Submitted:
18 July 2025
Posted:
18 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sampled Areas
2.2. Morphological Study of Aleurocanthus sp.
2.3. Field Survey
2.4. Data Analysis
2.5. Morphological Study of Parasitoids
2.6. Molecular Analysis
2.7. Phylogenetic Analysis
3. Results
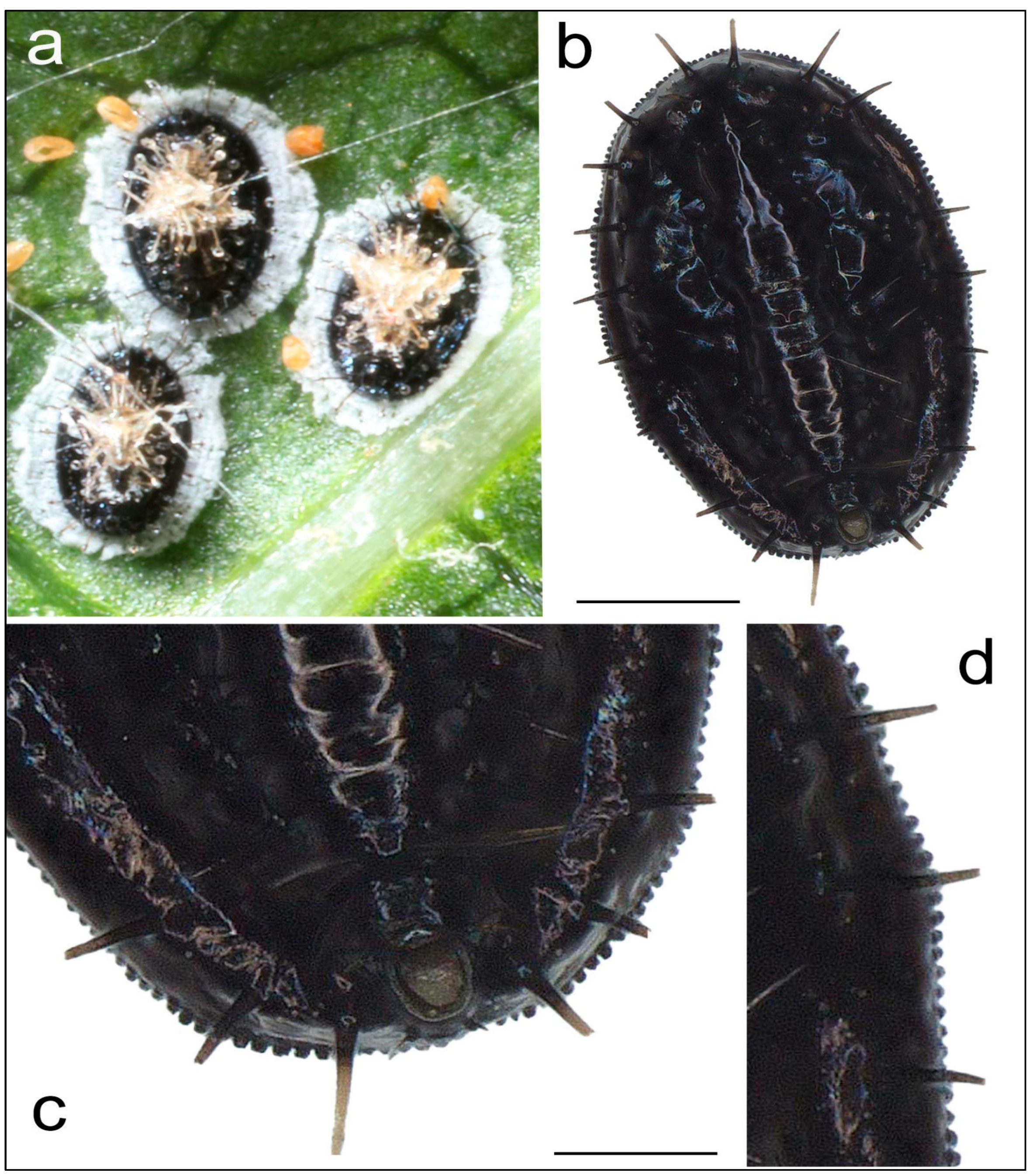
