Submitted:
01 August 2025
Posted:
04 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Extracellular Dps Detection (Western Blot Analysis)
2.3. RNA Purification and Sequencing
2.4. Pre-Processing of RNA-seq Datasets and Their Mapping to the Genome
2.5. Search for Oligonucleotides with Dps-Dependent Secretion
2.6. Search for Textual Deviations from the Genomic Sequences
2.7. Search for Oligonucleotides Containing Random Sequences at the 3’-End (RandExt Algorithm)
2.8. Datasets Used to Analyze LeuTVPQ Extentions Depending on Culture Media, the Presence of Competing Bacteria and Exogenous Oligonucleotides
2.9. Evaluation of the Effect of LeuTVPQ with Non-Template Nucleotides at the 3'-Ends on the E. coli Growth in Monoculture
2.10. Revival of Rat Fecal Bacteria in GMM and Their Treatment with Synthetic Oligonucleotides
2.11. Amplicon Sequencing of 16S rRNA Genes for Taxonomic Analysis
2.12. Taxonomic Analysis of Ex Vivo Cultured Bacterial Communities
2.13. Statistics
3. Results
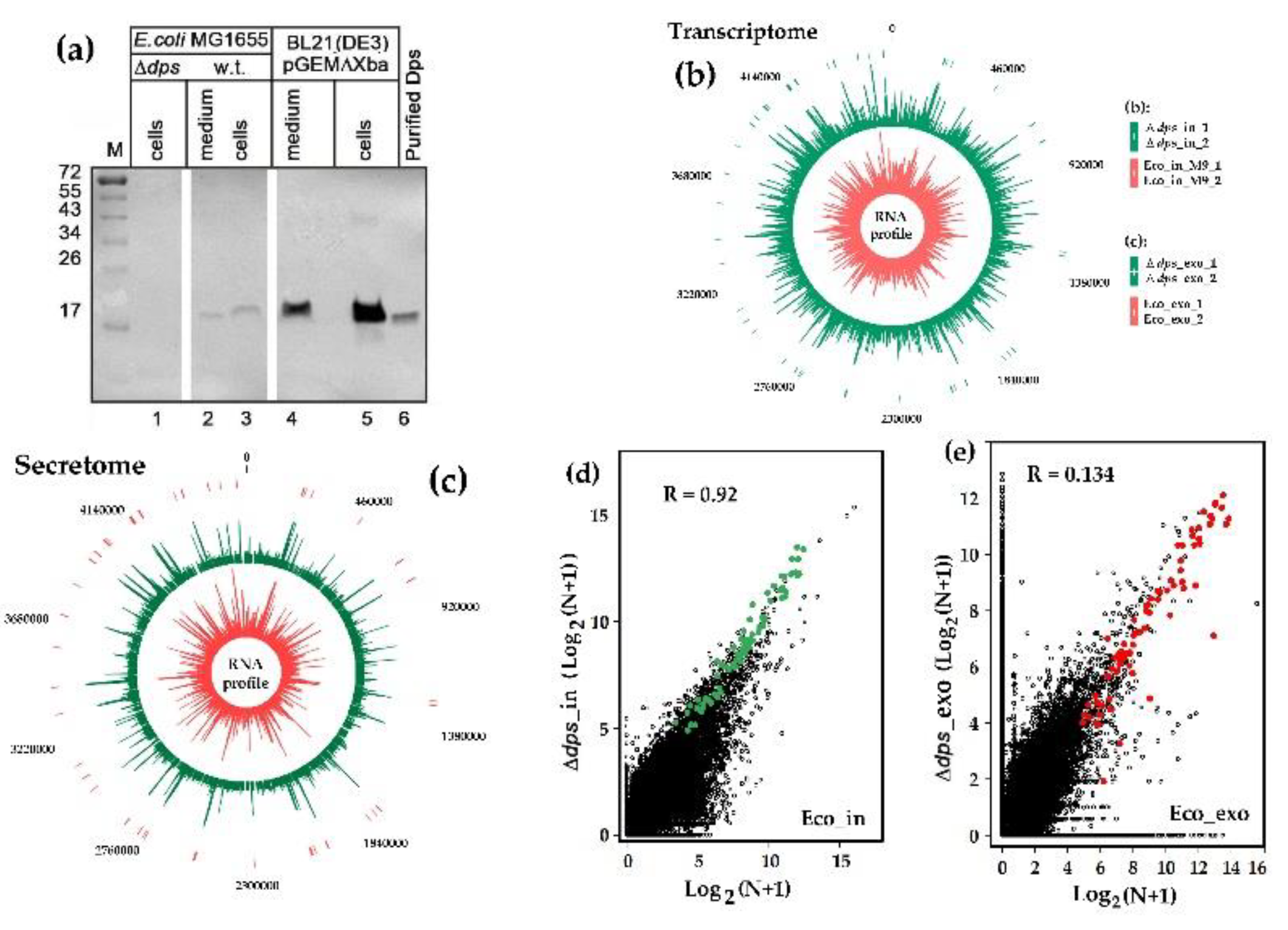
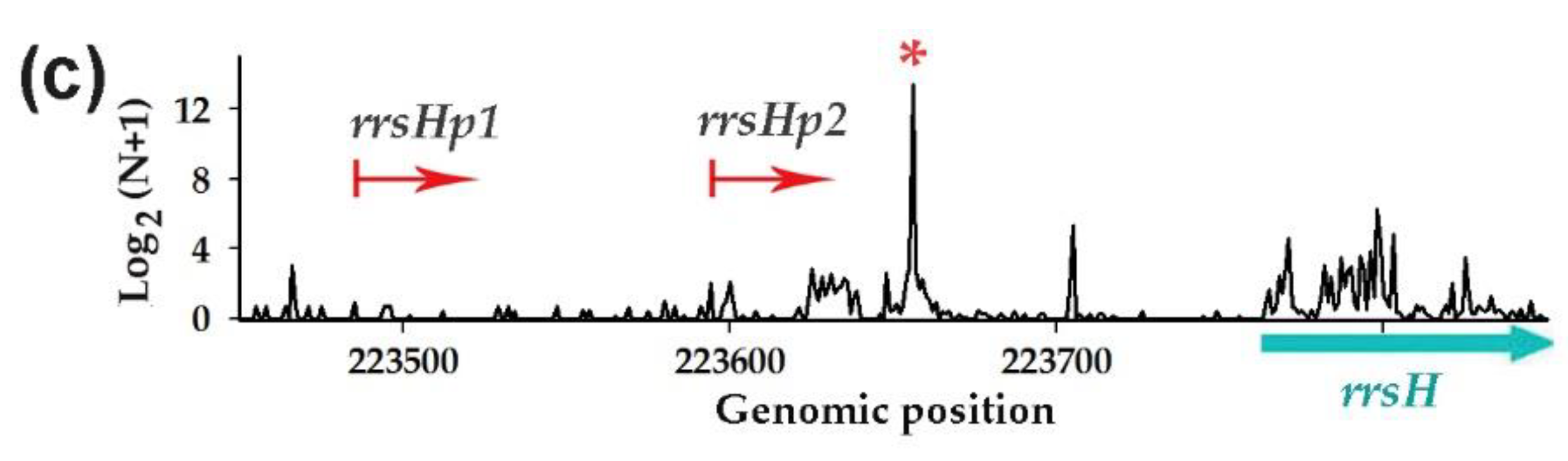
3.1. Dps Protein Found in Culture Milieu May Promote the Release of RNAs from Cells
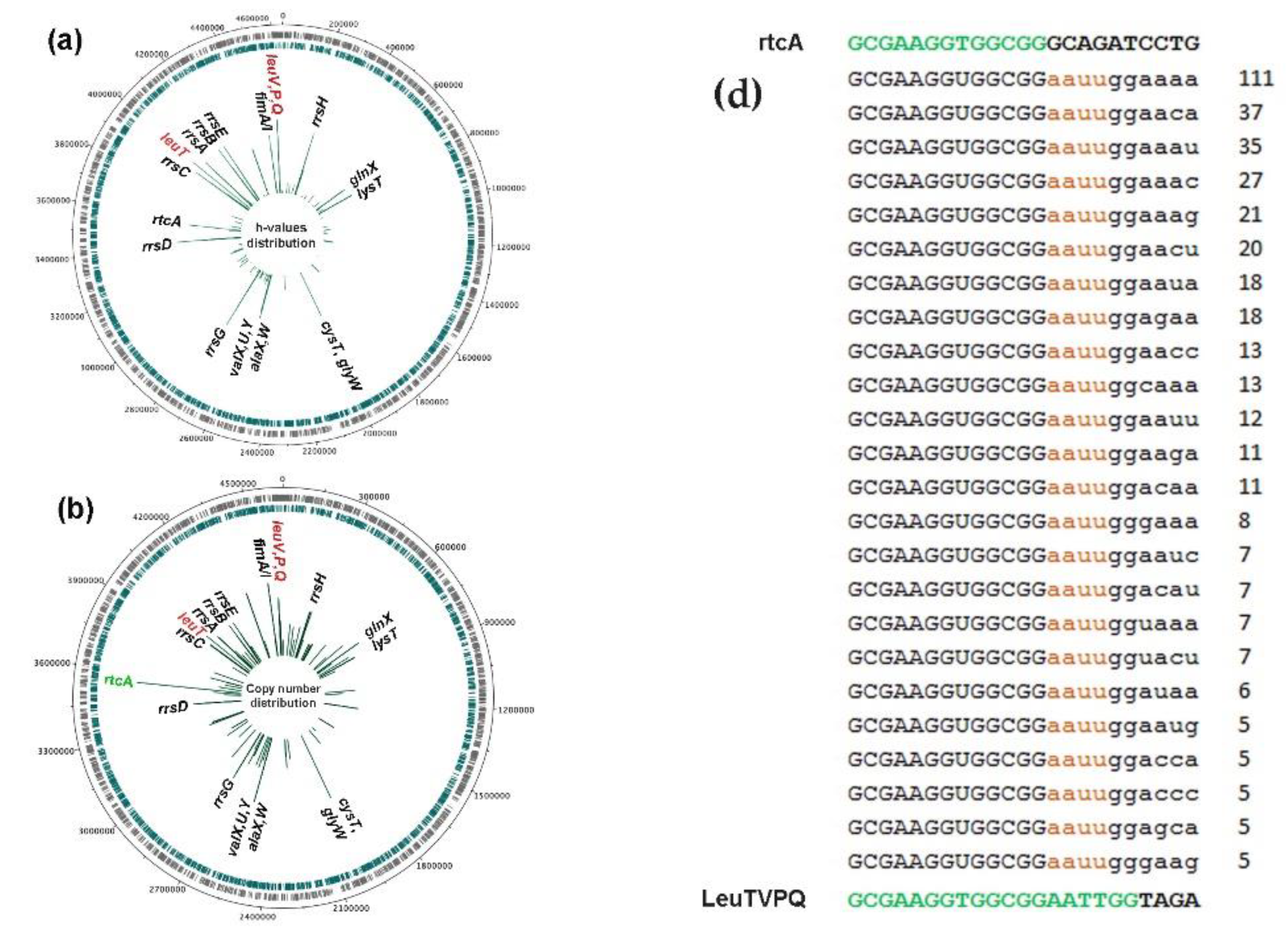
3.2. RNA-seq Analysis Revealed Oligonucleotides with Potential Dps-Dependent Secretion
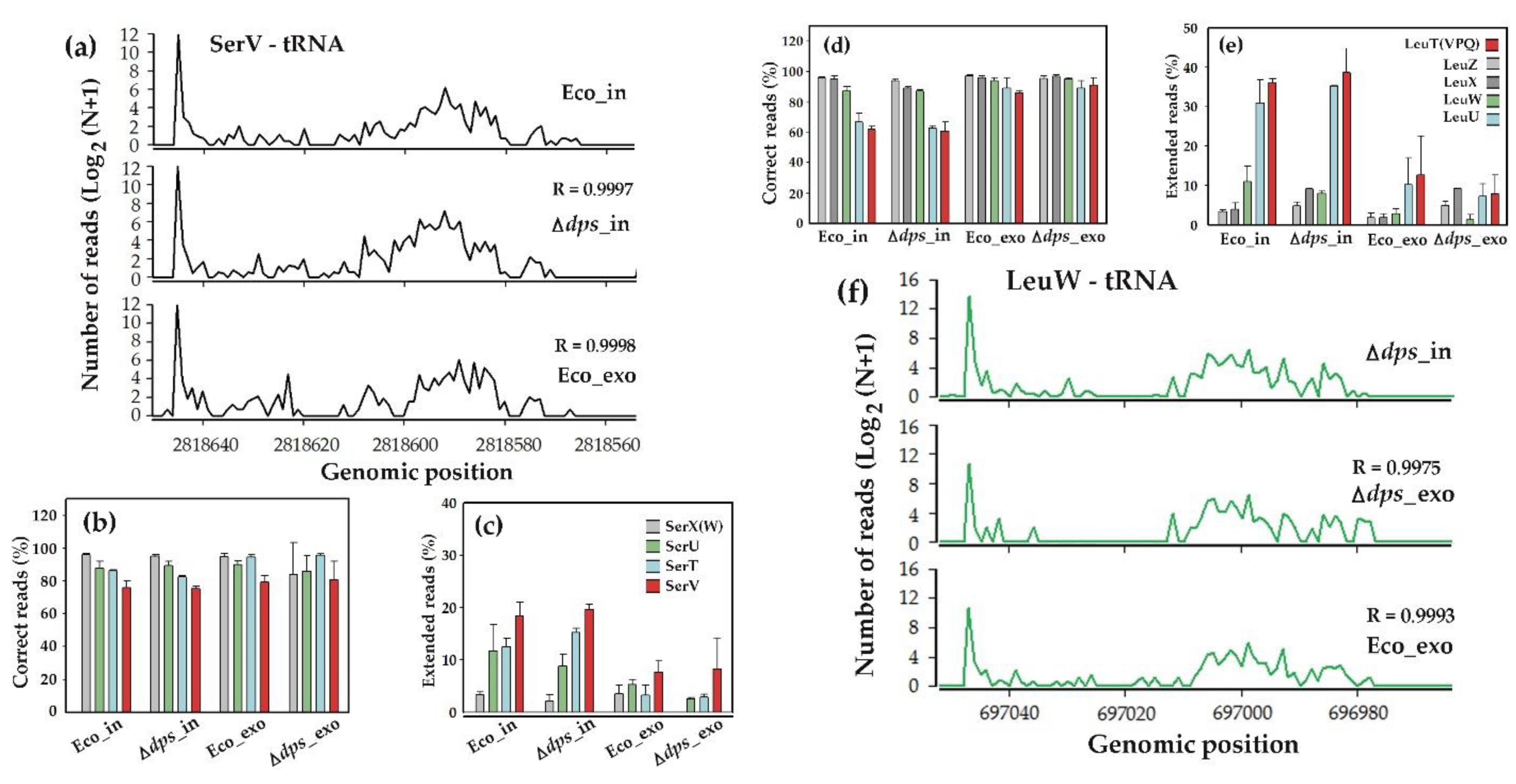
3.3. Fragments of Homologous Leucine and Serine tRNAs Exhibited Different Patterns of 3’-End Posttranscriptional Modifications
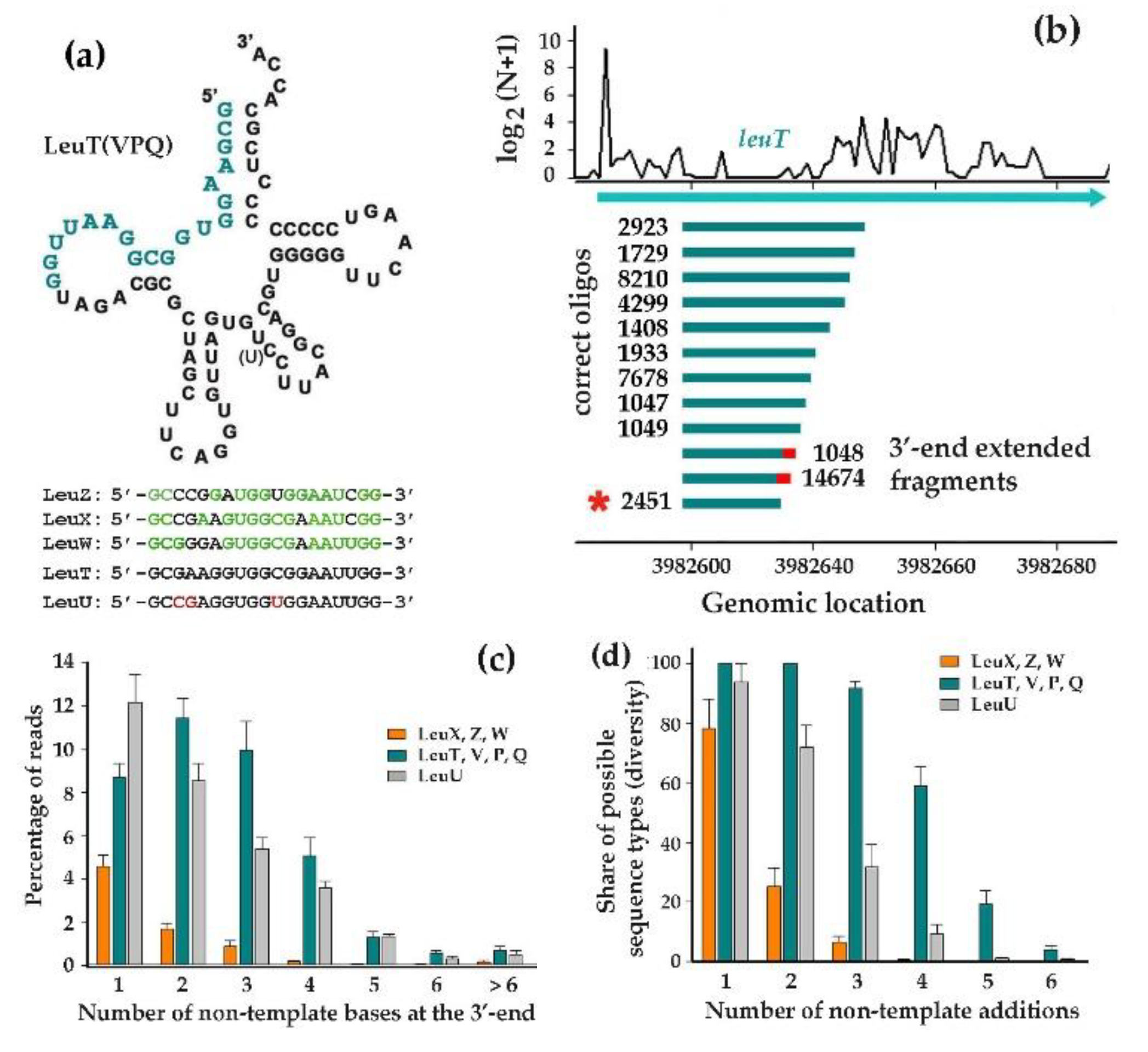
3.4. Fragments of Leucine tRNAs Exhibited Three Different Modes of 3’-End Modifications
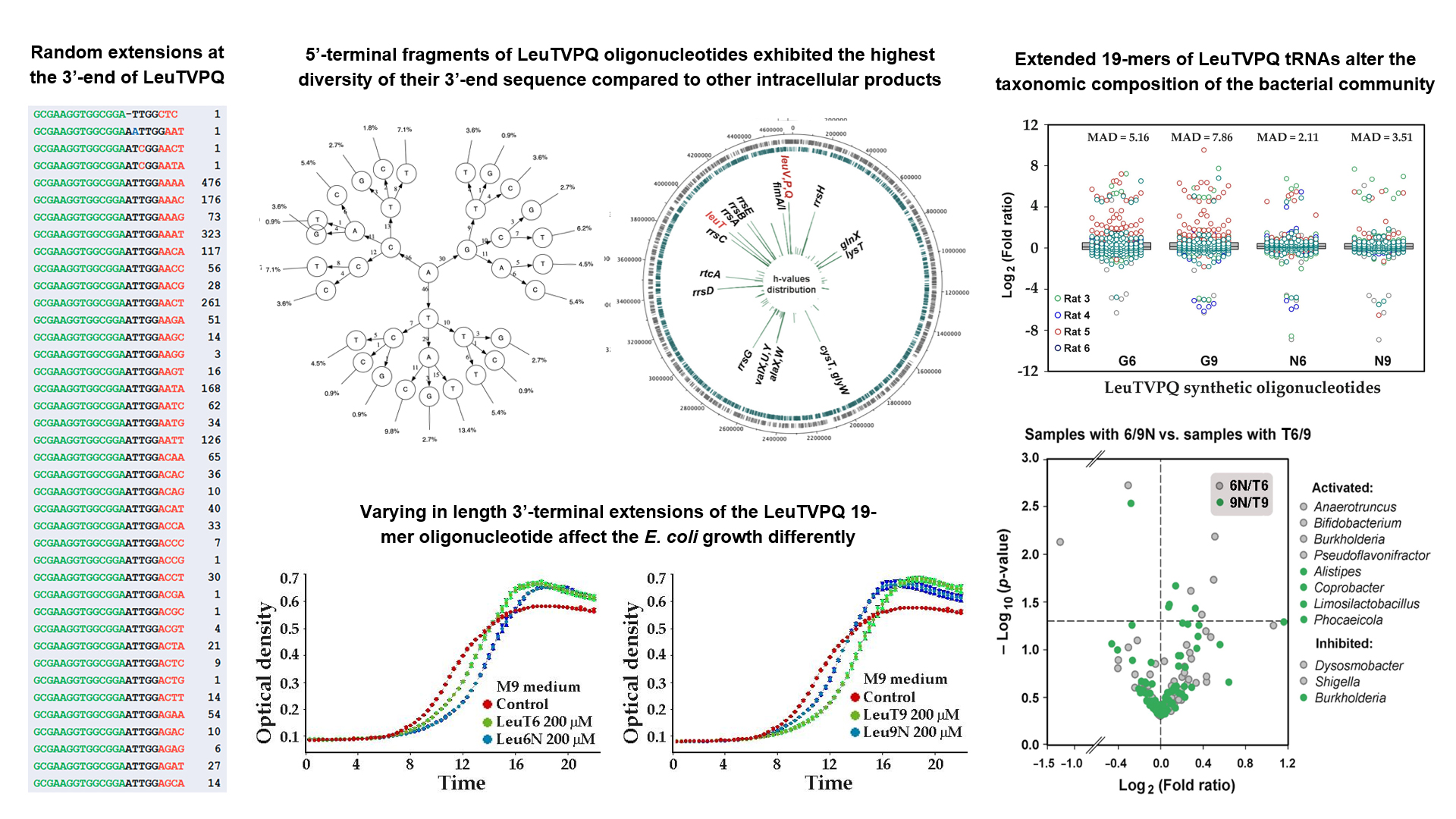
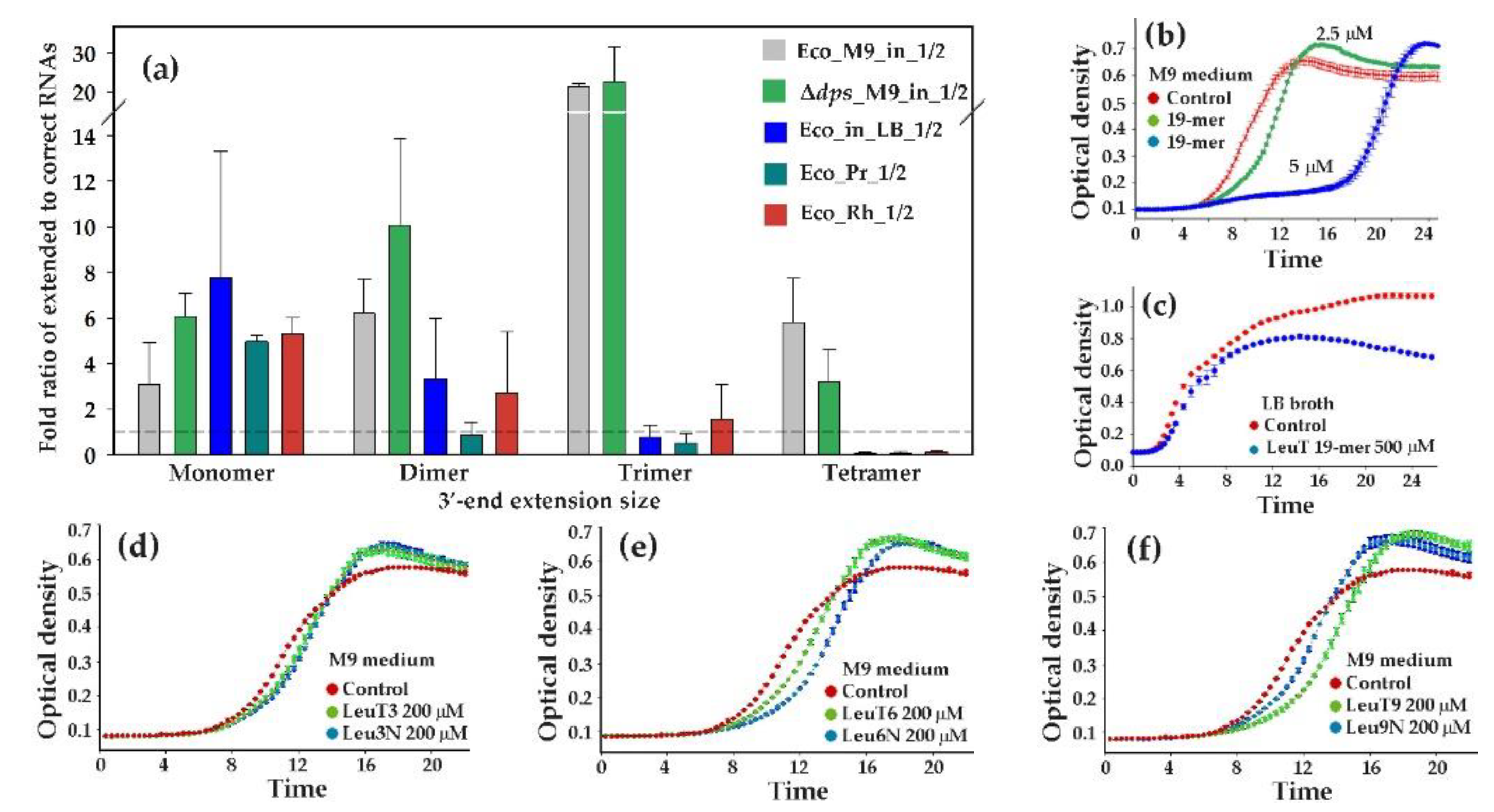
3.5. LeuTVPQ tRNA Fragments Show Exceptional 3’-End Extension Diversity but Are Not Exclusive Substrates for This Modification
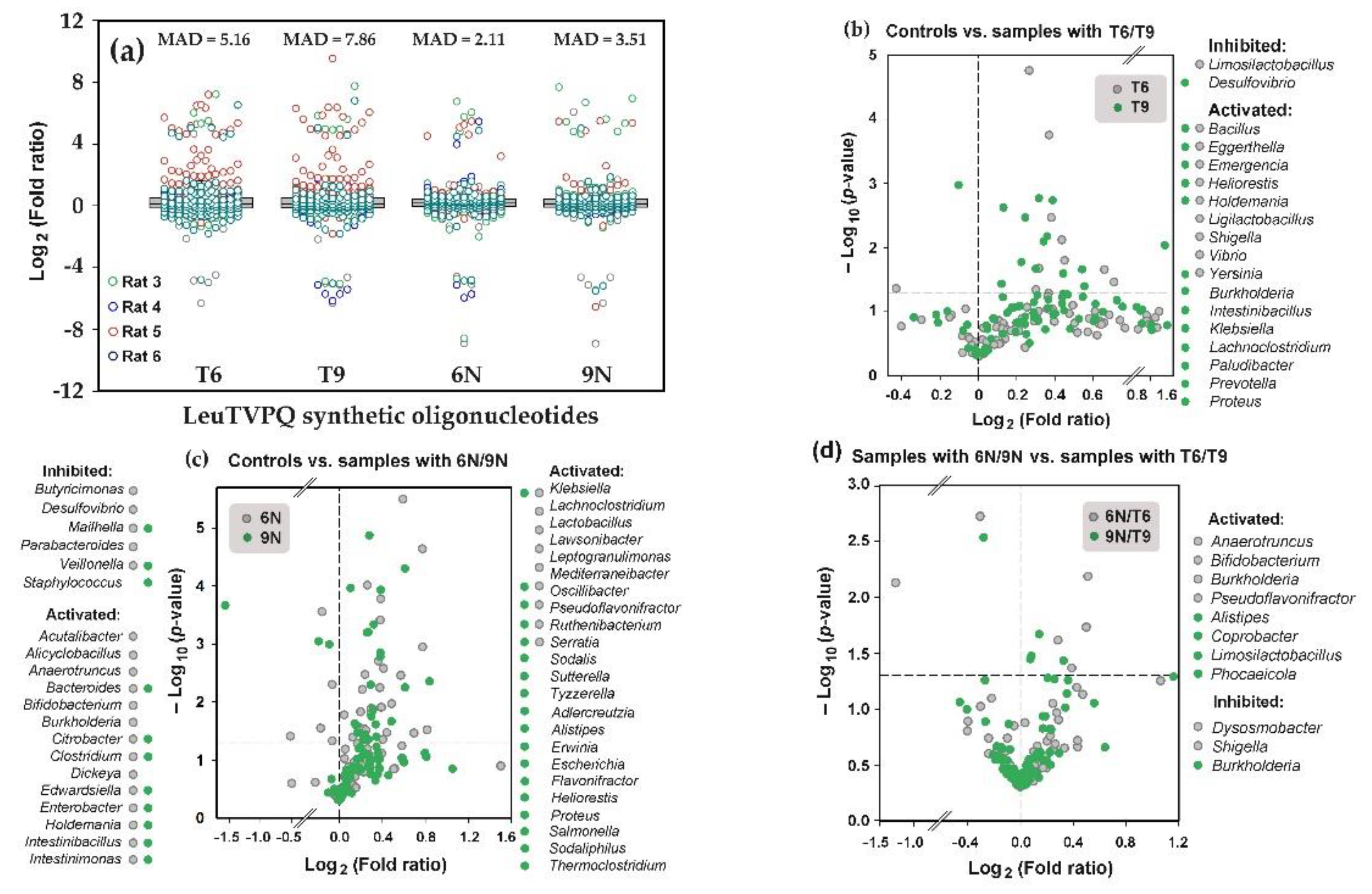
