have focused on the participation of host proteins in each critical stage of the WSSV life cycle from viral entry to progeny virion release.
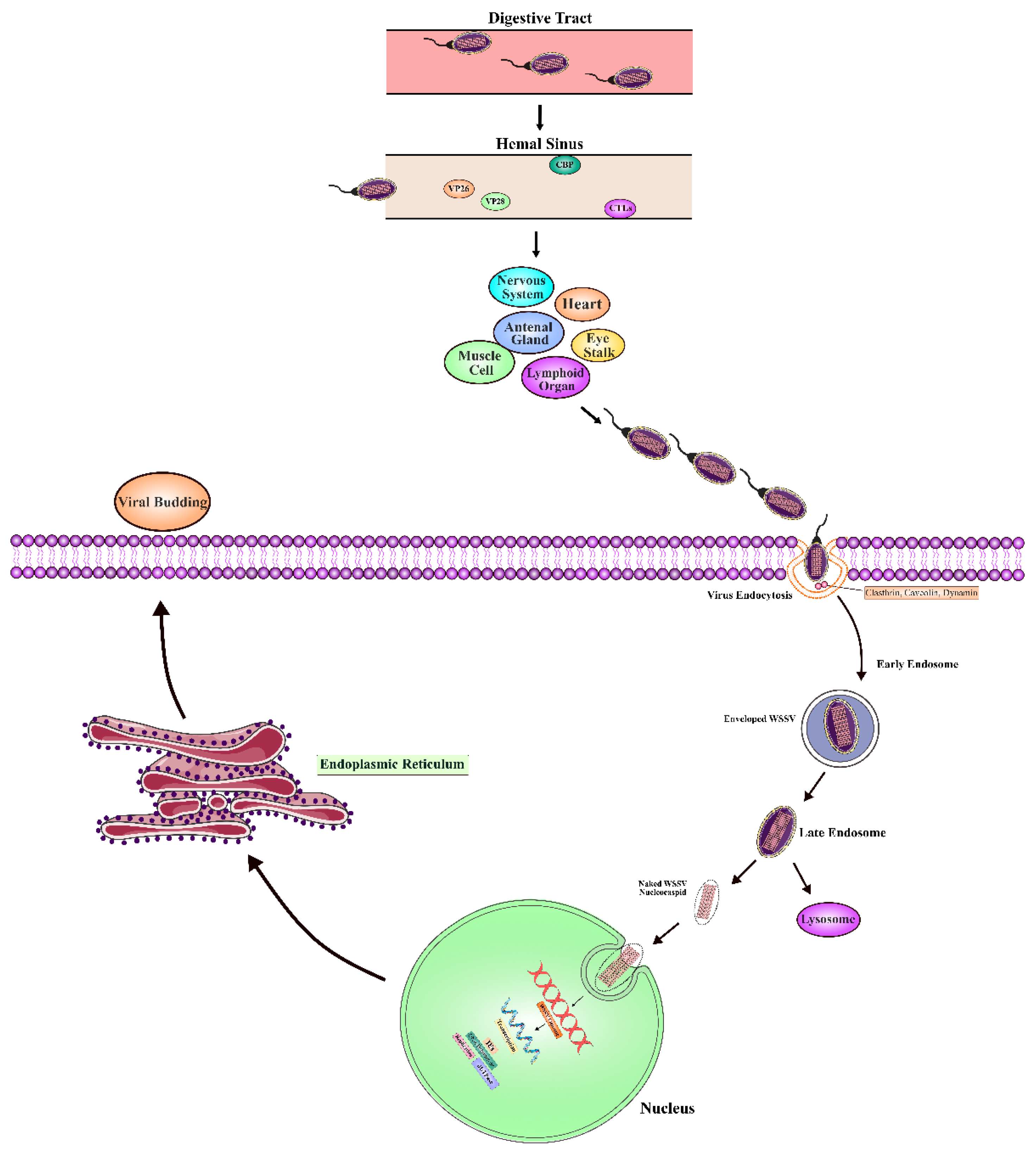
Entry of viruses into host cells: The process of infection is initiated when WSSV virions come in contact with host cells through the ingestion of infected or dead shrimp. Infection is mainly via the digestive tract, where the viral particles come into contact with and attach to the receptors of the host cells lining the epithelium (
Figure 4). These interactions are mediated by host proteins serving as receptors/co-receptors for the virus, which allow viral attachment to the surface of the cell. In this regard, one of the most studied receptor families involves the CBPs, more specifically the PmCBP in
Penaeus monodon, which is a critical participant in mediating WSSV attachment. Hence, this protein is capable of binding at least 11 envelope proteins of WSSV and establishes a stable interaction through which the virus can initiate entry into the host cell, such as VP24, VP32, VP39B, VP41A, VP51B, VP53A, and VP110 (Huang, Leu et al. 2014). Moreover, PTs play a vital role in the gut as receptors, especially in
L. vannamei, where LvPT interacts with viral proteins like VP32, VP38A, and VP39B (Verma, Gupta et al. 2017). These proteins are secreted into the stomach, possess potent chitin binding properties, and help in the transportation of viral particles across the epithelium of the digestive tract. Bound to these receptors, the WSSV particles penetrate the epithelial cells, cross over the basal membrane, and enter further into the circulatory system. Another major molecule that plays an important role in viral entry includes the glucose transporter 1 (Glut1) protein, expressed in almost all tissues, including the digestive tract, muscles, and pleopods. Glut1 plays a complementary role in identifying several envelope proteins of WSSV, such as VP28 and VP53A, during viral entry into cells (Huang, Chan et al. 2015). This protein binds to at least seven viral envelope proteins in an adjoining loop region, making it a key mediator of WSSV infection. More recent studies have proposed a complex of Glut1 forming with PmCBP, presenting a larger surface area to the virus by making viral binding and, therefore, attachment and internalization more effective (Encinas-García, Mendoza-Cano et al. 2023).
Endocytosis and intracellular trafficking: Attached to the host cell, the main mode of WSSV entry into the cell is through receptor-mediated endocytosis. This allows the virus to bypass the host cell membrane into the intracellular environment. So far, the best-characterized pathway of endocytosis exploited by WSSV is clathrin-mediated endocytosis, where the virus is engulfed into clathrin-coated vesicles that bud off from the plasma membrane (Pavelka and Roth 2010). These vesicles ferry the virus to early endosomes, in which the low pH allows viral uncoating to occur. This acidic environment is essential for initiating conformational changes in the virus, leading to the release of the viral genome and nucleocapsid into the host cytoplasm (Villanueva, Rouillé et al. 2005). Also, endocytosis depends on cholesterol and dynamin, evidenced by studies showing that WSSV invasion depends on these lipid components for membrane curvature and vesicle scission (Chen, Shen et al. 2016). The virus avoids lysosomal degradation via Rab GTPases, which regulate membrane trafficking events during endocytosis. Specifically, Rab5, during development in P. monodon and L. vannamei, mediates the maturation of early endosomes into late endosomes. Rab7 is involved in the later stages of endosome maturation and replaces Rab5, ensuring transport of viral nucleocapsid to the host cell nucleus without degradation (Attasart, Kaewkhaw et al. 2009).
Viral genome delivery and replication: Once inside the host's cytoplasm, the viral nucleocapsid has to reach the nucleus, where replication occurs. The viral envelope fuses with the host endosomal membrane and discharges the nucleocapsid into the cytoplasm, which then migrates to the nucleus. The nuclear pore complexes transport the viral genome into the nucleus. This marks the start of the replication phase. Inside the nucleus, WSSV expresses its immediate early genes, which are considered crucial for initiating the replication machinery. These early genes allow for the synthesis of mRNA, which is transported back to the cytoplasm for translation. The viral mRNA then undergoes translation in the cytoplasm by free ribosomes, encoding a major structural protein, VP664, which forms the backbone of the WSSV capsid. This occurs in parallel with the replication of the viral genome and the synthesis of viral proteins, enabling the assembly of new viral particles (Leu, Tsai et al. 2005). The involvement of host proteins continues to play a critical role in these processes. For instance, glucose transporter 1 (Glut1) mediates the trafficking of viral proteins between cellular compartments, while the tetraspanins like FcTetraspanin-3 bridge the connection between the inner and outer cell membranes to help mediate the intracellular locomotion of viral components (Wang, Li et al. 2010, Gui, Wang et al. 2012).
Viral assembly and egress: Assembly of the virus occurs near the host nucleus, where the major capsid proteins, including VP664, form the new structure of virions. These assembled virions incorporate viral envelope proteins like VP28, synthesized in the rough endoplasmic reticulum. These envelope proteins are then targeted to the inner nuclear membrane, where they associate with the assembling virions. VP28 is an essential protein for infection efficiency and has been shown to stabilize the viral envelope and increase the infectivity of the virions (van Hulten, Westenberg et al. 2000). As the viral load within the host cell increases, it builds up until the accumulation of viral particles overwhelms the cell. Finally, lysis of the host cell releases newly formed virions into the extracellular environment. Completion of the WSSV life cycle enables the virus to infect neighboring cells and to spread throughout the host organism. The lysis of infected cells is a pivotal event that permits the rapid propagation underlying the systemic infection that is typical of WSSV.
Systemic spread and organ targeting: Once internalized into the hemolymph, the virus is circulated via the open vascular system of the host to infect a wide array of tissues and organs of mesodermal and ectodermal origin. Integrin proteins on the surface of target cells, including those of the gonads, heart, muscles, and nervous system, constitute important docking points for WSSV. Integrins in L. vannamei bind viral proteins VP26, VP31, VP37, VP90, and VP136, thereby facilitating virus attachment and penetration of these vital tissues (Verma, Gupta et al. 2017). There exist specific viral motifs, such as RGD, YGL, and LDV, which enable the virus to recognize and bind integrins on the host cell surface through such interactions. The virus can either penetrate directly into the host cell membrane once bound to the integrins or be internalized by receptor-mediated endocytosis. WSSV research has shown that both clathrin-mediated and caveolae-mediated pathways are used for endocytosis depending on the host cell and tissue type (Leu, Tsai et al. 2005). Caveolae-mediated endocytosis is particularly cholesterol-dependent, and it promotes internalization within vesicles, thus bypassing lysosomal degradation of viral particles. The systemic spread of WSSV within the host is further facilitated by C-type lectins that bind viral proteins and facilitate the transportation of the virus through the circulatory system (Wang, Xu et al. 2009). In L. vannamei, LvCTL1 binds to viral proteins VP14, VP24, VP28, and VP95 and helps distribute them in the body via the hemolymph. These interactions initiate a systemic infection, with the virus targeting multiple organs and tissues, including hemal sinuses, gonads, and eyestalk (Zhao, Yin et al. 2009).
Host defense evasion mechanisms: WSSV expresses several mechanisms that enable the pathogen to evade host immune responses to establish a productive infection. This includes manipulation of host endosomal trafficking pathways to avoid degradation by lysosomes, in which the Rab GTPases, notably Rab5 and Rab7, play important roles (Verbruggen, Bickley et al. 2016). Rab5 controls endosomal maturation, thereby facilitating transport of viral nucleocapsids from early to late endosomes. Rab7 ensures that the viral particles are not targeted to lysosomes for degradation (Attasart, Kaewkhaw et al. 2009). In this way, the virus evades host immune responses and possibly persists in a latent state for longer periods. WSSV also manipulates immunomodulation within its host by interacting with immune-related proteins, such as tetraspanins and lectins, which act as receptors with important implications in immune signaling. The virus, through binding, may alter the immune response and inhibit processes leading to the production of antiviral mechanisms, thereby enhancing its survival and replication within the host.
5. Pathology
5.1. Gross Sign of WSSV
Gross clinical signs in animals infected with WSSV vary considerably at different stages of the infection process, however, white spots on the cuticle are pathognomonic signs for this viral disease in some shrimp species. These appear as white spots, 1-3 mm in diameter, and are calcium-rich viral accumulations on the exoskeleton especially on the cephalothorax, appendages, and abdomen (Sánchez-Paz 2010). However, white spots do not occur in all WSSV infections, and identical lesions due to non-viral etiology such as bacterial infection or shell mineralization disease are also possible, thereby making them an unsatisfactory single diagnostic feature. In species such as the Penaeus monodon, Penaeus vannamei, and Marsupenaeus japonicus, infected individuals in general become lethargic, exhibiting decreased feeding and erratic swimming before eventually succumbing to the disease (Walker and Mohan 2009). Infected shrimp usually move to the edge of the ponds or to the water surface, where they are preyed upon by predatory birds that can facilitate mechanical transmission of the virus (DAWR 2017). Characteristic symptoms of infection include soft shells, discoloration of the body to reddish, and loose appendages in worst cases. Similar signs are shown by Penaeus chinensis, with additional symptoms of soft bodies and gill necrosis (Rajendran, Vijayan et al. 1999). The signs of WSSV infection in Macrobrachium rosenbergii (giant freshwater prawn) include red bodies, gill necrosis, and lethargy (Sahul Hameed and Bonami 2012). Experimental studies have shown that infected shrimp can begin shedding viral DNA into the water within six hours of infection, with shedding peaking just before death. This greatly elevates the risk of infection to surrounding shrimp, particularly in closed systems like grow-out ponds (Arbon, Andrade Martinez et al. 2024, Cox, De Swaef et al. 2024, Kim, Shin et al. 2025). Other susceptible decapod crustaceans, like Metapenaeus ensis and Exopalaemon orientalis, also exhibit similar clinical manifestations of incapacitated mobility, reddening of the body, and lesions in the exoskeleton. WSSV is histologically characterised as an ectodermal and mesodermal tissue-attacking virus. Very severe degenerations are shown among infected gills, lymphoid organs, and antennal glands (Pradeep, Rai et al. 2012). WSSV infection in Scylla olivacea and Neohelice granulata causes lethargy, white spots on the carapace, and internal tissue necrosis (Moser and Marques 2023). The same type of white spots develops on lobsters and crayfish (Procambarus clarkia), infected with WSSV, along with erratic swimming, often accompanied by discolored or darkened exoskeletons (Jiang, Xiao et al. 2017). Even copepods and mollusks, which are less infected, may act as passive carrier or mechanical vectors of WSSV, especially after being exposed to high viral loads from contaminated environments (Chang, Chen et al. 2011). Although such organisms may occasionally harbor viral particles, the current evidence does not show that they allow productive viral replication, and therefore function primarily to propagate transmission but not disease expression (Matozzo, Ercolini et al. 2018). WSSV exerts its pathogenicity towards host species by virtue of tissue tropism for vital tissues such as cuticle and haemopoietic systems, inducing degeneration and death. Poor water quality may act as an environmental stressor that enhances disease progression through increased susceptibility and more rapid viral replication. In addition, certain factors such as rising or fluctuating temperature, salinity imbalance, and high stocking density are also found to impair the immune response of the shrimp, contributing to the progression and severity of WSSV outbreaks (Millard, Ellis et al. 2021). Behavioral changes, including convulsions and reduced mobility, can be observed in the late stages of infection, often in severely diseased populations.
Table 4.
Gross clinical signs of WSSV in various species.
Table 4.
Gross clinical signs of WSSV in various species.
| Species |
Clinical sign observed |
Reference |
| Penaeus monodon |
White spots, lethargy, soft shell, erratic swimming, reddish body |
(Walker and Mohan 2009) |
| Penaeus vannamei |
White patches, gill necrosis, soft cuticle, lethargy |
| Marsupenaeus japonicus |
White spots, disorientation, loose appendages, lethargy |
| Penaeus chinensis |
Soft body, white spots, gill necrosis, lethargy |
(Rajendran, Vijayan et al. 1999) |
| Scylla olivacea |
White spots on carapace, necrosis, lethargy |
(Moser and Marques 2023) |
| Neohelice granulata |
White spots, lethargy, body discoloration |
| Procambarus clarkii |
White spots, abnormal swimming, lethargy |
(Jiang, Xiao et al. 2017) |
| Macrobrachium rosenbergii |
Red body, gill necrosis, lethargy |
(Sahul Hameed and Bonami 2012) |
| Metapenaeus ensis |
Lethargy, body redness, white spots |
(Pradeep, Rai et al. 2012) |
| Exopalaemon orientalis |
White spots, soft exoskeleton, decreased mobility |
| Calappa lophos |
Lethargy, body discoloration, white spots |
5.2. Histopathology of WSSV
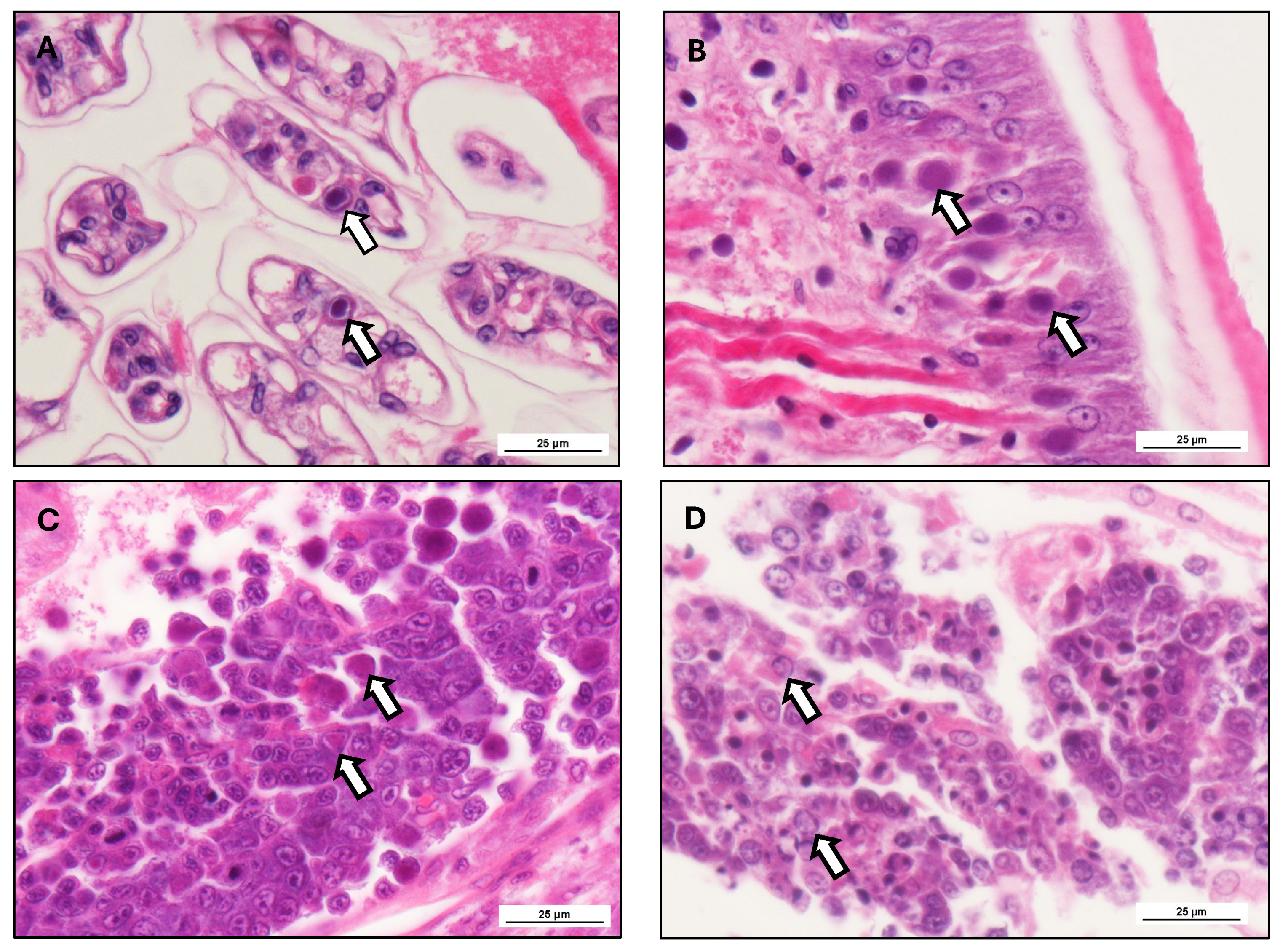
The histopathological changes of WSSV reflect the pathogenesis, disease course, and immunity associated with WSSV (
Figure 5). The main target cells of WSSV are the ectodermal and mesodermal tissues (Tang, Pantoja et al. 2013), including the cuticular epithelium, gills, lymphoid organ, foregut. In the hepatopancreas, WSSV infection has been observed predominantly in the haemocytes and connective tissues surrounding the tubules, but not in tubular epithelial cells themselves (Islam, Mou et al. 2023). The representation of WSSV infection features hypertrophied nuclei with intranuclear inclusion bodies of a basophilic nature, pyknosis, karyorrhexis, and cytoplasmic vacuolization (Rodríguez, Bayot et al. 2003). As infection progresses, these cellular changes result in widespread necrosis with attendant impaired physiologic function of infected organs (organ specific information available in
Table 5). Gill tissues exhibit epithelial sloughing and lamellar fusion, severely reducing respiratory efficiency, and the hepatopancreas, a principal organ of shrimp metabolism, degenerates, often complicated by secondary bacterial infections (Rajendran, Vijayan et al. 2005). The lymphoid organ plays a critical role in immunity as well, but in WSSV-infected shrimp, it undergoes lymphoid organ spheroid (LOS) formation, which is an attempt to limit viral replication but is unsuccessful because viral replication happens so rapidly (Sweet and Bateman 2016). Transmission electron microscopy (TEM) has revealed that WSSV virions are rod-like, approximately 275 nm in length and 120 nm in diameter (van Hulten, Westenberg et al. 2000), enveloped with a double-stranded DNA genome in a lipid envelope. Virus replication occurs in the nuclei of infected epithelial cells (Ng, Cheng et al. 2023). Compared to other viruses of shrimp such as Infectious Hypodermal and Hematopoietic Necrosis Virus and Taura Syndrome Virus, which essentially infect haemocytes, WSSV has a tropism for epithelial tissues that significantly aggravates systemic infections (Sánchez-Paz 2010). The progression of WSSV infection is inconsistent, taking either acute or chronic forms. From histopathological observation it also has been ascertained that massive tissue necrosis, intense virus replication, and extensive intranuclear inclusions happen in acute WSSV infections (Yin, Yan et al. 2023), resulting in explosive mortality of shrimp. In contrast, chronic infections consist of persistent, low-grade viral replication, with localized tissue damage and immune suppression, resulting in growth impairment and increased susceptibility to secondary infections.