3.1. Morphological Study of Aleurocanthus sp.
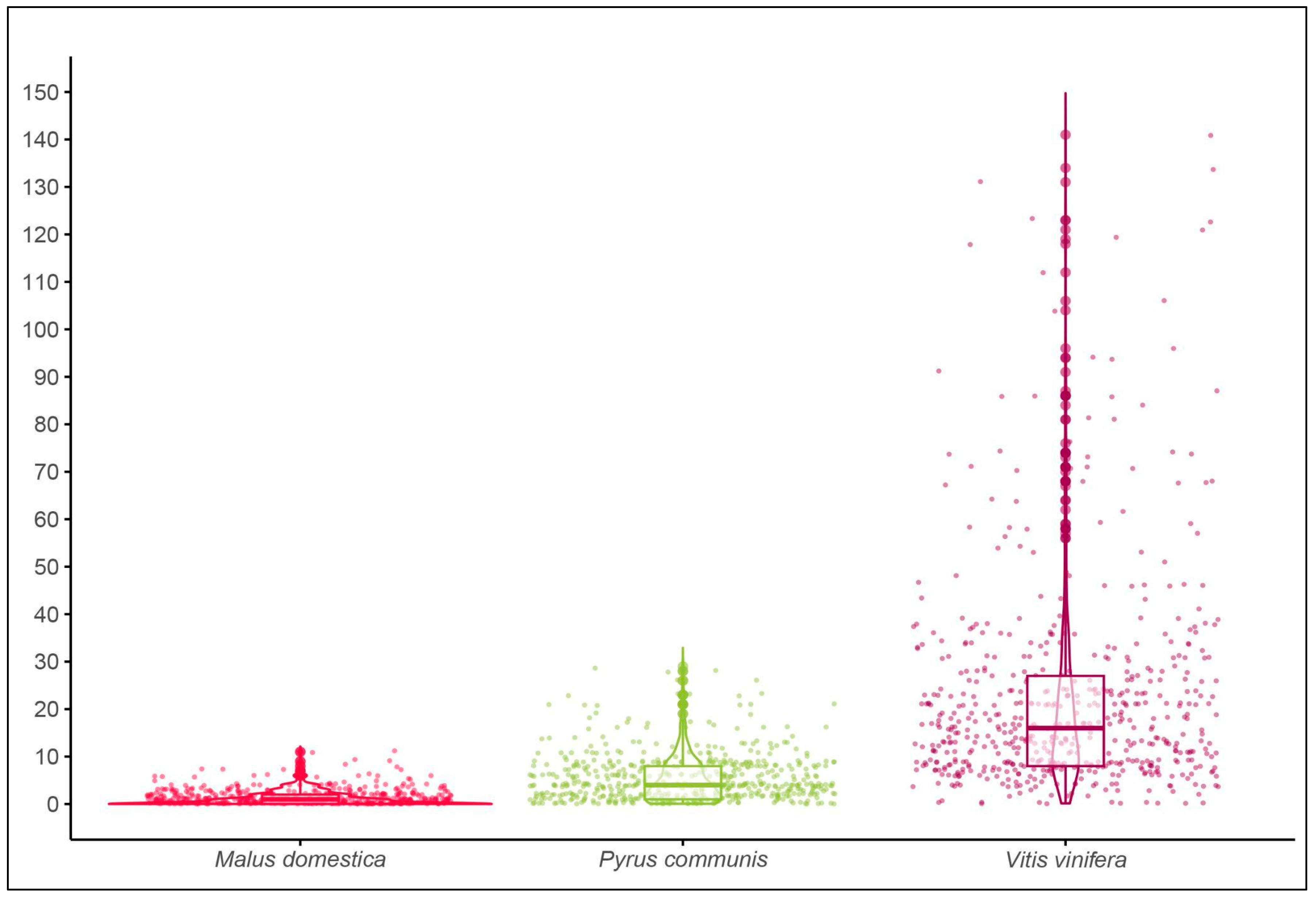
3.2. Field Survey
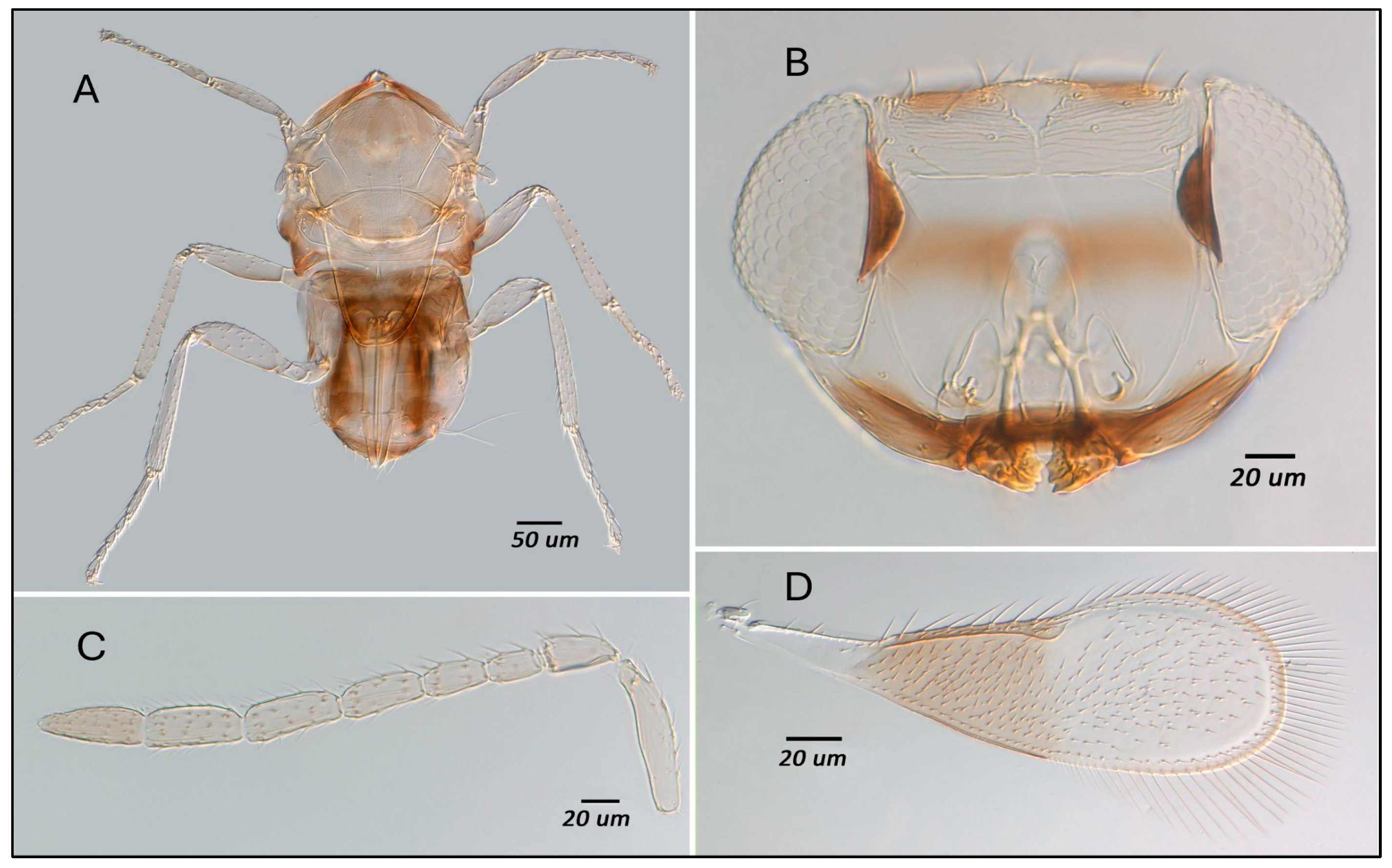