3.6. Presence of Modified LeuTVPQ Fragments in Transcriptomes and Their Influence on E. coli Growth in Monoculture are Medium-Dependent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Artika, I.M.; Arianti, R.; Demény, M.Á.; Kristóf, E. RNA modifications and their role in gene expression. Front. Mol. Biosci. 2025, 12, 1537861. [Google Scholar] [CrossRef]
- Arzumanian, V.A.; Dolgalev, G.V.; Kurbatov, I.Y.; Kiseleva, O.I.; Poverennaya, E.V. Epitranscriptome: review of top 25 most-studied RNA modifications. Int. J. Mol. Sci. 2022, 23(22), 13851. [Google Scholar] [CrossRef] [PubMed]
- Nachtergaele, S.; He, C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 2018, 52, 349–372. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yi, Y.; Gao, X.; Wang, X.; Zhao, D.; Wang, R.; Zhang, L.S.; Gao, B.; Zhang, Y.; Zhang, L.; et al. 2'-O-methylation at internal sites on mRNA promotes mRNA stability. Mol. Cell, 2320. [Google Scholar]
- Vandelli, A; Broglia, L.; Armaos, A.; Delli Ponti, R.; Tartaglia, G.G. Rationalizing the effects of RNA modifications on protein interactions. Mol. Ther. Nucleic Acids, 1023.