Histopathological grading is also an important technique for ascertaining WSSV severity and evaluating the extent of disease progression. The intensity of infection is classified into four grades: G0 (no infection), G1 (mild infection with nuclear hypertrophy in fewer than 10% of cells), G2 (moderate infection with inclusion bodies in 30-50% of infected cells and mild necrosis), G3 (severe infection with inclusion bodies in more than 50% of cells, together with extensive necrosis), and G4 (late infection with complete cell destruction in multiple organs) (Gholamhoseini, Afsharnasab et al. 2013, Kim, Kim et al. 2023). Molecular diagnostic techniques such as in situ hybridization (ISH) and polymerase chain reaction (PCR) significantly enhance the sensitivity of WSSV detection. Parallel to ISH, normal histology and electron microscopy permit precise localization of viral DNA in infected tissues, which usually reveals strong signals in connective tissue, gill lamellae, and reproductive organs, suggesting potential vertical transmission (Sánchez-Martínez, Aguirre-Guzmán et al. 2007, Pradeep, Rai et al. 2012). Histopathology remains a vital diagnostic tool for early detection of the disease, enforcement of biosecurity, and treatment evaluation. Preventive strategies such as probiotics, immunostimulants, and plant extracts have shown promise in reducing histopathological damage and improving survival rates in shrimp (Hoseinifar, Sun et al. 2018). Additionally, histological examination is employed to examine the efficacy of antiviral drugs in managing WSSV caused tissue pathology (Nilsen, Karlsen et al. 2017). The combination of histopathology and molecular diagnosis is more effective at disease detection and control. It is noteworthy that histopathological lesion provides a presumptive diagnosis of WSSV. In accordance with the World Organisation for Animal Health diagnostic manual, confirmatory diagnosis is required to be carried out with PCR and better sequencing to ascertain the presence of WSSV-specific genomic material, which is very much essential for accurate reporting and disease surveillance in accordance with international standards.
5.3. Co-Infection of WSSV and Other Disease of Shrimp
Prevalence of disease pattern in shrimp farms has shifted from previous simple infection to co-infection including complicated infections, mixed infections, super infections, polymicrobial diseases, secondary infections, multiple infections, dual infections, and concurrent infections of homologous or heterologous pathogens in recent years (Dai, Yu et al. 2018, Kooloth Valappil, Stentiford et al. 2021, Lee, Jeon et al. 2023). Simultaneous or sequential co-exposure of heterologous pathogen (parasite-bacteria, parasite-virus, virus-bacteria, fungus-bacteria) also termed as co-infection what further defined as the infection of host animal (shrimp) from two or more genetically distinct pathogens, each of which has pathogenic effects and harms the host in concert with other infections. Among different pattern of pathogens exposure to shrimp species (for details see
Table 6), homologous combinations of viruses (WSSV, Taura syndrome virus (TSV), hepatopancreatic parvovirus and infectious myonecrosis virus (IMNV), infectious hypodermal and haematopoietic necrosis virus (IHHNV), monodon baculovirus (MBV)) (Cavalli et al., 2013; Chayaburakul et al., 2004; Dewangan et al., 2017; Feijó et al., 2013; Flegel et al., 2004; Manivannan et al., 2002; Otta et al., 2014; Tan et al., 2009; Teixeira-Lopes et al., 2011; Thamizhvanan et al., 2019) and bacteria (
Bacillus cereus,
Bacillus flexus,
Shewanella decolorationis,
Aeromonas veronii, Shewanella amazonensis and
Kurthia gibsonii) (Dewangan et al., 2022) are most extensively investigated.
For shrimp species parasite-virus co-infections are relatively uncommon and were first reported involving a microsporidian parasite (Enterocytozoon hepatopenaei) and viral pathogens (Taura syndrome virus (Tang et al., 2017) and infectious myonecrosis virus (Jithendran et al., 2021)). It is now increasingly evident that EHP has become a substantial global threat to shrimp aquaculture with the economic loss from this microsporidian being approximately double that of WSSV for Indian shrimp industry in 2021 (Patil et al., 2021). Moreover, co-infection of EHP and WSSV (Thamizhvanan et al., 2019) from synergistic interaction between these genetically distinct pathogens has raised greater concern regarding the transmission dynamics of microsporidians in shrimp farming systems and adjacent ecosystems, which is an emerging threat to the shrimp industry in Bangladesh.
6. Immunological Responses of Shrimp to WSSV
Compared to the possession of real adaptive immune system in vertebrates, shrimp have a well-developed innate defense mechanism. The system is characterized by a non-specific immunological response, usually segregated into cellular and humoral components and activated pattern recognition receptors (PRPs) (Kulkarni, Krishnan et al. 2021). So far, several PRRs have been identified in Penaeid shrimp including toll-like receptors (TLRs), lectin, tetraspanin, and lipopolysaccharide and β-1,3-glucan binding protein (Li and Xiang 2013). These receptors play a central role in the recognition of pathogens and in triggering of immune responses in shrimp.
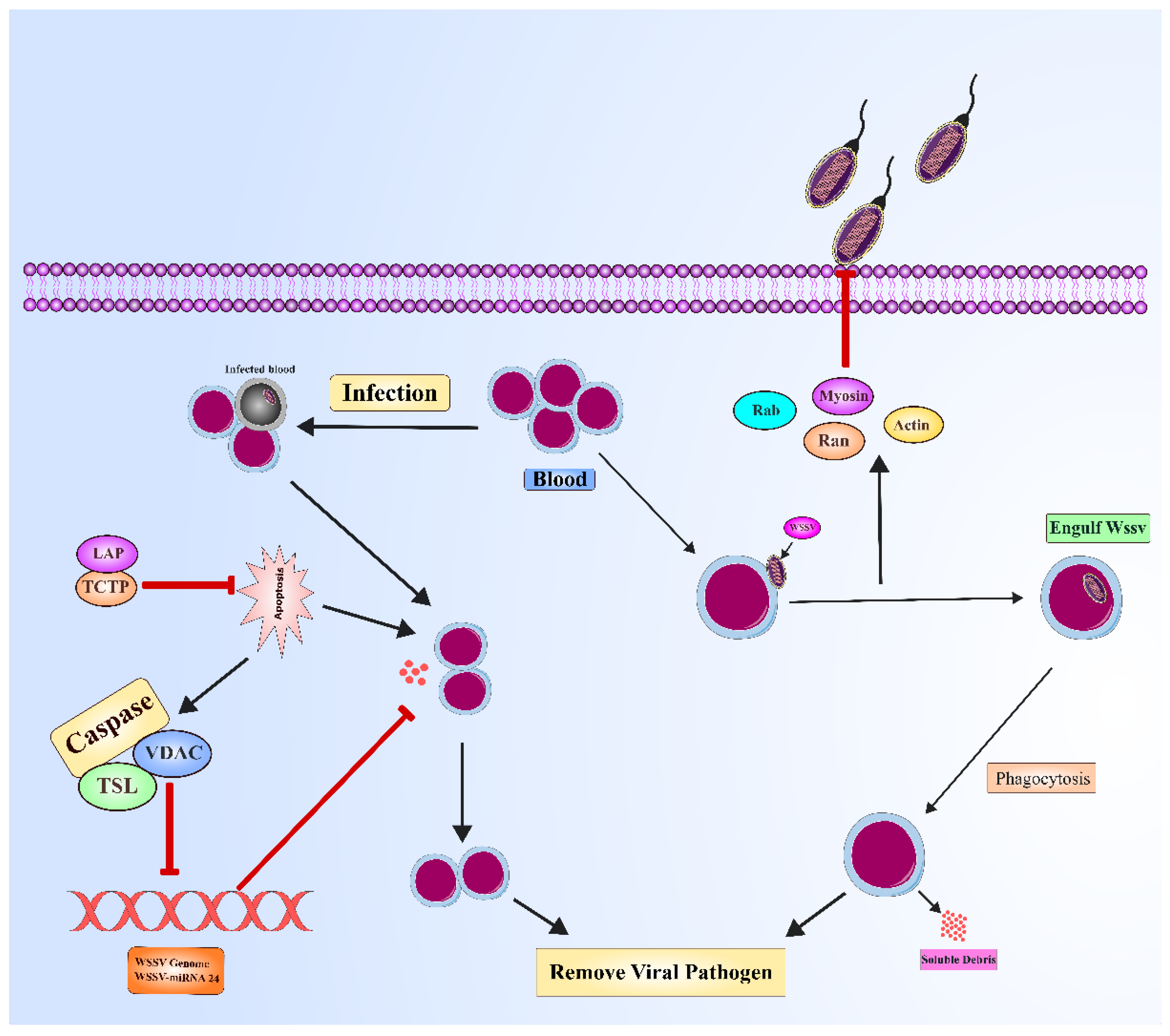
Cellular immune mechanisms in shrimp: Cellular immunity addresses the recognition and elimination of pathogens through various mechanisms, including phagocytosis, encapsulation, and apoptosis (
Figure 6). Hemocytes are circulating immune cells that essentially carry out the process in shrimp through phagocytosis, wherein pathogens are taken up and degraded. This also involves different small GTP-binding proteins such as Ran and Rab (Abubakar, Atmaca et al. 2015), reflecting the complexity of shrimp cellular immune responses. Crucially, hemocytes depend on immune mediators like lectins to improve their ability to recognize and respond to pathogens. Lectins are one of the most common classes of immune mediators, which are characterized by a carbohydrate recognition domain (Liu, Zheng et al. 2020). In shrimp, C-type lectins (CTLs) facilitate phagocytosis of microbial pathogens through opsonization, marking the pathogens for ingestion by immune cells. More directly, CTLs exhibit immunity by agglutination and inhibiting microbial growth, including gram-positive and gram-negative bacteria (Wang and Wang 2013). C-type lectins also show antiviral activity, especially against WSSV. A range of lactins, such as MjsvCL, LdlrLec1, LdlrLec2, LvAV, FmLC5, and FLdlr (found in
Fenneropenaeus merguiensis , Litopenaeus vannamei, and Marsupenaeus japonicus species) has been reported with anti-WSSV functions (Wang, Vasta et al. 2020). However, some lectin genes, for example LvCTL3 (found in
L. vannamei) and FmLC3 (found in
F. merguiensis), are paradoxically demonstrated to enhance their vulnerability to WSSV (Runsaeng, Kwankaew et al. 2018). These findings demonstrate the complexity of lectin immune modulation wherein their regulatory role may support or hinder resistance against pathogens. Recent studies have shown that the NFkB pathway could modulate CTLs expression in
L. vannamei. (Li, Li et al. 2014), again suggesting that in shrimp immune responses are controlled at every level in a highly ordered way.
Programmed cell death or apoptosis is another aspect of cellular immunity that helps shrimp eliminate cells harboring infectious agents (Cui, Liang et al. 2020). Apoptosis is typically brought about by various apoptosis-related genes such as caspases, inhibitor of apoptosis protein (IAP), apoptosis-inducing factor (AIF), cytochrome c, the mitochondrial voltage-dependent anion channel (VDAC), Fortilin or translationally controlled tumor protein (TCTP), gC1qR, BAX inhibitor-1 (BI-1), and apoptosis signal-regulating kinase 1 (ASK1) (Elmore 2007). These molecular components are important in coordinating the controlled and targeted destruction of cells, which is an important feature of the shrimp's immune response (Tang, Cui et al. 2019). Caspase are proteases that initiate the early phase of apoptosis in response to external signals. Apoptosis allows shrimp to limit pathogen dissemination in tissues by destroying infected cells in a timely manner (Clarke and Clem 2003). Shrimp were found to be resistant to WSSV by apoptosis-mediated blocking of viral propagation, hence hindering the virus from spreading in the host cells of shrimp (Huang, Cui et al. 2014). However, WSSV also evolved anti-apoptotic proteins such as AAP-1 (ORF390 or WSSV449), WSV222, VP38, WSSV134, and WSSV322 that delay or inhibit normal apoptosis, allowing completion of the viral replication cycle and further infection of other cells (Kulkarni, Krishnan et al. 2021).
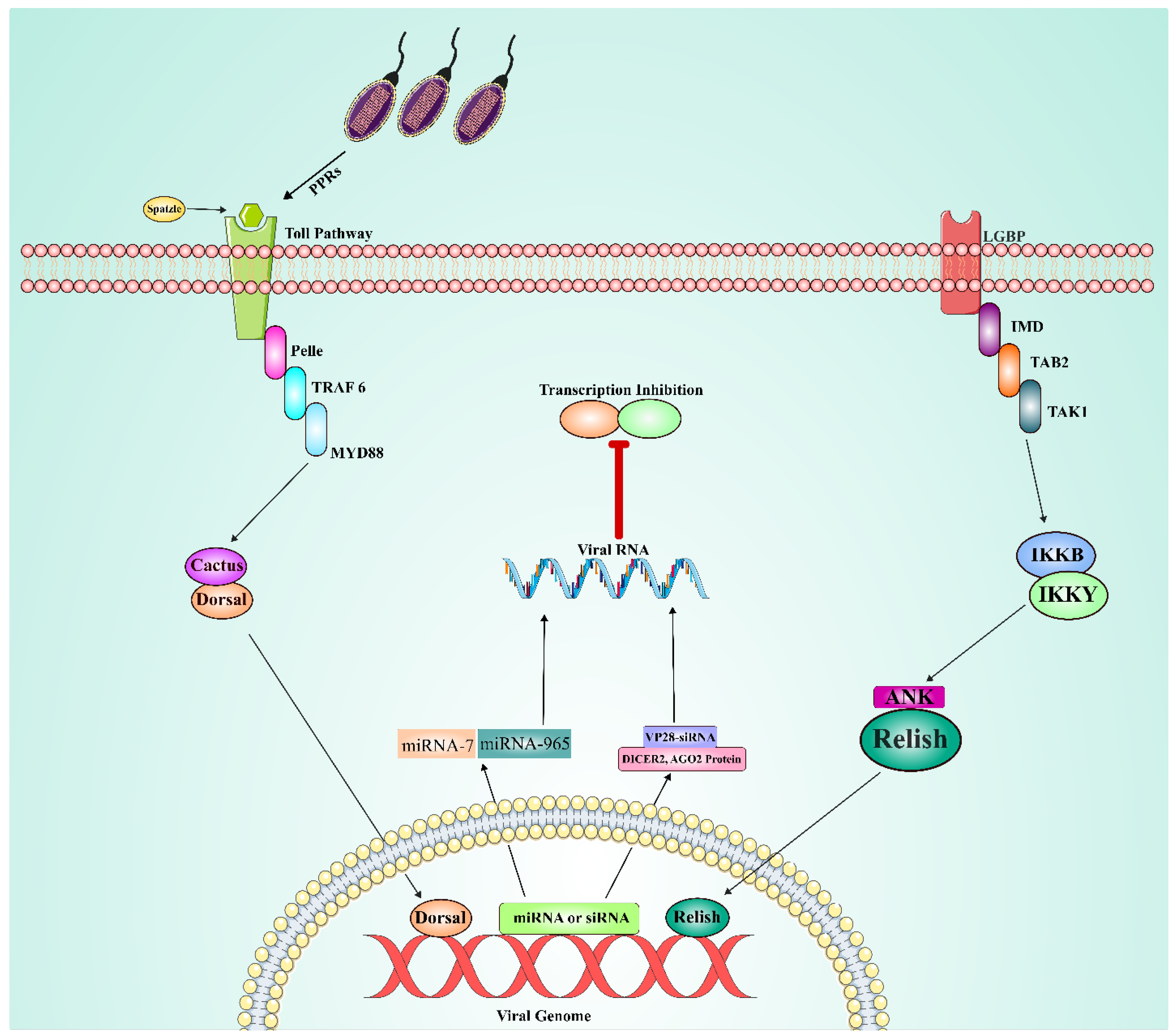
Humoral immune mechanisms in shrimp: In view of the absence of lymphocytes and immunoglobulins in shrimp, much dependence is placed on the humoral immune response as a complement to cellular immunity. Various biological macromolecules, including antimicrobial peptides, phosphatase, and lysozyme, are involved in mediating humoral immunity in shrimp against pathogen invasion (Kulkarni, Krishnan et al. 2021). It has been found that these molecules show crucial activity related to the recognition and neutralization of pathogens, thereby preventing their proliferation within the host (Wang and Zhang 2008). The Toll, IMD and JAK/STAT pathways are considered the main signaling pathways of the humoral response in shrimp, especially in their immune response to viral infections like WSSV (
Figure 7). Expression of the canonical Toll pathway has been well characterized in several shrimp species, including
L. vannamei, P. monodon, M. rosenbergii, P. clarkii, F. chinensis, and M
. japonicus (Mekata, Kono et al. 2008, Yang, Zhang et al. 2008, Feng, Zhao et al. 2016, Huang, Li et al. 2017, Liu, Xu et al. 2018, Yao, Su et al. 2018). Key molecules in this pathway include Spätzle, Toll, MyD88, Tube, Pelle, Pellino, TRAF6, Dorsal, Cactus, Tollip, SARM, Flightless-I, and b-arrestin, each of which plays subsequent roles in the activation of immune responses (Li, Wang et al. 2019). Interestingly, up to date, 25 Toll-like receptor genes have been identified in shrimp, with species-specific variations. These include LvToll1-9 from
L. vannamei, PmToll1 and PmToll9 from
P. monodon, FcToll from
F. chinensis, PcToll and PcToll1-5 from
P. clarkii, MjToll1-2 from
M. japonicas, as well as two MrTolls and MrToll1-3 from
M. rosenbergii (Yang, Yin et al. 2007, Mekata, Kono et al. 2008, Yang, Zhang et al. 2008, Wang, Tseng et al. 2010, Assavalapsakul and Panyim 2012, Wang, Liang et al. 2012, Hou, He et al. 2014, Srisuk, Longyant et al. 2014, Wang, Chen et al. 2015, Lan, Wei et al. 2016, Lan, Zhao et al. 2016, Guanzon and Maningas 2018). These findings highlight the evolutionary adaptation of the Toll pathway in shrimp to respond in a species-specific manner against pathogens.