3.3. Morphological Study of Parasitoids
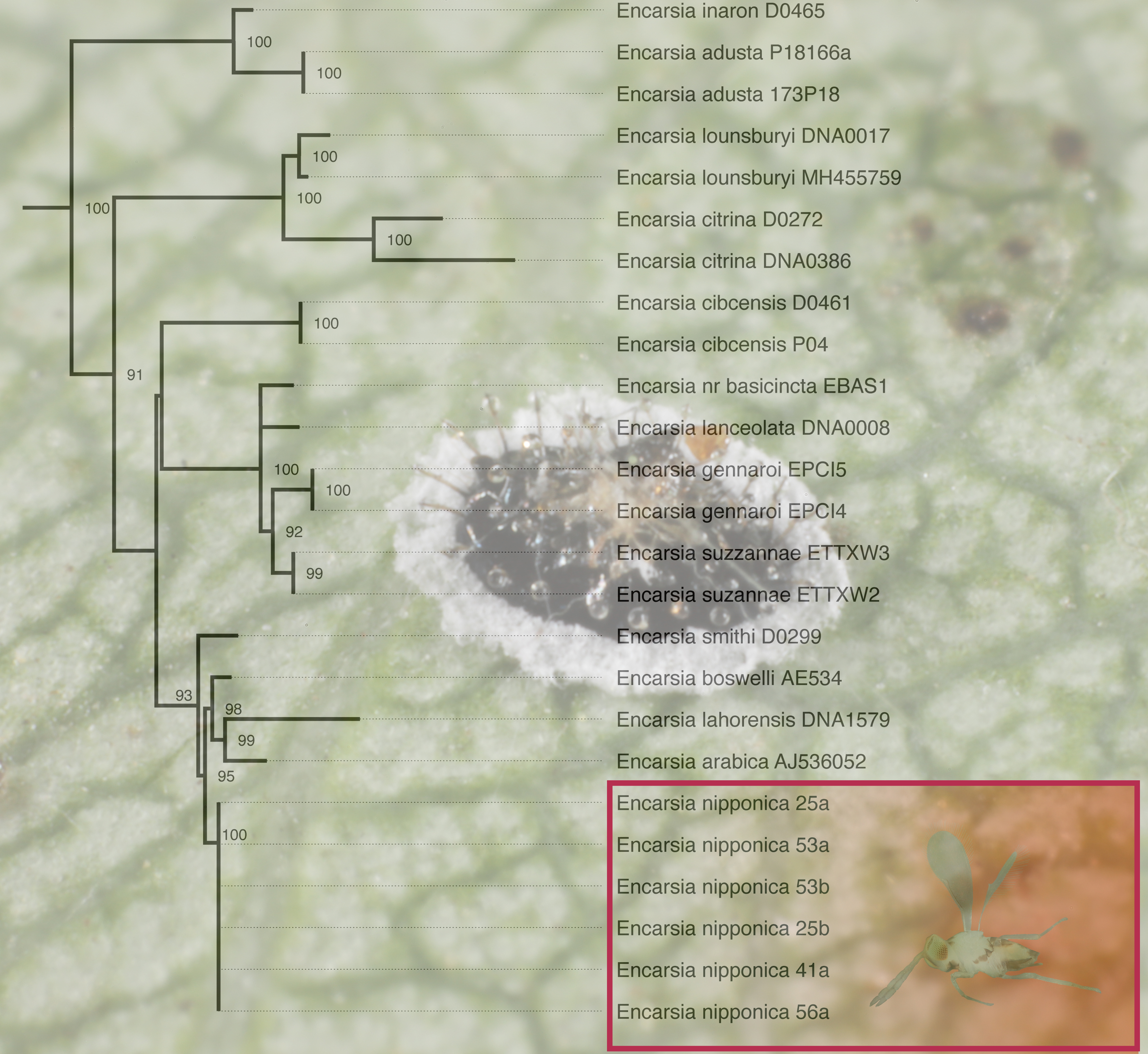