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361(6409), 1346–1349. [Google Scholar] [CrossRef]
- Zacco, E.; Broglia, L.; Kurihara, M.; Monti, M.; Gustincich, S.; Pastore, A.; Plath, K.; Nagakawa, S.; Cerase, A.; Sanchez de Groot, N.; Tartaglia, G.G. RNA: the unsuspected conductor in the orchestra of macromolecular crowding. Chem. Rev. 2024, 124(8), 4734–4777. [Google Scholar] [CrossRef]
- Xuan, J.; Chen, L.; Chen, Z.; Pang, J.; Huang, J.; Lin, J.; Zheng, L.; Li, B.; Qu, L.; Yang, J. RMBase v3. 0: decode the landscape, mechanisms and functions of RNA modifications, Nucleic Acids Res. 2024, 52, D273–D284. [Google Scholar]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Ge, J.; Yu, Y.-T. RNA pseudouridylation: new insights into an old modification. Trends in Bio-chemical Sciences.
- Agris, P.F.; Eruysal, E.R.; Narendran, A.; Väre, V.Y.P.; Vangaveti, S.; Ranganathan, S.V. Celebrating wobble decoding: Half a century and still much is new. RNA Biol.
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49(13), 7239–7255. [Google Scholar] [CrossRef]
- Zhao, B.; Roundtree, I.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3‘-end processing. Nucleic Acids Res. 2017, 45(19), 11356–11370. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 2018, 115(2), E325–E333. [Google Scholar] [CrossRef]
- Deng, X.; Chen, K.; Luo, G.-Z.; Weng, X.; Ji, Q.; Zhou, T.; He, C. Wide-spread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 6557. [Google Scholar]
- Szydlo., K.; Santos, L.; Christian, T.W.; Maharjan, S.; Dorsey, A.; Masuda, I.; Jia, J.; Wu, Y.; Tang, W.; Hou, Y.-M. Szydlo. K.; Santos, L.; Christian, T.W.; Maharjan, S.; Dorsey, A.; Masuda, I.; Jia, J.; Wu, Y.; Tang, W.; Hou, Y.-M.; et al. m6A modification is incorporated into bacterial mRNA without specific functional benefit. Nucleic Acids Res.
- Hajnsdorf, E.; Kaberdin, V.R. RNA polyadenylation and its consequences in prokaryotes. Phil. Trans. R. Soc. B. 2018, 373, 20180166. [Google Scholar] [CrossRef]
- Mofayezi, A.; Jadaliha, M.; Zangeneh, F.Z.; Khoddami, V. Poly(A) tale: from A to A; RNA polyadenylation in prokaryotes and eukaryotes. Wiley Interdiscip. Rev. RNA. 2024, 15(2), e1837. [Google Scholar] [CrossRef]
- Passmore, L.A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell. Biol. 2022, 23(2), 93–106. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.K.; Kushner, S.R. Bacterial/archaeal polyadenylation. Wiley Interdiscip. Rev. RNA 2016, 7(1), 31–44. [Google Scholar]
- Mohanty, B.K.; Kushner, S.R. New Insights into the relationship between tRNA processing and polyadenylation in Escherichia coli. Trends Genet. 2019, 35(6), 434–445. [Google Scholar] [CrossRef]
- Régnier, P.; Marujo, P.E. Polyadenylation and degradation of RNA in prokaryotes. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience 2000-2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK6253.