The immune deficiency (IMD) pathway, first identified in L. vannamei in 2009 (Wang, Gu et al. 2009), also contributes to antiviral humoral immunity. Several IMD homologs known to have conserved functions have become species-specific regarding tissue distribution and immune response (Lan, Zhou et al. 2013). For example, FcIMD is mainly expressed in the stomach and gills from F. chinensis, while PcIMD is highly expressed in the hepatopancreas, stomach and heart from P. clarkia (Wang, Gu et al. 2009). The IMD pathway contains several canonical gene components (Relish, TAK1, TAB1, and TAB2) shared with the Toll pathways and are involved in activating immune response (Li and Xiang 2013). L. vannamei has been found to express LvTAK1 and LvTAK2, where these Toll-like receptors regulate the expression of various antimicrobial peptides in vivo (Wang, Li et al. 2016). In addition, Lvb-TrCP, LvMKK6, LvAkirin, LvNKRF, LvRelish exhibit strong responses to WSSV viral infection from L. vannamei. Along with PmRelish, FcRelish, FcMKK4, and FcP38 also show activity against viral infections like WSSV in the M. japonicas, F. chinensis (Li, Wang et al. 2019).
Compared to the well-characterized Toll and IMD pathways, the role of the JAK/STAT pathway during WSSV infection remains unclear; however, recent studies have unraveled the dual function of the JAK/STAT pathway in WSSV infection. In the JAK/STAT pathway, LvSOCS2 activates the expression of antimicrobial peptides, exhibiting an antiviral role in L. vannamei. Conversely, LvJAK promoted infection with viral genes such as WSV069 during the early stages of WSSV infection (Li, Weng et al. 2019). From these observations, it seems that in general the JAK/STAT pathway plays a dual role (positive or negative) during a viral infection depending on the different stages of infection and specific immune factors involved.
RNA interference (RNAi) and microRNA (miRNA) in shrimp immunity: Another important aspect of shrimp immunity is that RNAi and miRNA are involved in regulating immune responses. RNAi mediated by siRNAs has been found to be an essential defense against viral infections (Xu, Han et al. 2007). Shrimps produce siRNAs specific to viruses, like vp28-siRNA against WSSV infection, lowering viral replication by targeting viral genes in hemocytes. The main RNAi machinery elements, Dicer2 and Argonaute2, take part in the generation and function of siRNAs in antiviral reactions (Zhu and Zhang 2011, Sabin and Cherry 2013). Along with RNAi, miRNAs have also become important controllers of shrimp immune reactions (Kaewkascholkul, Somboonviwat et al. 2016). Various miRNAs displaying differential expression resulting from infection by WSSV have been identified through miRNA microarray analysis. For example, miR-7 and miR-965 have been identified as down-regulators of WSSV early genes, such as wsv477 and wsv240 (Huang and Zhang 2012, Shu, Li et al. 2016) respectively, which ultimately inhibit viral replication and subsequent infection (He and Zhang 2012). In addition, miR-965 has been shown to enhance phagocytic activity via autophagy-related gene-5, a gene involved in autophagy – thereby aiding in the phagocytosis of viral pathogens. On the contrary, some other viral miRNAs, such as WSSV-miR-66 and WSSV-miR-68, enable viral infection via the enhancement of expression levels during the early stage in WSSV infection by enhancing the expression of viral genes (wsv094, wsv177, wsv248 and wsv309) (He, Yang et al. 2014). These findings illustrate the complex interplay between host and viral miRNAs in regulating immune responses during WSSV infection.
7. On-Going Research on Control Measures
The conventional control methods of WSSV involve the use of antibiotics, which have mostly proved ineffective and also cause harm due to bioaccumulation; hence the need for other strategies (Lulijwa, Rupia et al. 2020). Continuous research has shifted towards newer approaches involving the use of immunostimulants, dietary interventions, and advanced technologies such as CRISPR technology and nanotechnology (Govindaraju, Dilip Itroutwar et al. 2020, Mariot, Bolívar et al. 2021, Ferdous, Islam et al. 2022, Zhang, Shan et al. 2022, Gong, Pan et al. 2023, Kumar, Verma et al. 2023, Galib, Ghosh et al. 2024, Namitha, Santhiya et al. 2024, Pudgerd, Saedan et al. 2024). These new methodologies have promising applications in the control and possible eradication of the virus, thereby offering a ray of hope to shrimp farmers.
Immunostimulants have become a strong tool in the arsenal of enhancing immunity in shrimp against WSSV infection. Different natural substances, seaweed extracts, essential oils, probiotics, plant-based chemicals and animal-derived immunostimulants have proven their efficacy by enhancing the resistance of shrimp to viral infections through boosting innate and non-specific? immunity (Citarasu 2010, Huang and Zhang 2013, Bindhu, Velmurugan et al. 2014, Rajashekar Reddy, Dinesh et al. 2016, Xie, Liu et al. 2019, Salehpour, Biuki et al. 2021, Huang, He et al. 2022, Ghosh 2023). One such compound is fucoidan, a sulfated polysaccharide extracted from brown algae such as Fucus vesiculosus. In shrimp, it enhances hemocyte counts, respiratory activity, and prophenoloxidase activity (Sinurat, Saepudin et al. 2016). All these mechanisms are capable of preventing viral replication, thereby reducing WSSV infection intensity. Another example is the sulfated galactan from red algae, which interacts with viral proteins VP26 and VP28, thereby preventing virus attachment to shrimp cells and further severely impeding its entry and replication (Rudtanatip, Asuvapongpatana et al. 2014). A few other promising compounds that exhibit antiviral activity include epigallocatechin gallate from green tea and naringenin from citrus fruits (Sun, Chen et al. 2021, Zhang, Wen et al. 2022). These natural compounds upregulate important immune-related genes, such as those involved in viral replication, providing shrimp with enhanced immunity against WSSV. Medicinal herbs like Agathi grandiflora and Argemone mexicana have already shown their efficacy in increasing shrimp immunity against WSSV replication (Bindhu, Velmurugan et al. 2014, Palanikumar, Benitta et al. 2018). These immunostimulants are especially important, as no effective antiviral drugs are currently known that would boost shrimp defenses naturally. For developing effective antiviral drug, targeting conserved proteins such as ribonucleotide reductase in critical stages of the virus life cycle could be a viable strategy. Inhibitors against such conserved enzymes could be effectively utilized against a wide range of dsDNA viruses, including WSSV. For example, ribonucleotide reductase is an essential enzyme involved in nucleotide metabolism and DNA synthesis; interference with its activity might disrupt viral replication (Krishnan, Katneni et al. 2023).
The use of probiotics, which are beneficial bacteria improving shrimp gut health and strengthening immune responses, is another promising avenue in WSSV defense. Some of the cited probiotics include Pediococcus pentosaceus, Lactobacillus and Bacillus spp which have been shown to stimulate the shrimp's immune system. These probiotics enhance the activities of important immune-related enzymes such as phenoloxidase (PO) and prophenoloxidase (proPO), which play a critical role in shrimp immune response against viral infections (Dekham, Jones et al. 2023, Ghosh 2023). Probiotics like Bacillus subtilis and Vibrio alginolyticus enhance the activity of digestive enzymes, leading to improved growth and resistance against WSSV (Rodríguez, Espinosa et al. 2007, Sekar, Kim et al. 2019). Additionally, how probiotics enhance the gut microbial structure of shrimp to create an environment less permissive tor WSSV replication is a very promising avenue of research. Dietary interventions, such as the inclusion of bioactive compounds into shrimp feed, have been highly successful in conferring immunity in shrimp against WSSV. Quercetin, a bioactive flavonoid present in several fruits and vegetables, has been demonstrated to significantly upregulate immune-related genes such as TLR, ALF, and NF-κB, playing a key role in the control of viral infections and regulation of cell growth and death (Chen, Fan et al. 2023, Yang, Wang et al. 2024). Other dietary compounds, such as inulin, galactooligosaccharides, carotenoids, and polyphenols from chestnuts and olive extracts, have also been found effective in enhancing the immune response of shrimp and thereby reducing mortality due to WSSV (Luna-González, Almaraz-Salas et al. 2012, Mustafa, Buentello et al. 2019, Tan, Zhang et al. 2020). Another major advantage of dietary interventions is their efficiency in maintaining the health of the gut, thereby indirectly improving the immune system in shrimp. These dietary compounds promote nutrient digestibility and digestive enzyme secretion through maintenance of orderly gut microbiota, resulting in better growth and resilience to viral infections such as WSSV. Essential oils from plants such as Zanthoxylum tsihanimposa and Eucalyptus globulus were also recommended as effective natural treatments for WSSV (Babikian, Babikyan et al. 2020, Zafilaza, Andriantsimahavandy et al. 2020). The bioactive components of these oils, including terpinene and thymol, exhibit robust antiviral, antimodulatory, and antioxidant activities. Even essential oils, like thyme, when protected against deterioration by microencapsulation, were found to enhance the immune response of shrimp by boosting PO activity, thereby increasing survival upon WSSV infection (Tomazelli Júnior, Kuhn et al. 2018). Additionally, blending different essential oils can result in synergistic interactions among components, further enhancing antiviral properties (Tariq, Wani et al. 2019). This multilevel natural approach presents a hopeful alternative to chemical treatments and actively offers shrimp farmers a more sustainable and eco-friendly solution for WSSV control.
Apart from these nutritional and phytochemical based research, nanotechnology provides advanced solutions in the detection and prevention of WSSV. For example, the use of Surface Plasmon Resonance and DNA functionalized with gold nanoparticles enables more sensitive and accurate detection of WSSV in shrimp (Bai, He et al. 2023). These technologies support real-time monitoring of viral loads in shrimp populations, allowing for timely and efficient management of the virus. For prevention, PVP-coated silver nanoparticles have great potential in enhancing shrimp immunity, thereby increasing survival rates against WSSV infection (Namitha, Santhiya et al. 2024). These nanoparticles enhance the effectiveness of DNA vaccines and immunostimulants by enhancing delivery to target cells, offering better protection against the virus. Nanotechnology thus plays an important role in both detection and prevention of WSSV, raising new hope for shrimp aquaculture. In recent years, polyanhydride nanoparticles have gained attention as a novel method for delivering vaccine antigens and emerged as strong potential for encapsulating and controlling the release of dsRNA molecules to manage disease in shrimp aquaculture. Notably, research by Phanse et. al., (2022) advanced the use of dsRNA-based nano-vaccine to fight viral infections in shrimp species like L. vannamei. Their study highlights that nanoparticle made from copolymers of sebacic acid, 1,6-bis(p-carboxyphenoxy) hexane, and 1,8-bis(p-carboxyphenoxy)-3,6-dioxaoctane achieved an impressive ~80% protection rate in shrimp when challenged with WSSV. This high level of protection underscores the potential of these nanoparticles as effective dsRNA carriers, enhancing immune responses and providing a strong defense against viral threats. Alongside polyanhydride nanoparticles, virus-like particles (VLPs) present another promising tool for controlling diseases in shrimp, particularly for delivering dsRNA (Phanse, Puttamreddy et al. 2022). VLPs resemble viruses structurally but do not contain any viral genetic material, making them ideal for bypassing host defenses and safely transporting therapeutic dsRNA. Because they mimic the external properties of viruses, VLPs can stimulate immune responses without the risk of causing infection. Studies by Pudgerd, Saedan et al. (2024) and Weerachatyanukul, Pooljun et al. (2022) have shown that capsid proteins from viruses like infectious hypodermal and haematopoietic necrosis virus and Macrobracium rosenbergii nodavirus can be used to encapsulate and deliver dsRNA molecules, such as VP28 and VP37, which are known to boost immune responses and limit viral replication in shrimp. These findings suggest that VLPs not only protect dsRNA from breakdown within host cells but also improve the uptake and stability of these therapeutic molecules. Beyond VLPs, several other nanoparticle platforms are being explored to deliver dsRNA to shrimp. Plant virus-based particles like cowpea chlorotic mottle virus and brome mosaic virus, as well as non-viral nanoparticles such as liposome, chitosan and beta-glucan, are under study for their potential in dsRNA delivery (Itsathitphaisarn, Thitamadee et al. 2017, Abo-Al-Ela 2021, Ramos-Carreño, Giffard-Mena et al. 2021, Ruiz-Guerrero, Giffard-Mena et al. 2023, Jonjaroen, Charoonnart et al. 2024). Each of these systems offers unique benefits, like biocompatibility, high encapsulation efficiency, and controlled release capabilities.
One of the most promising recent developments in WSSV research involves the use of DNA vaccines, which work by encoding specific viral proteins such as VP28 into shrimp cells to trigger an immune response. Such vaccines have been shown to increase survival rates by enhancing enzymatic activities in shrimp, such as super-oxide dismutase (SOD) and alkaline phosphatase (AKP), which are generally induced during a viral infection (Du, Hu et al. 2022). These DNA vaccines targeting the VP28 gene (focusing on conserved regions, areas less likely to mutate in the WSSV genome) express specific immunity proteins that bind to and neutralize the virus, preventing further replication and spread (Krishnankutty Chandrika and Thavarool Puthiyedathu 2021, Mugunthan, Loganathan et al. 2025). Moreover, the global distribution and genetic diversity of WSSV (discussed earlier in section 4.2), along with its evolutionary trajectory (adaptation and mutation in response to climate change) will be helpful in identifying conserved regions in the viral genome – a promising candidate for vaccine development. Vaccines designed around these stable sequences would likely remain effective unless these regions undergo significant changes, providing a more reliable way to combat WSSV outbreaks. Other types of DNA vaccines coated with chitosan show high potential. The coating protects the vaccine from degradation and allows oral delivery, resulting in higher efficacy in preventing WSD (Feng, Wang et al. 2017, Wikumpriya, Prabhatha et al. 2023). Shrimp farmers may soon have a more reliable long-term solution for managing outbreaks with the recent development of DNA vaccine technology. Apart from DNA vaccines, RNA-based vaccines have also been shown to provide specific and robust protection against WSSV infection in cultured shrimp. However, the major challenge for field application of these vaccines is the limited stability of double-stranded RNA in aquatic systems (Phanse, Puttamreddy et al. 2022). Therefore, researchers have focused on developing RNA interference and nanoparticle-based RNA vaccines to inhibit WSSV. Since then, many WSSV genes (VP19, VP28, rr1 and rr2) have been used as targets for RNAi-mediated neutralization of WSSV (Sanjuktha, Stalin Raj et al. 2012, Rattanarojpong, Khankaew et al. 2016, Li, Hong et al. 2019, Krishnankutty Chandrika and Thavarool Puthiyedathu 2021). Additionally, RNAi targeting of non-WSSV genes (PmRab7, PmRab7/PmIAP and GFP) was also tested for their ability to prevent WSSV infection (Kulkarni, Caipang et al. 2014, Alenton, Kondo et al. 2016). In particular PmRab7 + rr2 showed high activity, reducing viral genome replication by approximately 95%.
CRISPR-Cas gene editing in WSSV research has introduced a new paradigm for improving shrimp resistance to viral infections. This technology enables researchers to edit the DNA of shrimp with precision, either directly at viral DNA or by modifying genes involved in immune suppression (Ferdous, Islam et al. 2022). For instance, some neuroendocrine hormones like GIH and MIH suppress the immune system, making shrimp more susceptible to WSSV infection (Wang, Li et al. 2019, Wei, Pan et al. 2020). These genes could be edited using the CRISPR-Cas technology to enhance natural immunity in shrimp against viral attack (Diwan, Ninawe et al. 2017). CRISPR-Cas biological systems can reproduce and insert small segments of DNA corresponding to WSSV as spacers between short repeat sequences into the genomes of host shrimp. This occurs during the invasion of WSSV in these shrimps. These spacers improve the immune response of shrimp by providing a template for rapid recognition and targeting of the same DNA sequence by RNA molecules during subsequent viral infections (Ferdous, Islam et al. 2022). Recent advances in transcriptome analyses also help researchers understand how shrimp respond to WSSV at the genetic level. These analyses identified key genes and pathways upregulated in response to WSSV infection, providing insight into how functional feeds might influence gene expression and improve shrimp survival. Feeds have also been reported to enhance antioxidant activity and immune responses, helping to moderate the effects of WSSV. Proteins identified in several proteome studies include proPO, lysozyme, and crustin, which showed increased expression during WSSV infection (Sun, Wang et al. 2017, Thamizhvanan, Nafeez Ahmed et al. 2021). Therefore, the genetic and proteomic changes discussed above may contribute to better design of specific treatments through diets and immunostimulants that enhance resistance to viral infections in shrimp. However, proteomics studies on WSSV are currently limited, highlighting the need for more extensive research to identify the functions of viral structural proteins and explore the potential of envelope proteins as subunit vaccines for host protection. Enhanced efforts in protein identification and structural characterization are essential to advance this area.
The growing threat of WSSV in shrimp farming has necessitated the development of alternative control strategies beyond traditional methods like antibiotics, which have proven ineffective and harmful. Recent advances in immunostimulants, dietary interventions, probiotics, essential oils, DNA vaccines, nanotechnology, and CRISPR-Cas gene editing offer promising solutions for enhancing shrimp immunity and preventing WSSV outbreaks. The continued integration of these technologies, along with a deeper understanding of shrimp immune responses through transcriptome and proteomic analyses, holds the key to future breakthroughs in combating WSSV.
8. Future Research Direction
WSSV is one such pathogen, and the battle against it in shrimp aquaculture underscores the urgency for novel means of control that are environmental-friendly and effective. Although WSSV was one of the first shrimp viruses to be studied in depth, it remains one of the major viral agents affecting shrimp farming worldwide, causing severe economic losses and food security concerns. In this review, we have illustrated the current knowledge of WSSV, with a special focus on virus sensing and manipulation, spread mechanisms, and the strategies being applied in aquaculture to ensure biosecurity, induce immunity, and apply biotechnology against WSSV. Yet, much more needs to be done to address unanswered questions and to develop scalable, long-lasting solutions.
Genome-editing approaches such as CRISPR/Cas9, specifically for WSSV resistance could facilitate the development of new shrimp lines with improved immunity. There is the need for more functional studies to identify not only resistance-related genes but also epigenetic traits that control resilience at a phenotypic level - traits that may confound outcomes related to WSSV susceptibility. Furthermore, exploring the interactions between environmental stressors such as water quality and temperature with genetic resistance could facilitate holistic management approaches that enhance disease resilience.
Shrimp immunology remains under-explored in the context of WSSV control. Since shrimp lack an adaptive immune system like vertebrates, innate immunity is essential. In conclusion, future studies on the molecular mechanism of immune stimulants and probiotics should focus on developing formulations that induce optimal and safe outcomes against WSSV. Despite advances in improving shrimp immunity, extensive research is still needed to determine the optimal doses, timing and combinations of plant extracts and marine probiotics as immunostimulants. Moreover, these natural immunostimulants alone, could be coupled with newly available genetic tools to create novel and complementary strategies for strengthening shrimp population against WSSV.
WSSV control through biotechnology still holds promise. Though DNA and RNA-type vaccines show great potential, the socio-economic hurdles remain for their large-scale commercial use. Newer generation vaccines, including nanoparticle and oral formulations, may overcome some of these constraints and make vaccination feasible in open aquaculture systems. Research should also explore delivery methods that are stable enough for use in the field, perhaps through embedding RNAi agents within feeds or devising a time-release nanoparticle system that can be disseminated into shrimp ponds. Additionally, studying WSSV genome evolution under the selective pressure of such control measures could help predict and prevent the development of viral resistance.
The introduction of nanotechnology, although still in its early stages for WSSV control, offers a range of possibilities for both detection and treatment. Detection tools that use nanoparticle sensors could provide rapid diagnostics in shrimp ponds, allowing farmers to respond before outbreaks develop. Moreover, nanoscale technology may also facilitate the delivery of antiviral compounds or RNAi-based therapeutics, increasing their stability and bioavailability. Nevertheless, studies are needed to assess the safety and environmental sustainability of nanomaterials in aquaculture systems, where biosecurity measures often impact the efficacy/success of transmission-related tools used for the mitigation of shrimp viruses at large scale.
Another challenge in WSSV control is the limited understanding of its transmission pathways, including the role of wild crustacean carriers and environmental reservoirs. Vertical transmission through waterborne exposure are known to be highly effective, but little is known about the environmental persistence of WSSV under varying conditions such as salinity, pH and temperature status. Information from field studies should be further exploited to assess how these variables affect viral persistence, enabling the design of biosecurity measures tailored to local climatic and ecological circumstances. Research into other polymicrobial environments involving WSSV and other pathogens may also uncover synergistic interactions that affect the severity and spread of the infection. This knowledge would also be useful for creating compartmentalized aquaculture systems that could limit disease spread without the use of antibiotics.
We believe that the path? towards well-informed WSSV management in shrimp aquaculture will be shaped by interdisciplinary efforts and ongoing challenges. Integration across genetic resistance, immunological studies, biotechnology and ecological research can result in multifaceted durable solutions to WSD. To ensure that these innovations translate into real-world impact, researchers, industry stakeholders, and policy makers must work together to bridge the gap between laboratory findings and field applications. . As shrimp aquaculture continues to expand globally, it must balance environmental sustainability with robust viral biosecurity practices. These two forces must operate in tandem, not in isolation, to maintain long-term stability and productivity.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Md. Iftehimul: Conceptualization, Methodology, Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Neaz A. Hasan: Conceptualization, Methodology, Supervision, Writing - original draft, Writing - review & editing. David Bass: Writing – original draft, Writing – review and editing. Abul Bashar, Mohammad Mahfujul Haque and Morena Santi: Writing - review & editing.
Funding
This study was supported by the Ocean Country Partnership Programme (OCPP) under the project “Shrimp Health in Coastal Aquaculture of Bangladesh (Project No, 2022/21/Other)”, funded through Official Development Assistance (ODA) as part of the UK’s Blue Planet Fund.
Acknowledgments
The authors would like to acknowledge Dr Kelly Bateman, Invertebrate Pathologist from Cefas, UK for her support with providing histopathological plate. We would also like to thank Dr Ronny van Aerle, Bioinformatician from Cefas, for providing valuable information and editing the manuscript.
Conflicts of Interest
All authors declare that there is no conflict about this study.
References
- CABI (2022). white spot disease, CABI International.
-
Abo-Al-Ela, H; 4343-4355; G. (2021). "RNA interference in aquaculture: a small tool for big potential." Journal of Agricultural and Food Chemistry 69(15).
-
Abubakar, M; 739783; , H. T. Atmaca, M. A. Zahoor and O. Kul (2015). "Cellular Immunity-Pathogen Interactions in Infectious Diseases." Journal of Immunology Research 2015.
-
Afsharnasab, M; 1021-1055; , S. Kakoolaki and F. Afzali (2014). "The Status of white spot syndrome virus (WSSV) in Islamic Republic of Iran." Iranian Journal of Fisheries Sciences 13(4).
-
Alenton, R; 65-70; R. R., H. Kondo, I. Hirono and M. B. B. Maningas (2016). "Gene silencing of VP9 gene impairs WSSV infectivity on Macrobrachium rosenbergii." Virus research 214.
-
Arbon, P; 923-941; , M. Andrade Martinez, D. Jerry and K. Condon (2024). "Towards a ‘systems’ approach for viral challenge experiments in shrimp: Reporting guidelines for publication." Reviews in Aquaculture 16(2).
-
Assavalapsakul, W; 484-493; and S. Panyim (2012). "Molecular cloning and tissue distribution of the Toll receptor in the black tiger shrimp, Penaeus monodon." Genet Mol Res 11(1).
-
Attasart, P; 127-133; , R. Kaewkhaw, C. Chimwai, U. Kongphom, O. Namramoon and S. Panyim (2009). "Inhibition of white spot syndrome virus replication in Penaeus monodon by combined silencing of viral rr2 and shrimp PmRab7." Virus Res 145(1).
- Babikian, H., Y. Babikian, H., Y. Babikyan, R. K. Jha, S. D. Wisoyo, Y. Asih and S. Srisombat (2020). "Performance of natural oil blend formulation (NOBF) against white spot syndrome virus (WSSV) agent in Penaeus vannamei boone, 1931.
-
Bai, Y; 738797; , L. He, M. Sun, X. Zhou and Z. Xu (2023). "Dark-field visual counting of white spot syndrome virus using gold nanoparticle probe." Aquaculture 562.
-
Bindhu, F; 482-492; , S. Velmurugan, M. B. S. Donio, M. Michaelbabu and T. Citarasu (2014). "Influence of Agathi grandiflora active principles inhibit viral multiplication and stimulate immune system in Indian white shrimp Fenneropenaeus indicus against white spot syndrome virus infection." Fish & Shellfish Immunology 41(2).
-
Cavalli, L; 1176-1179; S., L. A. Romano, L. F. Marins and P. C. Abreu (2011). "First report of White spot syndrome virus in farmed and wild penaeid shrimp from lagoa dos patos estuary, southern brazil." Braz J Microbiol 42(3).
-
Chamberlain, G; 1-34; (2010). "History of shrimp farming." The Shrimp Book Nottingham University Press, United Kingdom.
-
Chamberlain, G; impacts and management recommendations; , D. Lightner, R. Towner, P. van Wyk, M. Villarreal, N. Akazawa and A. Alvial (2013). "Case study of the outbreak of white spot syndrome virus at shrimp farms in Mozambique and Madagascar.
-
Chang, Y; 909-917; -S., T.-C. Chen, W.-J. Liu, J.-S. Hwang, G.-H. Kou and C.-F. Lo (2011). "Assessment of the Roles of Copepod Apocyclops royi and Bivalve Mollusk Meretrix lusoria in White Spot Syndrome Virus Transmission." Marine Biotechnology 13(5).
-
Chang, Y; 12555-12564; -S., W.-J. Liu, T.-L. Chou, Y.-T. Lee, T.-L. Lee, W.-T. Huang, G.-H. Kou and C.-F. Lo (2008). "Characterization of White Spot Syndrome Virus Envelope Protein VP51A and Its Interaction with Viral Tegument Protein VP26." Journal of Virology 82(24).
-
Chang, Y; 2963-2968; S., S. E. Peng, H. T. Yu, F. C. Liu, C. H. Wang, C. F. Lo and G. H. Kou (2004). "Genetic and phenotypic variations of isolates of shrimp Taura syndrome virus found in Penaeus monodon and Metapenaeus ensis in Taiwan." J Gen Virol 85(Pt 10).
-
Chanratchakool, P; 177-189; and M. J. Phillips (2002). "Social and economic impacts and management of shrimp disease among small-scale farmers in Thailand and Viet Nam." FAO Fisheries Technical Paper.
-
Chen, R; 28694; Y., K. L. Shen, Z. Chen, W. W. Fan, X. L. Xie, C. Meng, X. J. Chang, L. B. Zheng, J. Jeswin, C. H. Li, K. J. Wang and H. P. Liu (2016). "White spot syndrome virus entry is dependent on multiple endocytic routes and strongly facilitated by Cq-GABARAP in a CME-dependent manner." Sci Rep 6.
-
Chen, Z; 738887; , D. Fan, L. Pan, C. Su, Y. Ding and M. Lu (2023). "Study of effects of dietary quercetin (Que) on growth performance and disease resistance mechanism of Litopenaeus vannamei." Aquaculture 563.
-
Chou Hy, H; 165-173; C. Y., C. H. Wang, H. C. Chiang and C. F. Lo (1995). "Pathogenicity of a baculovirus infection causing white spot syndrome in cultured penaeid shrimp in Taiwan." Diseases of Aquatic Organisms 23(3).
-
Citarasu, T; 403-414; (2010). "Herbal biomedicines: a new opportunity for aquaculture industry." Aquaculture International 18(3).
-
Clarke, T; 401-424; E. and R. J. Clem (2003). "Insect defenses against virus infection: the role of apoptosis." International Reviews of Immunology 22(5-6).
-
Cock, J; 1-11; , T. Gitterle, M. Salazar and M. Rye (2009). "Breeding for disease resistance of Penaeid shrimps." Aquaculture 286(1-2).
-
Cox, N; 1824; , E. De Swaef, M. Corteel, W. Van Den Broeck, P. Bossier, J. J. Dantas-Lima and H. J. Nauwynck (2023). "The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp." Viruses 15(9).
-
Cox, N; 813; , E. De Swaef, M. Corteel, W. Van Den Broeck, P. Bossier, H. J. Nauwynck and J. J. Dantas-Lima (2024). "Experimental Infection Models and Their Usefulness for White Spot Syndrome Virus (WSSV) Research in Shrimp." Viruses 16(5).
-
Cui, C; 594390; , Q. Liang, X. Tang, J. Xing, X. Sheng and W. Zhan (2020). "Differential apoptotic responses of hemocyte subpopulations to white spot syndrome virus infection in Fenneropenaeus chinensis." Frontiers in Immunology 11.
-
Dai, W; 3755-3764; , W. Yu, L. Xuan, Z. Tao and J. Xiong (2018). "Integrating molecular and ecological approaches to identify potential polymicrobial pathogens over a shrimp disease progression." Applied Microbiology and Biotechnology 102.
-
Davies, R; //www; (2016). "Disease has cost Asia shrimp sector over $20bn." Retrieved 13 March, 2025, from https.
-
DAWR, D; 20; o. A. a. W. R. (2017). "Report into the cause of white spot syndrome virus outbreak in the Logan River area of Queensland – December 2016." Interim report.
-
Debnath, P; 71-77; , M. Karim and B. Belton (2014). "Comparative study of the reproductive performance and White Spot Syndrome Virus (WSSV) status of black tiger shrimp (Penaeus monodon) collected from the Bay of Bengal." Aquaculture 424-425.
-
Dekham, K; 21610; , S. M. Jones, S. Jitrakorn, P. Charoonnart, N. Thadtapong, R. Intuy, P. Dubbs, S. Siripattanapipong, V. Saksmerprome and S. Chaturongakul (2023). "Functional and genomic characterization of a novel probiotic Lactobacillus johnsonii KD1 against shrimp WSSV infection." Scientific Reports 13(1).
-
Desrina, S; 1843-1860; B. Prayitno, M. C. Verdegem, J. A. Verreth and J. M. Vlak (2022). "White spot syndrome virus host range and impact on transmission." Reviews in Aquaculture 14(4).
-
Dewangan, N; 1-7; K., G. Ayyaru, R. Kuzhanthaivel, S. Somasundaram Thirugnanasambandan, G. G. Martin, K. Daniel and R. S. Ramakrishna (2017). "Incidence of simultaneous infection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) and white spot syndrome virus (WSSV) in Litopenaeus vannamei." Aquaculture 471.
-
DISEASES-Cefas, I; //www; D. O. A. A. "DISEASE DATA WHITE SPOT SYNDROME VIRUS." from https.
-
Diwan, A; 65-72; , A. Ninawe and S. Harke (2017). "Gene editing (CRISPR-Cas) technology and fisheries sector." Canadian Journal of Biotechnology 1(2).
-
Du, Y; 1040336; , X. Hu, L. Miao and J. Chen (2022). "Current status and development prospects of aquatic vaccines." Frontiers in immunology 13.
-
Elmore, S; 495-516; (2007). "Apoptosis: a review of programmed cell death." Toxicologic pathology 35(4).
-
Encinas-García, T; 1-28; , F. Mendoza-Cano, A. Muhlia-Almazán, M. Porchas-Cornejo and A. Sánchez-Paz (2023). "A Review of Shrimp Cellular Receptors for WSSV: Potential Targets for Antiviral Strategies in Shrimp Aquaculture." Reviews in Fisheries Science & Aquaculture.
-
Escobedo-Bonilla, C; 1-18; M., V. Alday-Sanz, M. Wille, P. Sorgeloos, M. B. Pensaert and H. J. Nauwynck (2008). "A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus." J Fish Dis 31(1).
- Evan, Y. Evan, Y. and N. E. Putri (2021). Status of aquatic animal health in Indonesia. Proceedings of the International Workshop on the Promotion of Sustainable Aquaculture, Aquatic Animal Health, and Resource Enhancement in Southeast Asia, Aquaculture Department, Southeast Asian Fisheries Development Center.
-
Fauquet, C; VIIIth report of the International Committee on Taxonomy of Viruses; M., M. A. Mayo, J. Maniloff, U. Desselberger and L. A. Ball (2005). Virus taxonomy.
-
Fe, O; 77-82; M., T. N. Karlo, P. M. Veronica, G. A. Catalino, O. d. l. P. Â. a. Florian, O. M. Rolando, D. A. Juan, J. E. Cesar B. Nadala, C. L. Philip and M.-T. Lourdes (2000). "White spot syndrome virus (WSSV) in cultured Penaeus monodon in the Philippines." Diseases of Aquatic Organisms 42(1).
-
Feng, J; 148-159; , L. Zhao, M. Jin, T. Li, L. Wu, Y. Chen and Q. Ren (2016). "Toll receptor response to white spot syndrome virus challenge in giant freshwater prawns (Macrobrachium rosenbergii)." Fish & Shellfish Immunology 57.
-
Feng, S; 2923-2936; , C. Wang, S. Hu, Q. Wu and A. Li (2017). "Recent progress in the development of white spot syndrome virus vaccines for protecting shrimp against viral infection." Archives of Virology 162(10).
-
Ferdous, M; 2012; A., S. I. Islam, N. Habib, M. Almehmadi, M. Allahyani, A. A. Alsaiari and A. Shafie (2022). "CRISPR-Cas genome editing technique for fish disease management: current study and future perspective." Microorganisms 10(10).
-
Flegel, T; 433-442; W. (1997). "Major viral diseases of the black tiger prawn (Penaeus monodon) in Thailand." World Journal of Microbiology and Biotechnology 13(4).
-
Galavíz-Silva, L; 53-68; , Z. J. Molina-Garza, J. M. Alcocer-González, J. L. Rosales-Encinas and C. Ibarra-Gámez (2004). "White spot syndrome virus genetic variants detected in Mexico by a new multiplex PCR method." Aquaculture 242(1).
- Galib, M. R. H., A. Galib, M. R. H., A. K. Ghosh and W. Sabbir (2024). "Dietary impact of Ocimum tenuiflorum leaf extract on the growth metrics and immune responses of shrimp (Penaeus monodon) against white spot syndrome virus (WSSV)." Heliyon.
-
Gholamhoseini, B; 335-347; , M. Afsharnasab and A. A. Motallebi (2013). "Rate (ROI) and severity (SOI) of infection of white spot disease in cultured and captured Penaeidshrimps in the Persian Gulf using histopathology and polymerase chain reaction." Iranian Journal of Fisheries Sciences 12(2).
-
Ghosh, A; 108942; K. (2023). "Functionality of probiotics on the resistance capacity of shrimp against white spot syndrome virus (WSSV)." Fish & shellfish immunology.
-
Gong, J; 107931; , X. Pan, X. Zhou and F. Zhu (2023). "Dietary quercetin protects Cherax quadricarinatus against white spot syndrome virus infection." Journal of Invertebrate Pathology 198.
-
Govindaraju, K; 1163-1171; , P. Dilip Itroutwar, V. Veeramani, T. Ashok Kumar and S. Tamilselvan (2020). "Application of nanotechnology in diagnosis and disease management of white spot syndrome virus (WSSV) in aquaculture." Journal of Cluster Science 31.
-
Guanzon, D; 172-180; A. V. and M. B. B. Maningas (2018). "Functional elucidation of LvToll 3 receptor from P. vannamei through RNA interference and its potential role in the shrimp antiviral response." Developmental & Comparative Immunology 84.
-
Gui, L; 1008-1015; , B. Wang, F.-H. Li, Y.-M. Sun, Z. Luo and J.-H. Xiang (2012). "Blocking the large extracellular loop (LEL) domain of FcTetraspanin-3 could inhibit the infection of white spot syndrome virus (WSSV) in Chinese shrimp, Fenneropenaeus chinensis." Fish & Shellfish Immunology 32(6).
-
Ha, Y; 964-967; M., G. Soo-Jung, N. Thi-Hoai, C. H. Ra, K. H. Kim, Y. K. Nam and S. K. Kim (2008). "Vaccination of shrimp (Penaeus chinensis) against white spot syndrome virus (WSSV)." J Microbiol Biotechnol 18(5).
-
Hameed, A; 157-161; S., G. Balasubramanian, S. S. Musthaq and K. Yoganandhan (2003). "Experimental infection of twenty species of Indian marine crabs with white spot syndrome virus (WSSV)." Dis Aquat Organ 57(1-2).
-
Hasan, N; 735348; A., M. M. Haque, S. J. Hinchliffe and J. Guilder (2020). "A sequential assessment of WSD risk factors of shrimp farming in Bangladesh: Looking for a sustainable farming system." Aquaculture 526.
-
He, W; 164-173; , S. Rahimnejad, L. Wang, K. Song, K. Lu and C. Zhang (2017). "Effects of organic acids and essential oils blend on growth, gut microbiota, immune response and disease resistance of Pacific white shrimp (Litopenaeus vannamei) against Vibrio parahaemolyticus." Fish & shellfish immunology 70.
-
He, Y; 1104-1112; , K. Yang and X. Zhang (2014). "Viral MicroRNAs Targeting Virus Genes Promote Virus Infection in Shrimp In Vivo." Journal of Virology 88(2).
-
He, Y; 1019-1029; and X. Zhang (2012). "Comprehensive characterization of viral miRNAs involved in white spot syndrome virus (WSSV) infection." RNA Biology 9(7).
- Hoseinifar, S. H., Y. Hoseinifar, S. H., Y.-Z. Sun, A. Wang and Z. Zhou (2018). "Probiotics as Means of Diseases Control in Aquaculture, a Review of Current Knowledge and Future Perspectives." Frontiers in Microbiology 9.
-
Hossain, M; 1-11; S., A. Chakraborty, B. Joseph, S. Otta, I. Karunasagar and I. Karunasagar (2001). "Detection of new hosts for white spot syndrome virus of shrimp using nested polymerase chain reaction." Aquaculture 198(1-2).
-
Hou, F; 255-260; , S. He, Y. Liu, X. Zhu, C. Sun and X. Liu (2014). "RNAi knock-down of shrimp Litopenaeus vannamei Toll gene and immune deficiency gene reveals their difference in regulating antimicrobial peptides transcription." Developmental & Comparative Immunology 44(2).
-
Huang, A; 96-103; -G., W.-H. He, F.-L. Zhang, C.-S. Wei and Y.-H. Wang (2022). "Natural component geniposide enhances survival rate of crayfish Procambarus clarkii infected with white spot syndrome virus." Fish & shellfish immunology 126.
-
Huang, H; 305-314; -T., H.-L. Chan, T.-Y. Shih and L.-L. Chen (2015). "A study of the role of glucose transporter 1 (Glut1) in white spot syndrome virus (WSSV) infection." Fish & Shellfish Immunology 46(2).
-
Huang, P; 1799-1808; Y., J. H. Leu and L. L. Chen (2014). "A newly identified protein complex that mediates white spot syndrome virus infection via chitin-binding protein." J Gen Virol 95(Pt 8).
-
Huang, R; 1357-1361; , Y. Xie, J. Zhang and Z. Shi (2005). "A novel envelope protein involved in White spot syndrome virus infection." Journal of General Virology 86(5).
-
Huang, T; 2544-2554; , Y. Cui and X. Zhang (2014). "Involvement of Viral MicroRNA in the Regulation of Antiviral Apoptosis in Shrimp." Journal of Virology 88(5).
-
Huang, T; 12997-13004; and X. Zhang (2012). "Functional analysis of a crustacean microRNA in host-virus interactions." Journal of virology 86(23).
-
Huang, T; 137-146; and X. Zhang (2013). "Host defense against DNA virus infection in shrimp is mediated by the siRNA pathway." European journal of immunology 43(1).
-
Huang, Y; 140-147; , T. Li, M. Jin, S. Yin, K.-M. Hui and Q. Ren (2017). "Newly identified PcToll4 regulates antimicrobial peptide expression in intestine of red swamp crayfish Procambarus clarkii." Gene 610.