- Coburn, G.A.; Mackie, G.A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 1999, 62, 55–108. [Google Scholar] [PubMed]
- Spickler, C.; Mackie, G.A. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 2000, 182, 2422–2427. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Kushner, S.R. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006, 34, 5695–5704. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Kushner, S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999, 34, 1094–1108. [Google Scholar] [CrossRef]
- Mohanty, B.K.; Maples, V.F.; Kushner, S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004, 54, 905–920. [Google Scholar] [CrossRef]
- Raynal, L.C.; Carpousis, A.J. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 1999, 32, 765–775. [Google Scholar] [CrossRef]
- Ghosal, A.; Upadhyaya, B.B.; Fritz, J.V.; Heintz-Buschart, A.; Desai, M.S.; Yusuf, D.; Huang, D.; Baumuratov, A.; Wang, K.; Galas, D.; et al. The extracellular RNA complement of Escherichia coli. Microbiology 2015, 4, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Alikina, O.V.; Glazunova, O.A.; Bykov, A.A.; Kiselev, S.S.; Tutukina, M.N.; Shavkunov, K.S.; Ozoline, O.N. A cohabiting bacterium alters the spectrum of short RNAs secreted by Escherichia coli. FEMS Microbiol. Lett. 2018, 365, fny262. [Google Scholar] [CrossRef] [PubMed]
- Markelova, N.; Glazunova, O.; Alikina, O.; Panyukov, V.; Shavkunov, K.; Ozoline, O. Suppression of Escherichia coli growth dynamics via RNAs secreted by competing bacteria. Front. Mol. Biosci. 2021, 8, 609979. [Google Scholar] [CrossRef] [PubMed]
- Blenkiron, C.; Simonov, D.; Muthukaruppan, A.; Tsai, P.; Dauros, P.; Green, S.; Hong, J.; Print, C.J.; Swift, S.; Phillips, A.R. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS ONE 2016, 11, e0160440. [Google Scholar] [CrossRef]
- Ren, B.; Tibbelin, G.; Kajino, T.; Asami, O.; Ladenstein, R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J Mol Biol. 2003, 329(3), 467–477. [Google Scholar] [CrossRef]
- Wolf, S.G.; Frenkiel, D.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. DNA protection by stress-induced biocrystallization. Nature 1999, 400, 83–85. [Google Scholar] [CrossRef]
- Frenkiel-Krispin, D.; Levin-Zaidman, S.; Shimoni, E.; Wolf, S.G.; Wachtel, E.J.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 2001, 20, 1184–1191. [Google Scholar] [CrossRef]
- Janissen, R.; Arens, M.M.A.; Vtyurina, N.N.; Rivai, Z.; Sunday, N.D.; Eslami-Mossallam, B.; Gritsenko, A.A.; Laan, L.; de Ridder, D.; Artsimovitch, I.; et al. Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell, 1188. [Google Scholar]
- Bykov, A.A.; Shavkunov, K.S.; Panyukov, V.V.; Ozoline, O.N. Bacterial nucleoid protein Dps binds structured RNA molecules. Mat. Biol. Bioinform. 2017, 12(S), t1–t11. [Google Scholar]
- Park, C.; Jin, Y.; Kim, Y.J.; Jeong, H.; Seong, B.L. RNA-binding as chaperones of DNA binding proteins from starved cells. Biochem. Biophys. Res. Commun. 2020, 524(2), 484–489. [Google Scholar] [CrossRef] [PubMed]
- Lacqua, A.; Wanner, O.; Colangelo, T.; Martinotti, M.G.; Landini, P. Emergence of biofilm-forming subpopulations upon exposure of Escherichia coli to environmental bacteriophages. Appl. Environ. Microbiol. 2006, 72, 956–959. [Google Scholar] [CrossRef]
- Pang, B.; Hong, W.; Kock, N.D.; Swords, W.E. Dps promotes survival of nontypeable Haemophilus influenzae in biofilm communities in vitro and resistance to clearance in vivo. Front. Cell. Infect. Microbiol. 2012, 2, 58. [Google Scholar] [CrossRef] [PubMed]
- Shavkunov, K.S.; Markelova, N.Yu.; Alikina, O.V.; Glazunova, O.A.; Panyukov, V.V.; Kolzhetsov, N.P.; Kiselev, S.S.; Ozoline, O.N. Products of abortive transcription can prime synthesis of chimeric oligonucleotides. Mat. Biolog. Bioinform. 2024, 19(2), 453–471. [Google Scholar] [CrossRef]
- Antipov, S.S.; Tutukina, M.N.; Preobrazhenskaya, E.V.; Kondrashov, F.A.; Patrushev, M.V.; Toshchakov, S.V.; Dominova, I.; Shvyreva, U.S.; Vrublevskaya, V.V.; Morenkov, O.S.; et al. The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner. PLoS One 2017, 12(8), e0182800. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Melekhov, V.V.; Shvyreva, U.S.; Timchenko, A.A.; Tutukina, M.N.; Preobrazhenskaya, E.V.; Burkova, D.V.; Artiukhov, V.G.; Ozoline, O.N.; Antipov, S.S. Modes of Escherichia coli Dps interaction with DNA as revealed by atomic force microscopy. PloS One 2015, 10, e0126504. [Google Scholar] [CrossRef]
- Bykov, A.; Glazunova, O.; Alikina, O.; Sukharicheva, N.; Masulis, I.; Shavkunov, K.; Ozoline, O. Excessive promoters as silencers of genes horizontally acquired by Escherichia coli. Front. Mol. Biosci. 2020, 7, 28. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, 3–10. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G. 3rd.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science, 277, 1453-1462.