-
Inouye, K; 149-158; , S. Miwa, N. Oseko, H. Nakano, T. Kimura, K. Momoyama and M. Hiraoka (1994). "Mass Mortalities of Cultured Kuruma Shrimp Penaeus japonicus in Japan in 1993 : Electron Microscopic Evidence of the Causative Virus." Fish Pathology 29(2).
-
Inouye, K; 39-45; , K. Yamano, N. Ikeda, T. Kimura, H. Nakano, K. Momoyama, J. Kobayashi and S. Miyajima (1996). "The Penaeid Rod-shaped DNA Virus (PRDV), which Causes Penaeid Acute Viremia (PAV)." Fish Pathology 31(1).
-
Islam, S; 778-801; I., M. J. Mou, S. Sanjida and S. Mahfuj (2023). "A review on molecular detection techniques of white spot syndrome virus: Perspectives of problems and solutions in shrimp farming." Veterinary Medicine and Science 9(2).
-
Itsathitphaisarn, O; 76-85; , S. Thitamadee, W. Weerachatyanukul and K. Sritunyalucksana (2017). "Potential of RNAi applications to control viral diseases of farmed shrimp." Journal of invertebrate pathology 147.
-
Jang, I; 911-920; K., G. Qiao and S.-K. Kim (2014). "Effect of multiple infections with white spot syndrome virus and Vibrio anguillarum on Pacific white shrimp Litopenaeus vannamei (L.): mortality and viral replication." Journal of Fish Diseases 37(10).
-
Jassim, A; 88-100; A. R. and N. K. Al-Salim (2015). "Viral diseases of some species from Penaeid shrimp in Iraqi marine waters." Iraqi Journal of Aquaculture 12(2).
-
Jiang, L; 250-261; , J. Xiao, L. Liu, Y. Pan, S. Yan and Y. Wang (2017). "Characterization and prevalence of a novel white spot syndrome viral genotype in naturally infected wild crayfish, Procambarus clarkii, in Shanghai, China." VirusDisease 28(3).
-
Jithendran, K; 4701-4710; P., A. Navaneeth Krishnan, V. Jagadeesan, R. Anandaraja, P. Ezhil Praveena, S. Anushya, C. Bala Amarnath and T. Bhuvaneswari (2021). "Co-infection of infectious myonecrosis virus and Enterocytozoon hepatopenaei in Penaeus vannamei farms in the east coast of India." Aquaculture Research 52(10).
- Jonjaroen, V., P. Jonjaroen, V., P. Charoonnart, S. Jitrakorn, P. Payongsri, R. Surarit, V. Saksmerprome and N. Niamsiri (2024). "Nanoparticles-based double-stranded RNA delivery as an antiviral agent in shrimp aquaculture." Reviews in Aquaculture.
-
Joseph, T; 51-54; C., R. James, L. A. Rajan, P. Surendran and K. Lalitha (2015). "White spot syndrome virus infection: Threat to crustacean biodiversity in Vembanad Lake, India." Biotechnology Reports 7.
-
Kaewkascholkul, N; 191-201; , K. Somboonviwat, S. Asakawa, I. Hirono, A. Tassanakajon and K. Somboonwiwat (2016). "Shrimp miRNAs regulate innate immune response against white spot syndrome virus infection." Developmental & Comparative Immunology 60.
- Kalaimani, N., T. Kalaimani, N., T. Ravisankar, N. Chakravarthy, S. Raja, T. Santiago and A. Ponniah (2013). "Economic losses due to disease incidences in shrimp farms of India.
-
Karunasagar, I; 141-148; and L. Ababouch (2012). "Shrimp Viral Diseases, Import Risk Assessment and International Trade." Indian Journal of Virology 23(2).
-
Kim, M; 1676; -J., J.-O. Kim, G.-I. Jang, M.-G. Kwon and K.-I. Kim (2023). "Evaluation of the Horizontal Transmission of White Spot Syndrome Virus for Whiteleg Shrimp (Litopenaeus vannamei) Based on the Disease Severity Grade and Viral Shedding Rate." Animals 13(10).
-
Kim, M; 741597; J., D.-J. Shin, G. I. Jang, M.-G. Kwon and K. I. Kim (2025). "Influence of stocking density and interaction variability on disease progression of white spot syndrome virus-infected shrimp under different risk scenarios." Aquaculture 595.
-
Knibb, W; 26-29; , C. Le, M. Katouli, I. Bar and C. Lloyd (2018). "Assessment of the origin of white spot syndrome virus DNA sequences in farmed Penaeus monodon in Australia." Aquaculture 494.
-
Kooloth Valappil, R; 1888-1906; , G. D. Stentiford and D. Bass (2021). "The rise of the syndrome–sub-optimal growth disorders in farmed shrimp." Reviews in Aquaculture 13(4).
-
Krishnan, K; 72; , V. K. Katneni, S. K. Prabhudas, N. Kaikkolante, A. K. Jangam, U. K. Katneni, C. Hauton, L. Peruzza, S. S. Mudagandur and V. K. Koyadan (2023). "MRF: a tool to overcome the barrier of inconsistent genome annotations and perform comparative genomics studies for the largest animal DNA virus." Virology Journal 20(1).
-
Krishnankutty Chandrika, S; 734-743; and S. Thavarool Puthiyedathu (2021). "Challenges and prospects of Viral Envelope protein VP28-based control strategies to combat white spot syndrome virus in penaeid shrimps: a review." Reviews in Aquaculture 13(1).
-
Kulkarni, A; 431-459; , S. Krishnan, D. Anand, S. Kokkattunivarthil Uthaman, S. K. Otta, I. Karunasagar and R. Kooloth Valappil (2021). "Immune responses and immunoprotection in crustaceans with special reference to shrimp." Reviews in Aquaculture 13(1).
-
Kulkarni, A; 55-65; D., C. M. Caipang, V. Kiron, J. H. Rombout, J. M. Fernandes and M. F. Brinchmann (2014). "Evaluation of immune and apoptosis related gene responses using an RNAi approach in vaccinated Penaeus monodon during oral WSSV infection." Marine genomics 18.
-
Kumar, S; 25325-25343; , A. K. Verma, S. P. Singh and A. Awasthi (2023). "Immunostimulants for shrimp aquaculture: paving pathway towards shrimp sustainability." Environmental Science and Pollution Research 30(10).
-
Lan, J; 17-24; -F., S. Wei, Y.-Q. Wang, Y.-J. Dai, J.-G. Tu, L.-J. Zhao, X.-C. Li, Q.-W. Qin, N. Chen and L. Lin (2016). "PcToll3 was involved in anti-Vibrio response by regulating the expression of antimicrobial peptides in red swamp crayfish, Procambarus clarkii." Fish & Shellfish Immunology 57.
-
Lan, J; 59-66; -F., L.-J. Zhao, S. Wei, Y. Wang, L. Lin and X.-C. Li (2016). "PcToll2 positively regulates the expression of antimicrobial peptides by promoting PcATF4 translocation into the nucleus." Fish & shellfish immunology 58.
-
Lan, J; 608-617; -F., J. Zhou, X.-W. Zhang, Z.-H. Wang, X.-F. Zhao, Q. Ren and J.-X. Wang (2013). "Characterization of an immune deficiency homolog (IMD) in shrimp (Fenneropenaeus chinensis) and crayfish (Procambarus clarkii)." Developmental & Comparative Immunology 41(4).
-
Lee, C; 738922; , H. J. Jeon, B. Kim, S.-K. Choi, J. H. Kim and J. E. Han (2023). "Multiple infections of a new-type decapod hepanhamaparvovirus (DHPV) and Enterocytozoon hepatopenaei in Korea and DHPV infectivity in Penaeus vannamei." Aquaculture 563.
-
Lee, C; 737117; , J. H. Kim, S.-K. Choi, H. J. Jeon, S. H. Lee, B. K. Kim, Y. K. Kim, K.-J. Lee and J. E. Han (2021). "Detection of infectious white spot syndrome virus in red claw crayfish (Cherax quadricarinatus) and red swamp crayfish (Procambarus clarkii) imported into Korea." Aquaculture 544.
-
Leu, J; 140-149; -H., J.-M. Tsai, H.-C. Wang, A. H.-J. Wang, C.-H. Wang, G.-H. Kou and C.-F. Lo (2005). "The Unique Stacked Rings in the Nucleocapsid of the White Spot Syndrome Virus Virion Are Formed by the Major Structural Protein VP664, the Largest Viral Structural Protein Ever Found." Journal of Virology 79(1).
-
Li, C; 1785; , S. Wang and J. He (2019). "The two NF-κB pathways regulating bacterial and WSSV infection of shrimp." Frontiers in immunology 10.
-
Li, C; 558-571; , S. Weng and J. He (2019). "WSSV–host interaction: Host response and immune evasion." Fish & Shellfish Immunology 84.
-
Li, F; 11-26; and J. Xiang (2013). "Recent advances in researches on the innate immunity of shrimp in China." Developmental & Comparative Immunology 39(1).
-
Li, L; 197666; , Y. Hong, H. Qiu, F. Yang and F. Li (2019). "VP19 is important for the envelope coating of white spot syndrome virus." Virus research 270.
-
Li, M; 231-240; , C. Li, C. Ma, H. Li, H. Zuo, S. Weng, X. Chen, D. Zeng, J. He and X. Xu (2014). "Identification of a C-type lectin with antiviral and antibacterial activity from pacific white shrimp Litopenaeus vannamei." Developmental & Comparative Immunology 46(2).
-
Li, Q; 215-223; , Y. Chen and F. Yang (2004). "Identification of a collagen-like protein gene from white spot syndrome virus." Archives of Virology 149(2).
-
Li, Z; 1609-1620; , Q. Lin, J. Chen, J. L. Wu, T. K. Lim, S. S. Loh, X. Tang and C.-L. Hew (2007). "Shotgun identification of the structural proteome of shrimp white spot syndrome virus and iTRAQ differentiation of envelope and nucleocapsid subproteomes." Molecular & Cellular Proteomics 6(9).
-
Liang, Y; 81-85; , J. Huang, X.-L. Song, P.-J. Zhang and H.-S. Xu (2005). "Four viral proteins of white spot syndrome virus (WSSV) that attach to shrimp cell membranes." Diseases of Aquatic Organisms 66.
-
Lightner, D; 146-160; , R. Redman, B. Poulos, L. Nunan, J. Mari and K. Hasson (1997). "Risk of spread of penaeid shrimp viruses in the." Rev. Sci. Tech. Off. Int. Epiz 16(1).
-
Liu, Q; 27-33; , D. Xu, S. Jiang, J. Huang, F. Zhou, Q. Yang, S. Jiang and L. Yang (2018). "Toll-receptor 9 gene in the black tiger shrimp (Penaeus monodon) induced the activation of the TLR–NF-κB signaling pathway." Gene 639.
-
Liu, S; 268; , S.-C. Zheng, Y.-L. Li, J. Li and H.-P. Liu (2020). "Hemocyte-mediated phagocytosis in crustaceans." Frontiers in Immunology 11.
-
López-Téllez, N; 334-345; A., J. A. Corbalá-Bermejo, M. L. Bustamante-Unzueta, L. P. Silva-Ledesma, V. M. Vidal-Martínez and R. Rodriguez-Canul (2020). "History, impact, and status of infectious diseases of the Pacific white shrimp Penaeus vannamei (Bonne, 1831) cultivated in Mexico." Journal of the World Aquaculture Society 51(2).
-
Lotz, J; 199-209; M. and M. A. Soto (2002). "Model of white spot syndrome virus (WSSV) epidemics in Litopenaeus vannamei." Diseases of aquatic organisms 50(3).
-
Lulijwa, R; 640-663; , E. J. Rupia and A. C. Alfaro (2020). "Antibiotic use in aquaculture, policies and regulation, health and environmental risks: a review of the top 15 major producers." Reviews in Aquaculture 12(2).
-
Luna-González, A; 28-32; , J. C. Almaraz-Salas, J. A. Fierro-Coronado, M. del Carmen Flores-Miranda, H. A. González-Ocampo and V. Peraza-Gómez (2012). "The prebiotic inulin increases the phenoloxidase activity and reduces the prevalence of WSSV in whiteleg shrimp (Litopenaeus vannamei) cultured under laboratory conditions." Aquaculture 362.
-
Mariot, L; 737172; V., N. Bolívar, J. D. R. Coelho, P. Goncalves, S. M. Colombo, F. V. do Nascimento, D. D. Schleder and L. Hayashi (2021). "Diets supplemented with carrageenan increase the resistance of the Pacific white shrimp to WSSV without changing its growth performance parameters." Aquaculture 545.
-
Martorelli, S; 117-131; R., R. M. Overstreet and J. A. Jovonovich (2010). "First report of viral pathogens WSSV and IHHNV in Argentine crustaceans." Bulletin of marine science 86(1).
-
Matozzo, V; 165-179; , C. Ercolini, L. Serracca, R. Battistini, I. Rossini, G. Granato, E. Quaglieri, A. Perolo, L. Finos, G. Arcangeli, D. Bertotto, G. Radaelli, B. Chollet, I. Arzul and F. Quaglio (2018). "Assessing the health status of farmed mussels (Mytilus galloprovincialis) through histological, microbiological and biomarker analyses." Journal of Invertebrate Pathology 153.
-
Mekata, T; 122-133; , T. Kono, T. Yoshida, M. Sakai and T. Itami (2008). "Identification of cDNA encoding Toll receptor, MjToll gene from kuruma shrimp, Marsupenaeus japonicus." Fish & Shellfish Immunology 24(1).
-
Miao, H; 221-224; Z., S. L. Tong, B. Xu, M. Jiang and X. Y. Liu (2000). "[Multiplication of the shrimp baculovirus HHNBV with primary cell cultures from lymphoid organ of Penaeus chinensis]." Sheng Wu Gong Cheng Xue Bao 16(2).
-
Millard, R; 107369; S., R. P. Ellis, K. S. Bateman, L. K. Bickley, C. R. Tyler, R. van Aerle and E. M. Santos (2021). "How do abiotic environmental conditions influence shrimp susceptibility to disease? A critical analysis focussed on White Spot Disease." Journal of invertebrate pathology 186.
-
Moser, J; 975-996; R. and M. R. F. Marques (2023). "Susceptibility of Neohelice granulata (Decapoda, Varunidae) to white spot syndrome virus (WSSV)." Aquaculture International 31(2).
-
Muegue, M; 63-72; F. S., P. I. P. Padilla, M. R. A. C. Bermeo-Capunong, C. M. A. Caipang, R. J. Gestuveo, M. J. A. Amar and J. S. Geduspan (2023). "Histological Changes in the Hepatopancreas and Stomach of Litopenaeus vannamei Experimentally Induced with White Spot Syndrome Virus Infection." UTTAR PRADESH JOURNAL OF ZOOLOGY 44(16).
- Mugunthan, S. P., N. Mugunthan, S. P., N. Loganathan, B. Shanmugaraj and H. M. Chandra (2025). "A narrative review on the white spot syndrome virus and the perspective of vaccine development." Vacunas.
-
Mustafa, A; 662-675; , A. Buentello, D. Gatlin, D. Lightner, M. Hume and A. Lawrence (2019). "Dietary supplementation of galactooligosaccharides (GOS) in Pacific white shrimp, Litopenaeus vannamei, cultured in a recirculating system and its effects on gut microflora, growth, stress, and immune response." Journal of Immunoassay and Immunochemistry 40(6).
-
Nadala, E; 39-44; C. B., L. M. Tapay, S. Cao and P. C. Loh (1997). "Detection of yellowhead virus and Chinese baculovirus in penaeid shrimp by the Western blot technique." Journal of Virological Methods 69(1).
-
Namitha, R; 1-12; , P. Santhiya, I. B. Singh, G. Dharani, M. Kannan and K. Govindaraju (2024). "Preparation and characterization of silica nanoparticles using beach sand and their anti-viral activity against white spot syndrome virus (WSSV): in vitro and in silico studies." Aquaculture International.
-
Ng, Y; 546; S., C.-S. Cheng, M. Ando, Y.-T. Tseng, S.-T. He, C.-Y. Li, S.-W. Cheng, Y.-M. Chen, R. Kumar, C.-H. Liu, H. Takeyama, I. Hirono and H.-C. Wang (2023). "White spot syndrome virus (WSSV) modulates lipid metabolism in white shrimp." Communications Biology 6(1).
-
Nilsen, P; 1028; , M. Karlsen, K. Sritunyalucksana and S. Thitamadee (2017). "White spot syndrome virus VP28 specific double-stranded RNA provides protection through a highly focused siRNA population." Scientific Reports 7(1).
-
Ning, J; 1127-1135; -F., W. Zhu, J.-P. Xu, C.-Y. Zheng and X.-L. Meng (2009). "Oral delivery of DNA vaccine encoding VP28 against white spot syndrome virus in crayfish by attenuated Salmonella typhimurium." Vaccine 27(7).
-
Oakey, J; 2061-2082; , C. Smith, D. Underwood, M. Afsharnasab, V. Alday-Sanz, A. Dhar, S. Sivakumar, A. Sahul Hameed, K. Beattie and A. Crook (2019). "Global distribution of white spot syndrome virus genotypes determined using a novel genotyping assay." Archives of virology 164.
-
Oakey, J; 2061-2082; , C. Smith, D. Underwood, M. Afsharnasab, V. Alday-Sanz, A. Dhar, S. Sivakumar, A. S. Sahul Hameed, K. Beattie and A. Crook (2019). "Global distribution of white spot syndrome virus genotypes determined using a novel genotyping assay." Archives of Virology 164(8).
-
Otta, S; 67-70; , G. Shubha, B. Joseph, A. Chakraborty, I. Karunasagar and I. Karunasagar (1999). "Polymerase chain reaction (PCR) detection of white spot syndrome virus (WSSV) in cultured and wild crustaceans in India." Diseases of aquatic organisms 38(1).
-
Palanikumar, P; 243-252; , D. J. D. Benitta, C. Lelin, E. Thirumalaikumar, M. Michaelbabu and T. Citarasu (2018). "Effect of Argemone mexicana active principles on inhibiting viral multiplication and stimulating immune system in Pacific white leg shrimp Litopenaeus vannamei against white spot syndrome virus." Fish & shellfish immunology 75.
-
Panchal, V; 1168-1175; , S. Kumar, S. N. Hossain and D. Vasudevan (2021). "Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements." International Journal of Biological Macromolecules 167.
-
Park, J; 71-75; H., Y. S. Lee, S. Lee and Y. Lee (1998). "An infectious viral disease of penaeid shrimp newly found in Korea." Diseases of Aquatic Organisms 34(1).
-
Patil, P; 736231; K., R. Geetha, T. Ravisankar, S. Avunje, H. G. Solanki, T. J. Abraham, S. P. Vinoth, K. P. Jithendran, S. V. Alavandi and K. K. Vijayan (2021). "Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV)." Aquaculture 533.
-
Pavelka, M; 92-93; and J. Roth (2010). Receptor-Mediated Endocytosis Via Clathrin-Coated Vesicles and Virus Endocytosis. Functional Ultrastructure: Atlas of Tissue Biology and Pathology. Vienna, Springer Vienna.
-
Pazir, M; 708-726; K., M. Afsharnasab, B. Jalali Jafari, I. Sharifpour, A. A. Motalebi and A. Dashtiannasab (2011). "Detection and identification of white spot syndrome virus (WSSV) and infectious hypodermal and hematopoietic necrosis virus (IHHNV) of Litopenaus vannamei from Bushehr and Sistan and Baloochestan provinces (Iran), during 2009-2010." Iranian Journal of Fisheries Sciences 10(4).
-
Peña Navarro, N; 479-489; , R. Castro Vásquez and G. Dolz (2020). "White spot syndrome virus and Enterocytozoon hepatopenaei in shrimp farms in Costa Rica: WSSV and EPH in white shrimp farms in Costa Rica." Agronomy Mesoamerican 31(2).
-
Peng, S; 253-262; , C. Lo, C. Ho, C. Chang and G. Kou (1998). "Detection of white spot baculovirus (WSBV) in giant freshwater prawn, Macrobrachium rosenbergii, using polymerase chain reaction." Aquaculture 164(1-4).