- Panyukov, V.V.; Kiselev, S.S.; Shavkunov, K.S.; Masulis, I.S.; Ozoline, O.N. Mixed promoter islands as genomic regions with specific structural and functional properties. Mat. Biolog. Bioinform. 2013, 8(2), 432–448. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Shavkunov, K.S.; Markelova, N.Y.; Glazunova, O.A.; Kolzhetsov, N.P.; Panyukov, V.V.; Ozoline, O.N. The Fate and functionality of alien tRNA fragments in culturing medium and cells of Escherichia coli. Int. J. Mol. Sci. 2023, 24, 12960. [Google Scholar] [CrossRef] [PubMed]
- Kolzhetsov, N.; Markelova, N.; Frolova, M.; Alikina, O.; Glazunova, O.; Safonova, L.; Kalashnikova, I.; Yudin, V.; Makarov, V.; Keskinov, A.; et al. Enterotype-dependent probiotic-mediated changes in the male rat intestinal microbiome in vivo and in vitro. Int. J. Mol. Sci. 2024, 25, 4558. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Shavkunov, K.S.; Masulis, I.S.; Tutukina, M.N.; Deev, A.A.; Ozoline, O.N. Gains and unexpected lessons from genome-scale promoter mapping. Nucleic Acids Res. 2009, 37(15), 4919–4931. [Google Scholar] [CrossRef] [PubMed]
- Tierrafría, V.H.; Rioualen, C.; Salgado, H.; Lara, P.; Gama-Castro, S.; Lally, P.; Gómez-Romero, L.; Peña-Loredo, P.; López-Almazo, A.G.; Alarcón-Carranza, G.; et al. RegulonDB 11.0: Comprehensive high-throughput datasets on transcriptional regulation in Escherichia coli K-12. Microb. Genom. 2022, 2022 8, mgen000833. [Google Scholar] [CrossRef]
- Shi, Y.; Manley, J.L. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015, 29(9), 889–897. [Google Scholar] [CrossRef]
- Neve, J.; Patel, R.; Wang, Z.; Louey, A.; Furger, A.M. Cleavage and polyadenylation: Ending the message expands gene regulation. RNA Biol. 2017, 14(7), 865–890. [Google Scholar] [CrossRef]
- Kühn, U.; Gündel, M.; Knoth, A.; Kerwitz, Y.; Rüdel, S.; Wahle, E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 2009, 284(34), 22803–22814. [Google Scholar] [CrossRef]
- Weiner, A.M. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 2004, 14(20), R883–885. [Google Scholar] [CrossRef]

| Sample | Number of reads | Description | Data availability |
||
|---|---|---|---|---|---|
| Before QC | After QC | 16 ≤ L ≤ 50 | |||
| Eco_in_M9_1* Eco_in_M9_2 |
1,546,157 2,494,971 |
996,435 1,528,878 |
821,483 1,517,245 |
Intracellular RNAs isolated from the wild-type strain | GSM6892281 GSM6892282 |
|
dps_del_in_1* dps_del_in_2 |
2,093,000 1,771,628 |
1,294,678 1,082,635 |
1,292,272 1,072,005 |
Intracellular RNAs isolated from the Δdps mutant strain | GSM6892285 GSM6892286 |
| Eco_exo_1* Eco_exo_2 |
1,832,829 1,220,822 |
890,000 737,402 |
788,543 729,656 |
Extracellular RNAs isolated from the wild-type strain | GSM6892283 GSM6892284 |
|
dps_del_exo_1* dps_del_exo_2 |
608,759 917,604 |
182,984 258,455 |
158,810 180,502 |
Extracellular RNAs isolated from the Δdps mutant strain | GSM6892287 GSM6892288 |
| Sample | Number of reads | Growth conditions |
BioProject PRJNA687658 sample description and references |
|
|---|---|---|---|---|
| Before QC | After QC | |||
| Eco_in_LB_1 Eco_in_LB_2 |
1,320,485 2,305,864 |
299,167 1,589,066 |
Anaerobic growth | Intracellular RNAs of E. coli grown in monoculture [33] |
| Eco_Pr_1 Eco_Pr_2 |
2,629,571 864,537 |
1,563,678 652,497 |
Anaerobic growth | RNAs isolated from E. coli cells after their co-cultivation with Prevotella copri or Rhodospirillum rubrum in membrane-separated compartments within common chamber [52] |
| Eco_Rh_1 Eco_Rh_2 |
1,301,404 1,176,395 |
1,028,350 681,123 |
Anaerobic growth | |
| Name | 5’-terminal sequences of the fragments of four leucine tRNAs |
|---|---|
| LeuTVPQ | 5’- GCGAAGGUGGCGGAAUUGG - 3’ |
| LeuTVPQ_T3 | 5’-GCGAAGGUGGCGGAAUUGGUAG - 3’ |
| LeuTVPQ_3N* | 5’-GCGAAGGUGGCGGAAUUGGNNN - 3’ |
| LeuTVPQ_T6 | 5’-GCGAAGGUGGCGGAAUUGGUAGACG - 3’ |
| LeuTVPQ_6N | 5’-GCGAAGGUGGCGGAAUUGGNNNNNN - 3’ |
| LeuTVPQ_T9 | 5’-GCGAAGGUGGCGGAAUUGGUAGACGCGC - 3’ |
| LeuTVPQ_9N | 5’-GCGAAGGUGGCGGAAUUGGNNNNNNNNN - 3’ |
| Peak maximum | Strand | Number of reads* | Genomic locus | Strand | |||||
|---|---|---|---|---|---|---|---|---|---|
| Transcriptome | Secretome | Gene(s) | Borders | ||||||
| w.t. | Δdps | w.t. | Δdps | Right | Left | ||||
| 225381 | + | 941 | 638 | 2,318 | 966 | ileV | 225381 | 225457 | + |
| 3427152 | - | 618 | 718 | 1,204 | 1,109 | ileU | 3427076 | 3427152 | - |
| 4037141 | + | 618 | 718 | 1,159 | 966 | ileT | 4037141 | 4037217 | + |
| 564723 | + | 137 | 113 | 477 | 169 | argU | 564723 | 564799 | + |
| 2817860 | - | 422 | 386 | 1,311 | 1,662 | argQ(ZYV) | 2817784 | 2817860 | - |
| 3423655 | - | 1,387 | 2,127 | 4,380 | 4,053 | thrV | 3423580 | 3423655 | - |
| 696740 | - | 1,499 | 2,822 | 2,310 | 2,490 | metU(T) | 696664 | 696740 | - |
| 2947387 | + | 408 | 307 | 1,182 | 501 | metZ(WV) | 2947387 | 2947463 | + |
| 3318289 | - | 413 | 316 | 1,265 | 616 | metY | 3318213 | 3318289 | - |
| 697047 | - | 12,294 | 14,261 | 1,730 | 1,652 | leuW | 696963 | 697047 | - |
| 780765 | + | 1,046 | 941 | 473 | 357 | valT(Z) | 780765 | 780840 | + |
| 1031712 | - | 1,354 | 2,065 | 1,233 | 1,076 | serT | 1031625 | 1031712 | - |
| 1097652 | - | 2,279 | 3,837 | 818 | 724 | serX(W) | 1097565 | 1097652 | - |
| 2043557 | - | 432 | 624 | 198 | 148 | serU | 2043468 | 2043557 | - |
| 2521253 | + | 1,868 | 2,918 | 1,576 | 1,603 | lysV | 2521253 | 2521328 | + |
| 2729444 | - | 286 | 398 | 460 | 391 | gltW | 2729369 | 2729444 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