- Pereira Dantas Da Rocha Lima, J. J. Pereira Dantas Da Rocha Lima, J. J. (2013). Development of techniques to culture shrimp haemocytes and purify white spot syndrome virus (WSSV) in order to study WSSV-haemocyte interactions, Ghent University.
-
Phanse, Y; 1428; , S. Puttamreddy, D. Loy, J. V. Ramirez, K. A. Ross, I. Alvarez-Castro, M. Mogler, S. Broderick, K. Rajan and B. Narasimhan (2022). "RNA nanovaccine protects against white spot syndrome virus in shrimp." Vaccines 10(9).
-
Pradeep, B; 161-174; , P. Rai, S. A. Mohan, M. S. Shekhar and I. Karunasagar (2012). "Biology, Host Range, Pathogenesis and Diagnosis of White spot syndrome virus." Indian J Virol 23(2).
-
Pratapa, M; 108016; , S. Kumar, M. Bedekar, H. S. Kumar and K. Rajendran (2023). "Pathogenicity of white spot syndrome virus (WSSV) after multiple passages in mud crab, Scylla olivacea." Journal of Invertebrate Pathology 201.
-
Pudgerd, A; 26306; , S. Saedan, W. Santimanawong, W. Weerachatyanukul, P. Jariyapong, T. Chaijarasphong, K. Jongsomchai, K. Sritunyalucksana, R. Vanichviriyakit and C. Chotwiwatthanakun (2024). "Genome editing of WSSV CRISPR/Cas9 and immune activation extends the survival of infected Penaeus vannamei." Scientific Reports 14(1).
-
Quang, N; 69-72; D., P. T. P. Hoa, T. T. Da and P. H. Anh (2009). "Persistence of white spot syndrome virus in shrimp ponds and surrounding areas after an outbreak." Environmental Monitoring and Assessment 156(1).
-
Rajashekar Reddy, C; 2677-2681; B., S. Dinesh, N. Anusha, T. Itami, S. Rajasekhara Reddy and R. Sudhakaran (2016). "Antiviral activity of 3-(1-chloropiperidin-4-yl)-6-fluoro benzisoxazole 2 against White spot syndrome virus in Freshwater crab, Paratelphusa hydrodomous." Aquaculture Research(8).
-
Rajendran, K; 183-191; , K. Vijayan, T. Santiago and R. Krol (1999). "Experimental host range and histopathology of white spot syndrome virus (WSSV) infection in shrimp, prawns, crabs and lobsters from India." Journal of fish diseases 22(3).
-
Rajendran, K; 341-349; V., K. K. Vijayan, T. C. Santiago and J. J. S. Rajan (2005). "White spot syndrome virus (WSSV) infection in tiger shrimp Penaeus monodon: A non-lethal histopathological rapid diagnostic method using paraffin and frozen sections." Aquaculture International 13(4).
-
Ramos-Carreño, S; 1360-1373; , I. Giffard-Mena, J. N. Zamudio-Ocadiz, A. Nuñez-Rivera, R. Valencia-Yañez, J. Ruiz-Garcia, M. T. Viana and R. D. Cadena-Nava (2021). "Antiviral therapy in shrimp through plant virus VLP containing VP28 dsRNA against WSSV." Beilstein Journal of Organic Chemistry 17(1).
-
Rattanarojpong, T; 44-52; , S. Khankaew, P. Khunrae, R. Vanichviriyakit and K. Poomputsa (2016). "Recombinant baculovirus mediates dsRNA specific to rr2 delivery and its protective efficacy against WSSV infection." Journal of Biotechnology 229.
-
Rodríguez, J; 439-450; , B. Bayot, Y. Amano, F. Panchana, I. De Blas, V. Alday and J. Calderón (2003). "White spot syndrome virus infection in cultured Penaeus vannamei (Boone) in Ecuador with emphasis on histopathology and ultrastructure." Journal of Fish Diseases 26(8).
-
Rodríguez, J; 405-415; , Y. Espinosa, F. Echeverría, G. Cárdenas, R. Román and S. Stern (2007). "Exposure to probiotics and β-1, 3/1, 6-glucans in larviculture modifies the immune response of Penaeus vannamei juveniles and both the survival to White Spot Syndrome Virus challenge and pond culture." Aquaculture 273(4).
- Rosenberry, B. Rosenberry, B. (2002). World shrimp farming 2002, Shrimps news international.
-
Rudtanatip, T; 1126-1134; , S. Asuvapongpatana, B. Withyachumnarnkul and K. Wongprasert (2014). "Sulfated galactans isolated from the red seaweed Gracilaria fisheri target the envelope proteins of white spot syndrome virus and protect against viral infection in shrimp haemocytes." Journal of General Virology 95(5).
-
Ruiz-Guerrero, E; 15-28; A., I. Giffard-Mena, M. T. Viana, S. Ramos-Carreño and S. Sánchez-Serrano (2023). "Use of brome mosaic virus-like particles in feed, to deliver dsRNA targeting the white spot syndrome virus vp28 gene, reduces Penaeus vannamei mortality." Diseases of Aquatic Organisms 156.
-
Runsaeng, P; 200-213; , P. Kwankaew and P. Utarabhand (2018). "FmLC6: An ultimate dual-CRD C-type lectin from Fenneropenaeus merguiensis mediated its roles in shrimp defense immunity towards bacteria and virus." Fish & shellfish immunology 80.
-
Sabin, L; 27-33; R. and S. Cherry (2013). "Small creatures use small RNAs to direct antiviral defenses." European Journal of Immunology 43(1).
-
Sahul Hameed, A; 31-45; S., M. Anilkumar, M. L. Stephen Raj and K. Jayaraman (1998). "Studies on the pathogenicity of systemic ectodermal and mesodermal baculovirus and its detection in shrimp by immunological methods." Aquaculture 160(1).
-
Sahul Hameed, A; 134-140; S. and J. R. Bonami (2012). "White Tail Disease of Freshwater Prawn, Macrobrachium rosenbergii." Indian J Virol 23(2).
-
Sakowski, E; 15786-15791; G., E. V. Munsell, M. Hyatt, W. Kress, S. J. Williamson, D. J. Nasko, S. W. Polson and K. E. Wommack (2014). "Ribonucleotide reductases reveal novel viral diversity and predict biological and ecological features of unknown marine viruses." Proceedings of the National Academy of Sciences 111(44).
-
Salehpour, R; 84-95; , N. A. Biuki, M. Mohammadi, A. Dashtiannasab and P. Ebrahimnejad (2021). "The dietary effect of fucoidan extracted from brown seaweed, Cystoseira trinodis (C. Agardh) on growth and disease resistance to WSSV in shrimp Litopenaeus vannamei." Fish & Shellfish Immunology 119.
-
Sánchez-Martínez, J; 1339-1354; G., G. Aguirre-Guzmán and H. Mejía-Ruíz (2007). "White Spot Syndrome Virus in cultured shrimp: A review." Aquaculture Research 38(13).
-
Sánchez-Paz, A; an overview on an emergent concern; (2010). "White spot syndrome virus.
-
Sanjuktha, M; 993-998; , V. Stalin Raj, K. Aravindan, S. Alavandi, M. Poornima and T. Santiago (2012). "Comparative efficacy of double-stranded RNAs targeting WSSV structural and nonstructural genes in controlling viral multiplication in Penaeus monodon." Archives of virology 157.
-
Sass, C; //www; (2022). "Health Benefits of Shrimp." from https.
-
Sekar, A; 100663; , M. Kim, H. Jeon and K. Kim (2019). "Screening and selection of bacteria inhibiting white spot syndrome virus infection to Litopenaeus vannamei." Biochemistry and biophysics reports 19.
- Shankar, K. Shankar, K. and C. Mohan (1998). "Epidemiological aspects of shrimp viral diseases in India--a review." Journal of Aquaculture in the Tropics.
-
Shu, L; 427-434; , C. Li and X. Zhang (2016). "The role of shrimp miR-965 in virus infection." Fish & Shellfish Immunology 54.
-
Sinurat, E; 082-091; , E. Saepudin and S. Hudiyono (2016). "Immunostimulatory activity of brown seaweed-derived fucoidans at different molecular weights and purity levels towards white spot syndrome virus (WSSV) in shrimp Litopenaeus vannamei." Journal of Applied Pharmaceutical Science 6(10).
-
Srisuk, C; 552-562; , S. Longyant, S. Senapin, P. Sithigorngul and P. Chaivisuthangkura (2014). "Molecular cloning and characterization of a Toll receptor gene from Macrobrachium rosenbergii." Fish & Shellfish Immunology 36(2).
-
Stentiford, G; 302-306; D. and D. V. Lightner (2011). "Cases of White Spot Disease (WSD) in European shrimp farms." Aquaculture 319(1).
-
Stern, S; 207-232; and S. Sonnenholzner (2010). Semi-Intensive Shrimp Culture, the History of Shrimp Farming In Ecuador. The shrimp book, CABI GB.
-
Sun, B; 487-496; , Z. Wang and F. Zhu (2017). "The crustin-like peptide plays opposite role in shrimp immune response to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection." Fish & Shellfish Immunology 66.
-
Sun, Z; 1503-1513; C., C. Chen, F. F. Xu, B. K. Li, J. L. Shen, T. Wang, H. F. Jiang and G. X. Wang (2021). "Evaluation of the antiviral activity of naringenin, a major constituent of Typha angustifolia, against white spot syndrome virus in crayfish Procambarus clarkii." Journal of Fish Diseases 44(10).
-
Sunarto, A; Occurrence; , Widodo, Taukhid, I. Koesharyani, H. Supriyadi, L. Gardenia, B. Sugianti and D. Rukmono (2004). Current status of transboundary fish diseases in Indonesia.
-
Sweet, M; 28-44; J. and K. S. Bateman (2016). "Reprint of ‘Diseases in marine invertebrates associated with mariculture and commercial fisheries’." Journal of Sea Research 113.
-
Taengchaiyaphum, S; 56-64; , H. Nakayama, J. Srisala, R. Khiev, D. J. Aldama-Cano, S. Thitamadee and K. Sritunyalucksana (2017). "Vaccination with multimeric recombinant VP28 induces high protection against white spot syndrome virus in shrimp." Developmental & Comparative Immunology 76.
-
Talukder, A; 107535; S., N. J. Punom, M. M. E. Eshik, M. K. Begum, H. M. R. Islam, Z. Hossain and M. S. Rahman (2021). "Molecular identification of white spot syndrome virus (WSSV) and associated risk factors for white spot disease (WSD) prevalence in shrimp (Penaeus monodon) aquaculture in Bangladesh." Journal of Invertebrate Pathology 179.
-
Tan, K; 3041; , H. Zhang, L.-S. Lim, H. Ma, S. Li and H. Zheng (2020). "Roles of carotenoids in invertebrate immunology." Frontiers in Immunology 10.
-
Tang, K; 179-185; F. J., S. A. Navarro, C. R. Pantoja, F. L. Aranguren and D. V. Lightner (2012). "New genotypes of white spot syndrome virus (WSSV) and Taura syndrome virus (TSV) from the Kingdom of Saudi Arabia." Diseases of Aquatic Organisms 99(3).
-
Tang, K; 82-85; F. J., C. R. Pantoja, R. M. Redman and D. V. Lightner (2013). "A histological variant of white spot syndrome virus (WSSV) from the Kingdom of Saudi Arabia." Journal of Invertebrate Pathology 113(1).
-
Tang, X; 907-915; , C. Cui, Q. Liang, X. Sheng, J. Xing and W. Zhan (2019). "Apoptosis of hemocytes is associated with the infection process of white spot syndrome virus in Litopenaeus vannamei." Fish & Shellfish Immunology 94.
-
Tariq, S; 103580; , S. Wani, W. Rasool, K. Shafi, M. A. Bhat, A. Prabhakar, A. H. Shalla and M. A. Rather (2019). "A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens." Microbial pathogenesis 134.
-
Thamizhvanan, S; 573-584; , A. Nafeez Ahmed, D. Vinoth Kumar, S. Vimal, S. A. Majeed, G. Taju, C. Hauton and A. Sahul Hameed (2021). "Silencing of prophenoloxidase (proPO) gene in freshwater prawn, Macrobrachium rosenbergii, makes them susceptible to white spot syndrome virus (WSSV)." Journal of Fish Diseases 44(5).
-
Tidbury, H; 105200; , D. Ryder, M. Thrush, F. Pearce, E. Peeler and N. Taylor (2020). "Comparative assessment of live cyprinid and salmonid movement networks in England and Wales." Preventive veterinary medicine 185.
-
Tomazelli Júnior, O; 1459-1468; , F. Kuhn, P. Mendonça Padilha, C. Nunes Nesi, M. Mestres, J. Dal Magro and S. De Lamo Castellví (2018). "Effect of microencapsulated thyme essential oil on white spot virus-infected Litopenaeus vannamei." Aquaculture international 26.
-
Tsai, J; 11360-11370; M., H. C. Wang, J. H. Leu, H. H. Hsiao, A. H. Wang, G. H. Kou and C. F. Lo (2004). "Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus." J Virol 78(20).
-
Tsai, J; 3021-3029; M., H. C. Wang, J. H. Leu, A. H. Wang, Y. Zhuang, P. J. Walker, G. H. Kou and C. F. Lo (2006). "Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion." J Virol 80(6).
-
Tuyen, N; 286-294; , J. Verreth, J. Vlak and M. De Jong (2014). "Horizontal transmission dynamics of White spot syndrome virus by cohabitation trials in juvenile Penaeus monodon and P. vannamei." Preventive Veterinary Medicine 117(1).
-
Valdez, A; 347-357; , G. Yepiz-Plascencia, E. Ricca and J. Olmos (2014). "First Litopenaeus vannamei WSSV 100% oral vaccination protection using CotC::Vp26 fusion protein displayed on Bacillus subtilis spores surface." Journal of Applied Microbiology 117(2).
-
van Hulten, M; 227-236; C. W., M. Westenberg, S. D. Goodall and J. M. Vlak (2000). "Identification of Two Major Virion Protein Genes of White Spot Syndrome Virus of Shrimp." Virology 266(2).
-
van Hulten, M; 7-22; C. W., J. Witteveldt, S. Peters, N. Kloosterboer, R. Tarchini, M. Fiers, H. Sandbrink, R. K. Lankhorst and J. M. Vlak (2001). "The White Spot Syndrome Virus DNA Genome Sequence." Virology 286(1).
-
Van Thuong, K; 39; , V. Van Tuan, W. Li, P. Sorgeloos, P. Bossier and H. Nauwynck (2016). "Per os infectivity of white spot syndrome virus (WSSV) in white-legged shrimp (Litopenaeus vannamei) and role of peritrophic membrane." Veterinary Research 47(1).
-
Verbruggen, B; 23; , L. K. Bickley, R. Van Aerle, K. S. Bateman, G. D. Stentiford, E. M. Santos and C. R. Tyler (2016). "Molecular Mechanisms of White Spot Syndrome Virus Infection and Perspectives on Treatments." Viruses 8(1).
-
Verma, A; 141-146; K., S. Gupta, S. P. Singh and N. S. Nagpure (2017). "An update on mechanism of entry of white spot syndrome virus into shrimps." Fish Shellfish Immunol 67.
-
Vijayan, K; 108058; , P. S. Anand, C. Balasubramanian, J. S. Rajan, P. E. Praveena, R. Aravind, N. Sudheer, B. Francis, A. Panigrahi and S. Otta (2024). "Vertical transmission and prevalence of white spot syndrome virus (WSSV) in the wild spawning population of the Indian white shrimp, Penaeus indicus." Journal of Invertebrate Pathology 203.
-
Vijayan, K; 97-106; K., C. P. Balasubramanian, K. P. Jithendran, S. V. Alavandi and T. C. Santiago (2003). "Histopathology of Y-organ in Indian white shrimp Fenneropenaeus indicus, experimentally infected with white spot syndrome virus." Aquaculture 221(1).
-
Vijayaram, S; 294-308; , Y.-Z. Sun, A. Zuorro, H. Ghafarifarsani, H. Van Doan and S. H. Hoseinifar (2022). "Bioactive immunostimulants as health-promoting feed additives in aquaculture: A review." Fish & Shellfish Immunology 130.
-
Villanueva, R; 171-244; A., Y. Rouillé and J. Dubuisson (2005). "Interactions between virus proteins and host cell membranes during the viral life cycle." Int Rev Cytol 245.
-
Walker, P; 125-154; J. and C. V. Mohan (2009). "Viral disease emergence in shrimp aquaculture: origins, impact and the effectiveness of health management strategies." Rev Aquac 1(2).
-
Wang, B; 15-29; , F. Li, J. Xiang, L. Gui, Z. Luo and H. Yan (2010). "Three tetraspanins from Chinese shrimp, Fenneropenaeus chinensis, may play important roles in WSSV infection." Journal of Fish Diseases 33(1).
-
Wang, F; 2160; , S. Li and F. Li (2021). "Different Immune Responses of the Lymphoid Organ in Shrimp at Early Challenge Stage of Vibrio parahaemolyticus and WSSV." Animals 11(8).
-
Wang, F; 1-14; , S. Li, J. Xiang and F. Li (2019). "Transcriptome analysis reveals the activation of neuroendocrine-immune system in shrimp hemocytes at the early stage of WSSV infection." BMC genomics 20.
-
Wang, H; 1053-1054; -C., I. Hirono, M. B. B. Maningas, K. Somboonwiwat, G. Stentiford and I. R. Consortium (2019). "ICTV Virus Taxonomy Profile: Nimaviridae." Journal of General Virology 100(7).
-
Wang, K; 49-58; H.-C., C.-W. Tseng, H.-Y. Lin, I.-T. Chen, Y.-H. Chen, Y.-M. Chen, T.-Y. Chen and H.-L. Yang (2010). "RNAi knock-down of the Litopenaeus vannamei Toll gene (LvToll) significantly increases mortality and reduces bacterial clearance after challenge with Vibrio harveyi." Developmental & Comparative Immunology 34(1).
-
Wang, P; 1897-1904; -H., Z.-H. Gu, X.-D. Huang, B.-D. Liu, X.-x. Deng, H.-S. Ai, J. Wang, Z.-X. Yin, S.-P. Weng, X.-Q. Yu and J.-G. He (2009). "An immune deficiency homolog from the white shrimp, Litopenaeus vannamei, activates antimicrobial peptide genes." Molecular Immunology 46(8).
-
Wang, P; 359-371; -H., J.-P. Liang, Z.-H. Gu, D.-H. Wan, S.-P. Weng, X.-Q. Yu and J.-G. He (2012). "Molecular cloning, characterization and expression analysis of two novel Tolls (LvToll2 and LvToll3) and three putative Spätzle-like Toll ligands (LvSpz1–3) from Litopenaeus vannamei." Developmental & Comparative Immunology 36(2).
-
Wang, S; 278-288; , H. Li, K. Lǚ, Z. Qian, S. Weng, J. He and C. Li (2016). "Identification and characterization of transforming growth factor β-activated kinase 1 from Litopenaeus vannamei involved in anti-bacterial host defense." Fish & shellfish immunology 52.
- Wang, S., H. Wang, S., H. Li, S. Weng, C. Li and J. He (2020). "White Spot Syndrome Virus Establishes a Novel IE1/JNK/c-Jun Positive Feedback Loop to Drive Replication." iScience 23(1).
-
Wang, W; 1-23; , J. Sun, C. Liu and Z. Xue (2017). "Application of immunostimulants in aquaculture: current knowledge and future perspectives." Aquaculture Research 48(1).
-
Wang, W; 522-527; and X. Zhang (2008). "Comparison of antiviral efficiency of immune responses in shrimp." Fish & Shellfish Immunology 25(5).
-
Wang, X; 103708; -W., G. R. Vasta and J.-X. Wang (2020). "The functional relevance of shrimp C-type lectins in host-pathogen interactions." Developmental & Comparative Immunology 109.
-
Wang, X; 27-38; -W. and J.-X. Wang (2013). "Diversity and multiple functions of lectins in shrimp immunity." Developmental & Comparative Immunology 39(1).
-
Wang, X; 556-562; W., W. T. Xu, X. W. Zhang, X. F. Zhao, X. Q. Yu and J. X. Wang (2009). "A C-type lectin is involved in the innate immune response of Chinese white shrimp." Fish Shellfish Immunol 27(4).
-
Wang, Y; 221-231; -C., C.-F. Lo, P.-S. Chang and G.-H. Kou (1998). "Experimental infection of white spot baculovirus in some cultured and wild decapods in Taiwan." Aquaculture 164(1-4).
-
Wang, Y; 1-11; G., M. D. Hassan, M. Shariff, S. M. Zamri and X. Chen (1999). "Histopathology and cytopathology of white spot syndrome virus (WSSV) in cultured Penaeus monodon from peninsular Malaysia with emphasis on pathogenesis and the mechanism of white spot formation." Diseases of Aquatic Organisms 39(1).
-
Wang, Z; 219-229; , Y.-H. Chen, Y.-J. Dai, J.-M. Tan, Y. Huang, J.-F. Lan and Q. Ren (2015). "A novel vertebrates Toll-like receptor counterpart regulating the anti-microbial peptides expression in the freshwater crayfish, Procambarus clarkii." Fish & shellfish immunology 43(1).
-
Weerachatyanukul, W; 53-61; , C. Pooljun, I. Hirono, H. Kondo, C. Chotwiwatthanakun and P. Jariyapong (2022). "Infectious hypodermal and hematopoietic necrosis virus-like particle (IHHNV-VLP) induces peroxiredoxin expression and activity in Fenneropenaeus merguiensis." Fish & Shellfish Immunology 121.
-
Wei, C; 145115; , L. Pan, X. Zhang and R. Tong (2020). "Comparative transcriptome analysis of eyestalk from the white shrimp Litopenaeus vannamei after the injection of dopamine." Gene 763.
-
Wikumpriya, G; 1682; C., M. W. S. Prabhatha, J. Lee and C.-H. Kim (2023). "Epigenetic modulations for prevention of infectious diseases in shrimp aquaculture." Genes 14(9).
-
Wongteerasupaya, C; 69-77; , J. Vickers, S. Sriurairatana, G. Nash, A. Akarajamorn, V. Boonsaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnkul and T. Flegel (1995). "A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon." Diseases of aquatic organisms 21(1).
-
Xiao, B; 390-412; , Q. Fu, S. Niu, P. Zhu, J. He and C. Li (2020). "Penaeidins restrict white spot syndrome virus infection by antagonizing the envelope proteins to block viral entry." Emerg Microbes Infect 9(1).
-
Xiao, Y; 9012; , H. Wang, C. Wang, H. Gao, Y. Wang and J. Xu (2023). "Trends in and Future Research Direction of Antimicrobial Resistance in Global Aquaculture Systems: A Review." Sustainability 15(11).
-
Xie, Q; 82-89; , Y. Liu, F. Luo, Q. Yi, Y. Wang, L. Deng, J. Dai and T. Feng (2019). "Antiviral activity of cathelicidin 5, a peptide from Alligator sinensis, against WSSV in caridean shrimp Exopalaemon modestus." Fish & Shellfish Immunology 93.
-
Xie, X; 10615-10623; , L. Xu and F. Yang (2006). "Proteomic Analysis of the Major Envelope and Nucleocapsid Proteins of White Spot Syndrome Virus." Journal of Virology 80(21).
-
Xu, J; 126-131; , F. Han and X. Zhang (2007). "Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA." Antiviral Res 73(2).
-
Yang, C; 564-574; , J. Zhang, F. Li, H. Ma, Q. Zhang, T. J. Priya, X. Zhang and J. Xiang (2008). "A Toll receptor from Chinese shrimp Fenneropenaeus chinensis is responsive to Vibrio anguillarum infection." Fish & shellfish immunology 24(5).
-
Yang, L; 1999-2008; -S., Z.-X. Yin, J.-X. Liao, X.-D. Huang, C.-J. Guo, S.-P. Weng, S.-M. Chan, X.-Q. Yu and J.-G. He (2007). "A Toll receptor in shrimp." Molecular immunology 44(8).
-
Yang, X; 109579; , B. Wang, K. Jiang, K. Xu, C. Zhong, M. Liu and L. Wang (2024). "The combined analysis of transcriptomics and metabolomics reveals the mechanisms by which dietary quercetin regulates growth and immunity in Penaeus vannamei." Fish & Shellfish Immunology 149.
-
Yao, D; 410-417; , H. Su, J. Zhu, X. Zhao, J. J. Aweya, F. Wang, M. Zhong and Y. Zhang (2018). "SNPs in the Toll1 receptor of Litopenaeus vannamei are associated with immune response." Fish & shellfish immunology 72.
-
Yi, G; 726-734; , Z. Wang, Y. Qi, L. Yao, J. Qian and L. Hu (2004). "Vp28 of shrimp white spot syndrome virus is involved in the attachment and penetration into shrimp cells." BMB Reports 37(6).
-
Yin, B; 739747; , X. Yan, S. Li, D. Liu, J. Liu, J. He and C. Li (2023). "WSSV latency is maintained by a dynamic balance of host immune factors and disturbed by exposure to bacterial infection and low salinity." Aquaculture 575.
- Zafilaza, A., A. Zafilaza, A., A. Andriantsimahavandy and R. H. Randrianarivo (2020). "Use Essential Oil And Power Of Antihelium Tsihanimposa To Eliminate The White Spot Syndrome Virus (WSSV) In Organic Shrimp Culture In Madagascar." European Journal of Medical and Health Sciences 2(4).
-
Zhan, W; 405-410; -B., Y.-H. Wang, J. L. Fryer, K.-K. Yu, H. Fukuda and Q.-X. Meng (1998). "White Spot Syndrome Virus Infection of Cultured Shrimp in China." Journal of Aquatic Animal Health 10(4).
-
Zhang, K; 233-237; , K. Koiwai, H. Kondo and I. Hirono (2018). "White spot syndrome virus (WSSV) suppresses penaeidin expression in Marsupenaeus japonicus hemocytes." Fish & Shellfish Immunology 78.
-
Zhang, X; 2731; , L.-P. Shan, Q. Zhao, L. Liu, X. OuYang, Y. Hu, C.-J. Fei and J. Chen (2022). "Taxifolin inhibits WSSV infection and transmission by increasing the innate immune response in Litopenaeus vannamei." Viruses 14(12).
-
Zhang, Y; 445-450; , J. Wen, Y. Xu, H. Wang, L. Lu, R. Song and J. Zou (2022). "Epigallocatechin-3-gallate inhibits replication of white spot syndrome virus in the freshwater crayfish Procambarus clarkii." Journal of Fish Diseases 45(3).
-
Zhao, Z; 347-356; Y., Z. X. Yin, X. P. Xu, S. P. Weng, X. Y. Rao, Z. X. Dai, Y. W. Luo, G. Yang, Z. S. Li, H. J. Guan, S. D. Li, S. M. Chan, X. Q. Yu and J. G. He (2009). "A novel C-type lectin from the shrimp Litopenaeus vannamei possesses anti-white spot syndrome virus activity." J Virol 83(1).
-
Zhu, F; 311-314; and X. Zhang (2011). "The antiviral vp28-siRNA expressed in bacteria protects shrimp against white spot syndrome virus (WSSV)." Aquaculture 319(3).
Figure 1.
Global distribution of white spot syndrome virus (WSSV) disease. The biology of WSSV provides insight into how efficiently it wreaks havoc as a pathogen. It is a double-stranded DNA virus from the Nimaviridae family, and expresses a large number of proteins that facilitate immune evasion, manipulation of host physiology, and rapid replication–characteristics (Wang, Hirono et al. 2019). The ability of the virus to infect a range of crustaceans including shrimp, crabs and even non-crustacean carriers (Liang, Huang et al. 2005), highlights its adaptability and cosmopolitanism. The structural proteins of WSSV, especially the envelope proteins VP28 and VP26, are critical in the infection process by interacting with receptors on host cells to facilitate entry (Xiao, Fu et al. 2020). Once the virus enters, it exploits the host cell machinery to replicate itself and quickly spread through shrimp tissues, leading to systemic infection (Verbruggen, Bickley et al. 2016). Due to the absence of adaptive immunity in shrimp, this pathogen readily overwhelms the immune physiology of its crustacean host (Walker and Mohan 2009). Infected organisms quickly die as the virus multiplies, and the disease spreads through aquaculture systems. One of the biggest hindrances in controlling outbreaks of WSSV are their complex transmission dynamics. Moreover, the virus can remain viable over a wide range of salinities and temperatures, which makes control more difficult, especially in areas of environmental fluctuations (Millard, Ellis et al. 2021). WSSV causes USD 19 billion of losses annually worldwide across small- and commercial-scale shrimp farms (Millard, Ellis et al. 2021). For countries such as Thailand, India or China, which depend heavily on shrimp trading for their revenues, the economic impacts from crop failures have been severe and in many cases resulted in bankruptcy. WSSV disease outbreaks also caused global shifts in the shrimp supply chain; nations often stopped domestic shrimp export for a period of time and/or increased their imports to maintain supplies to local markets (Karunasagar and Ababouch 2012, Oakey, Smith et al. 2019). WSSV also imposes an environmental cost as some farmers – despite the ineffectiveness of antibiotics against viruses – use large quantities of antibiotics and other chemicals in bids to control outbreaks. This inappropriate use leads to the accumulation of antibiotic residues in the environment and the rise of antimicrobial resistance (Xiao, Wang et al. 2023). This emphasizes the need for rapid development of environmentally sustainable strategies for controlling WSSV in aquaculture. Scientists and industry have tested various approaches over several decades to control WSSV, but a single solution has not yet been found. Conventional efforts like selective breeding of disease-resistant shrimp and better management practices have had limited success (Cock, Gitterle et al. 2009). Nevertheless, additional development of immunostimulants, probiotics and essential oils has shown potential in stimulating natural immunity of the shrimp (Wang, Sun et al. 2017). Some immunostimulants from algae, plants, even fungi also enhance the innate immune response of shrimp, thus reducing viral infection (Vijayaram, Sun et al. 2022). Beneficial bacteria that help overall gut health, probiotics have also entered into assessments for use within shrimp diets as a means to decrease infectivity of WSSV (He, Rahimnejad et al. 2017). These methods are signs of an evolution to more natural and sustainable measures that improve shrimp resistance to diseases, as opposed to application of chemical treatments. Recent advances in some innovative technologies, such as DNA and RNA vaccines, and nanotechnology offer further tools for effective WSSV mitigation (Islam, Mou et al. 2023). DNA vaccines utilize specific genes from the virus to provoke an immune response in the animal; targeting critical viral proteins (e.g., VP28) has been shown to be effective in vitro (Ning, Zhu et al. 2009). Moreover, RNA interference (RNAi) technology has been used to target specific WSSV genes, enabling gene silencing and significantly inhibiting viral replication in the shrimp (Xu, Han et al. 2007). The specificity and controllability of nanotechnology may open new avenues for precise delivery of drugs for targeted therapies against WSSV (Phanse, Puttamreddy et al. 2022), thus improving treatment efficiency and reducing instability under variable environmental conditions. These advancements showcase the promise of biotechnology applications as tools for a sustainable, effective disease management plan in shrimp aquaculture, paving the way to a more robust shrimp supply chain. Considering both the severity of WSSV selective pressure and the multitude of mechanisms involved in its proliferation and circulation, we posit that a multifaceted strategy is required to reach durable control over this pathogen in shrimp aquaculture. Taking a broad perspective from the biology and transmission of WSSV to new control strategies, this review is intended as a one-stop-shop for researchers, farmers and policymakers struggling to keep pace with the diverse aspects of WSSV research. Drawing on material across disciplines, this review highlights the opportunity for concerted action to produce resilient and sustainable options that will provide solutions capable of future-proofing shrimp aquaculture. With the growing aquaculture industry and demand for shrimp there is an urgent need to solve the threat posed by WSSV.
Figure 1.
Global distribution of white spot syndrome virus (WSSV) disease. The biology of WSSV provides insight into how efficiently it wreaks havoc as a pathogen. It is a double-stranded DNA virus from the Nimaviridae family, and expresses a large number of proteins that facilitate immune evasion, manipulation of host physiology, and rapid replication–characteristics (Wang, Hirono et al. 2019). The ability of the virus to infect a range of crustaceans including shrimp, crabs and even non-crustacean carriers (Liang, Huang et al. 2005), highlights its adaptability and cosmopolitanism. The structural proteins of WSSV, especially the envelope proteins VP28 and VP26, are critical in the infection process by interacting with receptors on host cells to facilitate entry (Xiao, Fu et al. 2020). Once the virus enters, it exploits the host cell machinery to replicate itself and quickly spread through shrimp tissues, leading to systemic infection (Verbruggen, Bickley et al. 2016). Due to the absence of adaptive immunity in shrimp, this pathogen readily overwhelms the immune physiology of its crustacean host (Walker and Mohan 2009). Infected organisms quickly die as the virus multiplies, and the disease spreads through aquaculture systems. One of the biggest hindrances in controlling outbreaks of WSSV are their complex transmission dynamics. Moreover, the virus can remain viable over a wide range of salinities and temperatures, which makes control more difficult, especially in areas of environmental fluctuations (Millard, Ellis et al. 2021). WSSV causes USD 19 billion of losses annually worldwide across small- and commercial-scale shrimp farms (Millard, Ellis et al. 2021). For countries such as Thailand, India or China, which depend heavily on shrimp trading for their revenues, the economic impacts from crop failures have been severe and in many cases resulted in bankruptcy. WSSV disease outbreaks also caused global shifts in the shrimp supply chain; nations often stopped domestic shrimp export for a period of time and/or increased their imports to maintain supplies to local markets (Karunasagar and Ababouch 2012, Oakey, Smith et al. 2019). WSSV also imposes an environmental cost as some farmers – despite the ineffectiveness of antibiotics against viruses – use large quantities of antibiotics and other chemicals in bids to control outbreaks. This inappropriate use leads to the accumulation of antibiotic residues in the environment and the rise of antimicrobial resistance (Xiao, Wang et al. 2023). This emphasizes the need for rapid development of environmentally sustainable strategies for controlling WSSV in aquaculture. Scientists and industry have tested various approaches over several decades to control WSSV, but a single solution has not yet been found. Conventional efforts like selective breeding of disease-resistant shrimp and better management practices have had limited success (Cock, Gitterle et al. 2009). Nevertheless, additional development of immunostimulants, probiotics and essential oils has shown potential in stimulating natural immunity of the shrimp (Wang, Sun et al. 2017). Some immunostimulants from algae, plants, even fungi also enhance the innate immune response of shrimp, thus reducing viral infection (Vijayaram, Sun et al. 2022). Beneficial bacteria that help overall gut health, probiotics have also entered into assessments for use within shrimp diets as a means to decrease infectivity of WSSV (He, Rahimnejad et al. 2017). These methods are signs of an evolution to more natural and sustainable measures that improve shrimp resistance to diseases, as opposed to application of chemical treatments. Recent advances in some innovative technologies, such as DNA and RNA vaccines, and nanotechnology offer further tools for effective WSSV mitigation (Islam, Mou et al. 2023). DNA vaccines utilize specific genes from the virus to provoke an immune response in the animal; targeting critical viral proteins (e.g., VP28) has been shown to be effective in vitro (Ning, Zhu et al. 2009). Moreover, RNA interference (RNAi) technology has been used to target specific WSSV genes, enabling gene silencing and significantly inhibiting viral replication in the shrimp (Xu, Han et al. 2007). The specificity and controllability of nanotechnology may open new avenues for precise delivery of drugs for targeted therapies against WSSV (Phanse, Puttamreddy et al. 2022), thus improving treatment efficiency and reducing instability under variable environmental conditions. These advancements showcase the promise of biotechnology applications as tools for a sustainable, effective disease management plan in shrimp aquaculture, paving the way to a more robust shrimp supply chain. Considering both the severity of WSSV selective pressure and the multitude of mechanisms involved in its proliferation and circulation, we posit that a multifaceted strategy is required to reach durable control over this pathogen in shrimp aquaculture. Taking a broad perspective from the biology and transmission of WSSV to new control strategies, this review is intended as a one-stop-shop for researchers, farmers and policymakers struggling to keep pace with the diverse aspects of WSSV research. Drawing on material across disciplines, this review highlights the opportunity for concerted action to produce resilient and sustainable options that will provide solutions capable of future-proofing shrimp aquaculture. With the growing aquaculture industry and demand for shrimp there is an urgent need to solve the threat posed by WSSV.
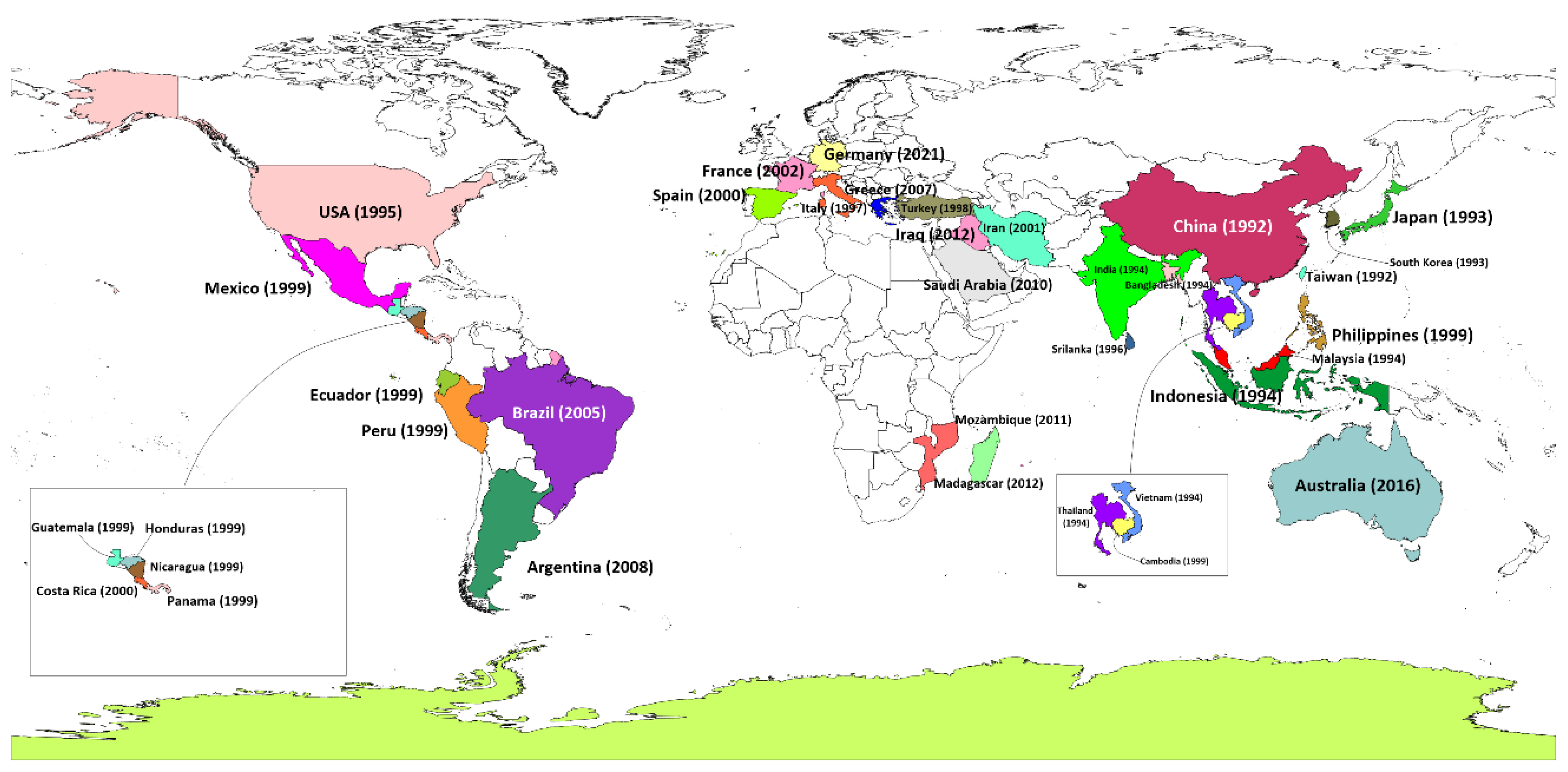
Figure 2.
Geographical genetic diversity of WSSV across major affected countries.
Figure 2.
Geographical genetic diversity of WSSV across major affected countries.
Figure 3.
Transmission pathways and influencing factors for White Spot Syndrome Virus (WSSV) in shrimp aquaculture.
Figure 3.
Transmission pathways and influencing factors for White Spot Syndrome Virus (WSSV) in shrimp aquaculture.
Figure 4.
Life cycle of WSSV.
Figure 4.
Life cycle of WSSV.
Figure 5.
White Spot Syndrome Virus (WSSV) within Penaeus monodon tissues. A) WSSV in gill tissues, enlarged nuclei with marginalised chromatin and containing eosinophilic inclusions (arrows) can be observed distributed throughout the cuticular epithelium of the gill filaments of an infected shrimp. (B) Cuticular epithelial cells of the stomach showing hypertrophied nuclei with eosinophilic staining (arrows). C) WSSV infection within the haematopoietic tissue. Affected cells displaying enlarged nuclei with eosinophilic inclusions (arrows). D) WSSV infected lymphoid organ tissues displaying loss of structure of tubules. Enlarged nuclei with marginalised chromatin can be seen throughout (arrows). Lymphoid organ spherules will develop as tissue structure is lost. All images H&E Stain. Scale bars = 25 µm.
Figure 5.
White Spot Syndrome Virus (WSSV) within Penaeus monodon tissues. A) WSSV in gill tissues, enlarged nuclei with marginalised chromatin and containing eosinophilic inclusions (arrows) can be observed distributed throughout the cuticular epithelium of the gill filaments of an infected shrimp. (B) Cuticular epithelial cells of the stomach showing hypertrophied nuclei with eosinophilic staining (arrows). C) WSSV infection within the haematopoietic tissue. Affected cells displaying enlarged nuclei with eosinophilic inclusions (arrows). D) WSSV infected lymphoid organ tissues displaying loss of structure of tubules. Enlarged nuclei with marginalised chromatin can be seen throughout (arrows). Lymphoid organ spherules will develop as tissue structure is lost. All images H&E Stain. Scale bars = 25 µm.
Figure 6.
Cellular immune response mechanisms in shrimp against White Spot Syndrome Virus (WSSV) infection.
Figure 6.
Cellular immune response mechanisms in shrimp against White Spot Syndrome Virus (WSSV) infection.
Figure 7.
Humoral immune response mechanisms in shrimp against White Spot Syndrome Virus (WSSV) infection.
Figure 7.
Humoral immune response mechanisms in shrimp against White Spot Syndrome Virus (WSSV) infection.
Table 2.
Host species reported to be naturally or experimentally infected with WSSV. (Hameed, Balasubramanian et al. 2003, Escobedo-Bonilla, Alday-Sanz et al. 2008, Pradeep, Rai et al. 2012).
Table 2.
Host species reported to be naturally or experimentally infected with WSSV. (Hameed, Balasubramanian et al. 2003, Escobedo-Bonilla, Alday-Sanz et al. 2008, Pradeep, Rai et al. 2012).
| Scientific name |
Common name |
Type |
Detection method |
| Alpheus brevicristatus |
Snapping shrimp |
N |
Nested PCR |
| Alpheus lobidens |
Apping shrimp |
N |
Nested PCR |
|
Aristeussp.
|
Red shrimp |
N |
Nested PCR |
Exopalaemon
orientalis
|
Oriental prawn |
N, E |
Nested PCR, DNA Probe |
| Penaeus aztecus |
Northern brown shrimp |
N, E |
PCR, Histo |
| Penaeus duorarum |
Pink shrimp |
N, E |
Histo |
| penaeus penicillatus |
Rod Tail Shrimp |
N |
Nested PCR |
| penaeus chinensis |
Chinese white shrimp |
N |
Histo, Electron microscope |
| Penaeus vannamei |
Whiteleg shrimp |
N, E |
TEM, PCR, Histo |
| Penaeus setiferus |
Atlantic white shrimp |
N, E |
PCR, Histo |
Macrobrachium
rosenbergii
|
Giant freshwater
shrimp |
N, E
|
Nested PCR, Southern blot, Histo |
| Macrobrachium idella |
Sunset shrimp |
E |
Southern blot, Histo |
| Marsupenaeus japonicus |
Kuruma shrimp |
N, E |
TEM, PCR, Histo |
| Metapenaeus ensis |
Greasyback shrimp |
N, E |
PCR, DNA Probe |
| Metapenaeus dobsoni |
Kadal shrimp |
N, E |
PCR, DNA Probe |
| Metapenaeus lysianassa |
Bird shrimp |
N |
PCR, Histo |
| Metapenaeus monoceros |
Speckled shrimp |
N, E |
Nested PCR, DNA Probe, Histo |
| Metapenaeus elegans |
Fine shrimp |
N |
Nested PCR |
| Palaemon adspersus |
Baltic prawn |
E |
TEM, Dot Blots, ISH, 1-step PCR |
| Palaemon styliferus |
Grass shrimp |
N |
Nested PCR, DNA Probe, Histo |
| Parapenaeopsis stylifera |
Kiddi shrimp |
N |
Nested PCR, DNA Probe |
| Penaeus monodon |
Giant tiger shrimp |
N, E |
TEM, PCR, Histo |
| Penaeus indicus |
Indian white prawn |
N, E |
TEM, Histo |
| Penaeus merguiensis |
Banana prawn |
N |
ISH, Histo |
| Penaeus semiculcatus |
Green tiger prawn |
N, E |
PCR |
| Penaeus schmitti |
Southern white shrimp |
E |
Histo, ISH |
| Penaeus duorarum |
Northern pink shrimp |
N/E |
Histo |
| Penaeus stylirostris |
Northern white shrimp |
N/E |
TEM, PCR, Histo |
| Solenocera indica |
Coastal mud shrimp |
N |
PCR, DNA Probe |
| Trachypenaeus curvirostris |
Southern rough shrimp |
N, E |
LAMP, 2-step PCR |
| Atergatis integerrimus |
Bashful crab |
E |
PCR, Histo |
| Cancer pagurus |
Edible or rock crab |
E |
TEM, ISH, Histo, 1-step PCR |
| Calappa lophos |
Box crab |
N, E |
PCR |
| Calappa philargius |
Box crab |
E |
PCR, Histo |
| Callinectes arcuatus |
Swimming crab |
N |
PCR |
| Callinectes sapidus |
Blue crab |
N |
ISH, PCR |
| Carcinus maenas |
Littoral crab |
E |
TEM, DNA hybridization, PCR |
| Charybdis annulata |
Swimming crab |
N, E |
Histo, PCR |
| Charybdis cruciata |
Red sea crab |
N |
PCR |
| Charybdis feriata |
Coral crab |
E |
Nested PCR |
| Charybdis granulata |
Swimming crab |
E |
Nested PCR |
| Charybdis hoplites |
Swimming crab |
N |
PCR |
| Charybdis lucifera |
Swimming crab |
N, E |
Histo, PCR |
| Charybdis natator |
Hairyback crab |
N |
Histo, PCR |
| Demania splendida |
|
E |
Histo, PCR |
| Doclea hybrida |
|
E |
Histo, PCR |
| Gelasimus marionis nitidus |
|
N |
PCR |
| Grapsus albolineatus |
Rock crab |
E |
Histo, PCR |
| Halimede ochtodes |
Hairy crab |
E |
Histo, PCR |
| Helice tridens |
Shore crab |
N |
2-step PCR |
| Liocarcinus depurator |
Harbour crab |
E |
TEM, Dot-blots, ISH, PCR |
| Liocarcinus puber |
Velvet swimming
crab |
E |
TEM, Dot-blots, ISH, PCR |
| Lithodes maja |
Deepsea king crab |
E |
Histo, PCR |
| Macrophthalmus sulcatus |
Ghost/fiddler crab |
N |
PCR, DNA Probe |
| Mantura sp. |
|
N |
PCR |
| Matuta miersi |
Moon crab |
E |
Histo, PCR |
| Matuta planipes |
Moon crab |
N |
PCR |
| Metopograpsus messor |
Purple climber crab |
N |
PCR, DNA Probe |
| Menippe rumphii |
Stone crab |
E |
Histo, PCR |
| Paradorippe granulata |
|
E |
Histo, PCR |
| Parthenope prensor |
Elbow crab |
E |
Histo, PCR |
| Parathelphusa hydrodomous |
|
E |
PCR, Histo |
| Parathelphusa pulvinata |
|
E |
PCR, Histo |
| Philyra syndactyla |
Purse crab |
E |
PCR, Histo |
| Podophthalmus vigil |
Long-eyed swimming crab |
E |
PCR, Histo |
| Portunus pelagicus |
Sand crab |
N, E |
PCR |
| Portunus sanguinolentus |
Blood spot crab |
N, E |
PCR, Histo |
| Pseudograpsus intermedius |
Mosaic crab |
N |
Nested PCR, DNA Probe, Histo |
| Scylla serrata |
Mud crab |
N, E |
PCR, Histo |
| Scylla tranquebarica |
Mangrove crab |
N, E |
PCR, TEM |
| Scylla olivacea |
Orange mud crab |
E |
qPCR |
| Sesarma sp. |
Marsh crabs |
N, E |
PCR, Histo |
| Somanniathelphusa sp. |
Black rice crab |
E |
PCR, Histo |
| Thalamita danae |
Swimming crab |
E |
PCR, Histo |
| Acetes sp. |
Krill |
E |
PCR, Histo |
| Panulirus homarus |
Scalloped spiny lobster |
E |
Histo, Bioassay |
| Panulirus longipes |
Longlegged spiny lobster |
E |
Nested PCR |
| Panulirus ornatus |
Ornata spiny lobster |
E |
Histo, Bioassay |
| Panulirus penicillatus |
Pronghorn spiny lobster |
N, E |
PCR |
| Panulirus polyphagus |
Mud spiny lobster |
E |
Histo, Bioassay |
| Panulirus versicolor |
Painted spiny lobster |
E |
Nested PCR |
| Scyllarus arctus |
Small European locust lobster |
E |
TEM, Dot Blots, ISH, PCR |
| Artemia |
|
E |
nested-PCR |
| Artemia franciscana |
|
E |
nested-PCR |
| Schmackeria dubia |
Copepoda |
N |
PCR |
| Squilla mantis |
Mantis shrimp |
N |
Nested PCR, DNA Probe |
| Marphysa gravelyi |
Polychaeta |
N |
2-step PCR |
| Brachionus urceus |
Rotifera |
E |
Nested PCR |
|
Ephydridaesp.
|
Shore fly |
N |
Nested PCR |
| Astacus leptodactylus |
Turkish crayfish |
E |
TEM, Dot Blots, ISH, 1-step PCR |
| Astacus astacus |
Broad-fingered crayfish |
E |
PCR |
| Cherax destructor albidus |
Yabby |
E |
DNA Probe, Histo |
| Cherax quadricarinatus |
Australian redclaw |
E |
TEM, ISH, Nested PCR |
| Orconectes limosus |
Spinycheek crayfish |
E |
TEM, Dot Blots, ISH, 1-step PCR |
| Orconectes punctimanus |
Spothanded Crayfish |
N |
DNA Probe, southern blot, PCR |
| Pacifastacus leniusculus |
Signal crayfish |
E |
Histo, PCR, ISH |
| Procambarus clarkii |
Red swamp crayfish |
E |
Histo, PCR |
Table 3.
Identified WSSV proteins and their gene origins.
Table 3.
Identified WSSV proteins and their gene origins.
| Protein Names |
A.A residues size |
Apparent size (kDa) |
Location in
WSSV virion
|
WSSV-CN ORF |
References |
| VP187 |
1606 |
174 |
Envelope |
wsv209 |
(Tsai, Wang et al. 2004) |
| VP180 |
1684 |
169 |
Envelope |
wsv001 |
| VP150 |
1301 |
144 |
Envelope |
wsv011 |
| VP136B |
1243 |
136 |
Envelope |
wsv465 |
| VP124 |
1219 |
136 |
Envelope |
wsv216 |
| VP110 |
972 |
110 |
Envelope |
wsv035 |
| VP90 |
856 |
96 |
Envelope |
wsv327 |
| VP75 |
786 |
75 |
Envelope |
wsv332 |
| VP56 (VP60A) |
465 |
60 |
Envelope |
wsv325 |
| VP55 |
448 |
55 |
Envelope |
wsv526 |
| VP53A |
1301 |
144 |
Envelope |
wsv011 |
| VP53B |
968 |
53 |
Envelope |
wsv115 |
| VP52A |
486 |
51 |
Envelope |
wsv238 |
| VP38 |
283 |
32 |
Envelope |
wsv259 |
| VP33 (VP36B) |
281 |
32 |
Envelope |
wsv254 |
| VP32 |
278 |
32 |
Envelope |
wsv198 |
| VP28 |
204 |
28 |
Envelope |
wsv421 |
| VP22 |
891 |
100 |
Envelope |
wsv303 |
| VP19 |
121 |
19 |
Envelope |
wsv414 |
| VP13A |
100 |
13 |
Envelope |
wsv284 |
| VP12 (VP12A) |
95 |
11 |
Envelope |
wsv009 |
| VP12B |
68 |
7 |
Envelope |
wsv386 |
| VP11 |
433 |
11 |
Envelope |
wsv338 |
| WSSV189 |
- |
- |
ND* |
wsv134 |
| WSSV471 |
- |
- |
ND* |
wsv412 |
| VP95 |
95 |
11 |
Tegument |
wsv442 |
| VP39A |
419 |
39 |
Tegument |
wsv306 |
| VP36A |
297 |
36 |
Tegument |
wsv077 |
| VP26 |
204 |
26 |
Tegument |
wsv311 |
| VP24 |
208 |
24 |
Tegument |
wsv002 |
| WSSV458 |
- |
- |
ND* |
wsv399 |
| WSSV186 |
- |
- |
ND* |
wsv131 |
| VP160B |
1280 |
143.8 |
Nucleocapsid |
wsv037 |
(Tsai, Wang et al. 2006) |
| VP53C |
489 |
53 |
Envelope |
wsv269 |
(Xie, Xu et al. 2006) |
| VP52B (VP51B) |
384 |
46 |
Envelope |
wsv256 |
| VP41A |
292 |
33 |
Envelope |
wsv237 |
| VP41B |
300 |
34 |
Envelope |
wsv242 |
| VP39 (VP39B) |
419 |
39 |
Envelope |
wsv339 |
| VP38B |
309 |
35 |
Envelope |
wsv390 |
| VP31 |
261 |
31 |
Envelope |
wsv340 |
| VP13 (VP13B) |
117 |
13 |
Envelope |
wsv321 |
| VP14 |
97 |
11 |
Envelope |
wsv293a |
| VP664 |
6077 |
664 |
Nucleocapsid |
wsv360 |
| VP190 |
1565 |
174 |
Nucleocapsid |
wsv289 |
| VP136 |
1218 |
135 |
Nucleocapsid |
wsv271 |
| VP60 (VP60B) |
544 |
62 |
Nucleocapsid |
wsv415 |
| VP51 (VP51C) |
466 |
62 |
Nucleocapsid |
wsv308 |
| VP15 |
80 |
15 |
Nucleocapsid |
wsv214 |
| VP76 (VP73) |
674 |
76 |
Nucleocapsid |
wsv220 |
(Huang, Xie et al. 2005) |
Table 5.
Histopathological degeneration of different organs in WSSV affected shrimp.
Table 5.
Histopathological degeneration of different organs in WSSV affected shrimp.
| Target organ |
Histopathological degeneration |
Reference |
| Epidermis and cuticular epithelium |
Nuclear hypertrophy, intranuclear inclusion bodies, necrosis, epithelial detachment |
(Pazir, Afsharnasab et al. 2011) |
| Gills |
Lamellar sloughing, epithelial fusion, necrosis, presence of viral inclusion bodies |
(Dewangan, Ayyaru et al. 2017) |
| Lymphoid organ and haematopoietic tissues |
Formation of LOS, haemocyte infiltration, apoptotic bodies |
(Wang, Li et al. 2021) |
| Hepatopancreas and digestive system |
Hepatopancreatic tubular degeneration, bacterial co-infection, severe necrosis in midgut epithelium |
(Muegue, Padilla et al. 2023) |
| Y-organ (endocrine gland) |
Destruction of moulting gland cells, hypertrophic changes affecting growth and reproduction |
(Vijayan, Balasubramanian et al. 2003) |
Table 6.
Interactions between homologous and heterologous pathogens that co-infect different shrimp species.
Table 6.
Interactions between homologous and heterologous pathogens that co-infect different shrimp species.
| Host species |
First pathogen |
Second pathogen |
Type of infection |
Reference |
| Homologous co-infection |
| |
Viral co-infections |
| Litopenaeus vannamei |
Infectious hypodermal and haematopoietic necrosis virus (IHHNV) |
White spot syndrome virus (WSSV) |
Synergistic |
Dewangan et al., (2017); Nunan et al., (2001) |
| Penaeus monodon |
IHHNV |
WSSV |
Synergistic |
Flegel et al., (2004); Saravanan et al., (2021) |
| Penaeus monodon |
WSSV |
Monodon baculovirus |
Antagonistic |
Anshary et al., (2017; Flegel et al., (2004); Orosco and Lluisma, (2017) |
| Penaeus monodon |
Monodon baculovirus |
IHHNV |
| Penaeus monodon |
Monodon baculovirus |
Hepatopancreatic parvovirus |
Synergistic |
Anshary et al., (2017); Chayaburakul et al., (2004); Flegel et al., (2004) |
| Penaeus monodon |
WSSV |
Penaeus stylirostris densovirus |
Synergistic |
Orosco and Lluisma, (2017) |
| Litopenaeus vannamei |
Infectious myonecrosis virus |
WSSV |
Synergistic |
Feijó et al., (2013) |
| Penaeus monodon |
Hepatopancreatic parvovirus |
WSSV |
Antagonistic |
Flegel et al., (2004) |
| Penaeus monodon |
Hepatopancreatic parvovirus |
IHHNV |
Antagonistic |
| |
Bacterial co-infections |
| Litopenaeus vannamei |
Vibrio parahaemolyticus |
Vibrio harveyi |
Synergistic |
Muthukrishnan et al., (2019) |
| |
Parasitic co-infections
|
| Penaeus monodon |
Microsporidian |
Gregarine |
Synergistic |
Chayaburakul et al., (2004) |
| Heterologous co-infection |
| |
Parasitic and bacterial co-infections |
| Macrobrachium rosenbergii |
Metschnikowia bicuspidata |
Enterococcus faecium |
Synergistic |
Chen et al., (2003) |
| Penaeus vannamei |
Enterocytozoon hepatopenaei |
Vibrio parahaemolyticus |
Synergistic |
Aranguren et al., (2017) |
| Penaeus vannamei |
Enterocytozoon hepatopenaei |
V. campbellii |
Antagonistic |
| |
Parasitic and viral co-infections |
| Penaeus vannamei |
Enterocytozoon hepatopenaei |
WSSV |
Synergistic |
Thamizhvanan et al., (2019) |
| Penaeus vannamei |
Infectious myonecrosis virus |
Enterocytozoon hepatopenaei |
Synergistic |
Jithendran et al., (2021) |
| Penaeus vannamei |
Enterocytozoon hepatopenaei |
Taura syndrome virus |
Synergistic |
Tang et al., (2017) |
| Penaeus vannamei |
Enterocytozoon hepatopenaei |
Hepatopancreatic parvovirus |
Synergistic |
Singaravel et al., (2021) |
| Penaeus monodon |
Monodon baculovirus |
Microsporidian |
Synergistic |
Chayaburakul et al., (2004) |
| Monodon baculovirus |
Gregarine |
| Hepatopancreatic parvovirus |
Microsporidian |
| |
Bacterial and viral co-infections |
| Penaeus monodon |
Vibrio parahaemolyticus |
WSSV |
Synergistic |
(Babu et al., 2021) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).