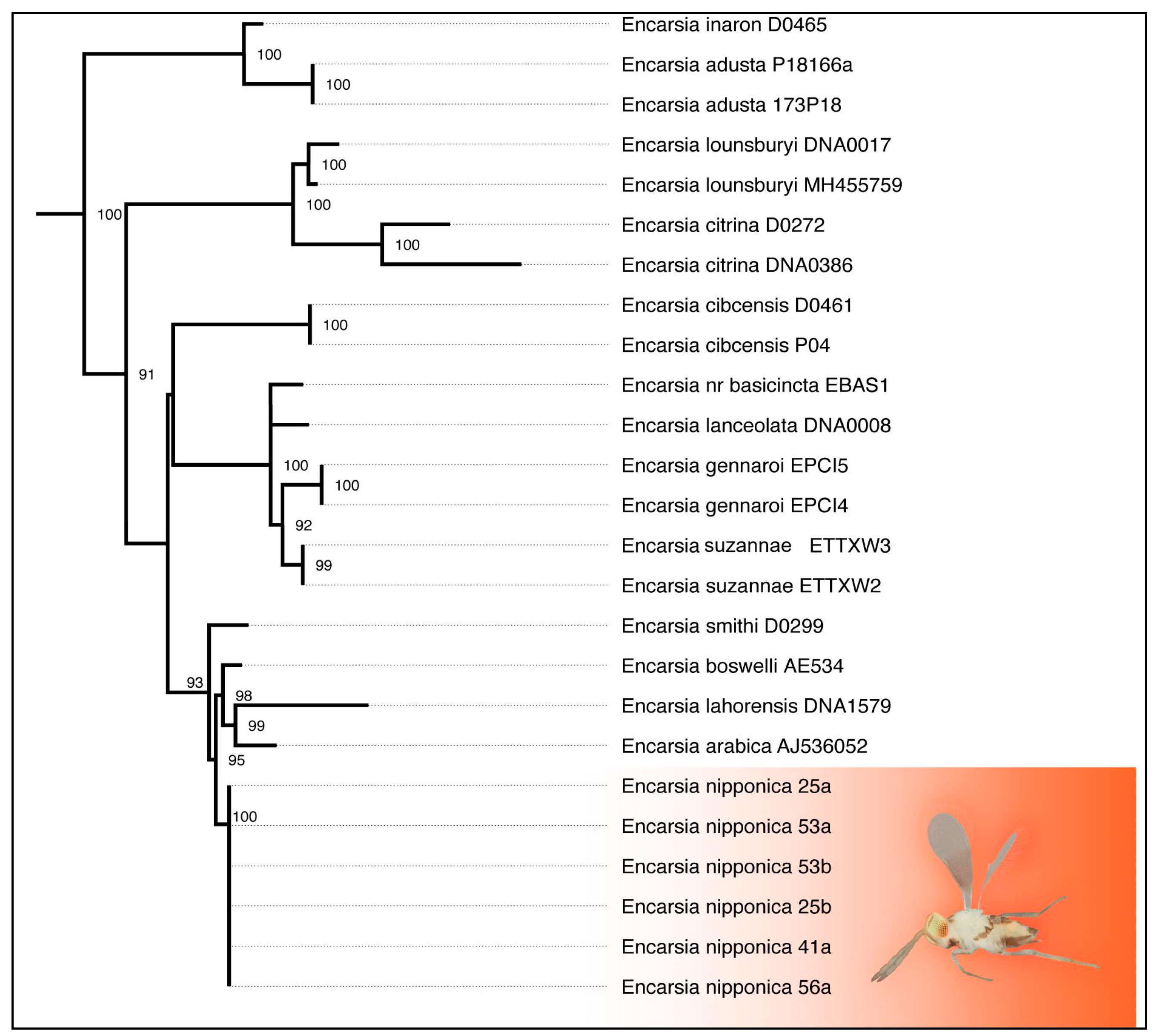
3.4. Phylogenetic Analysis
3.5. Molecular Analysis
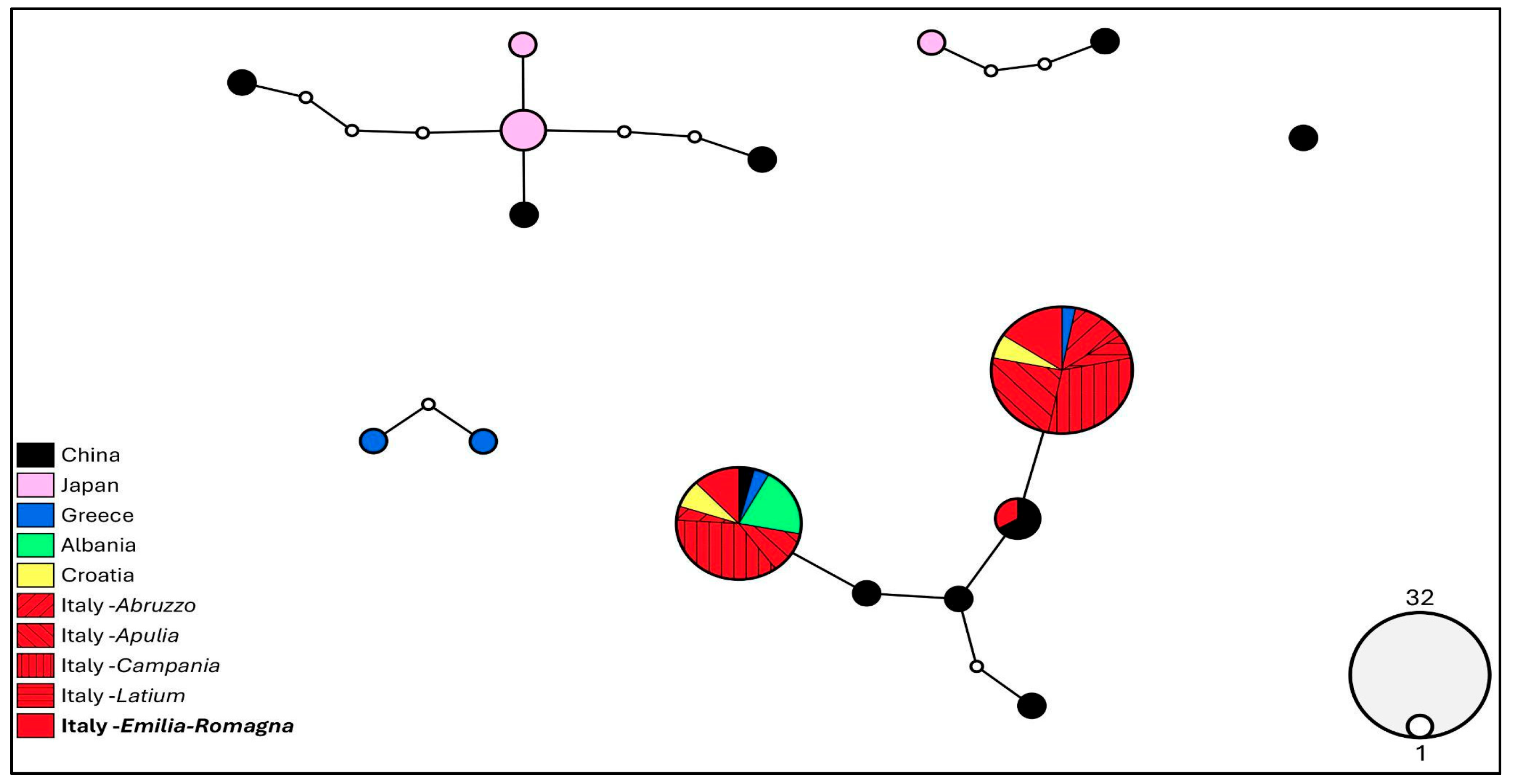
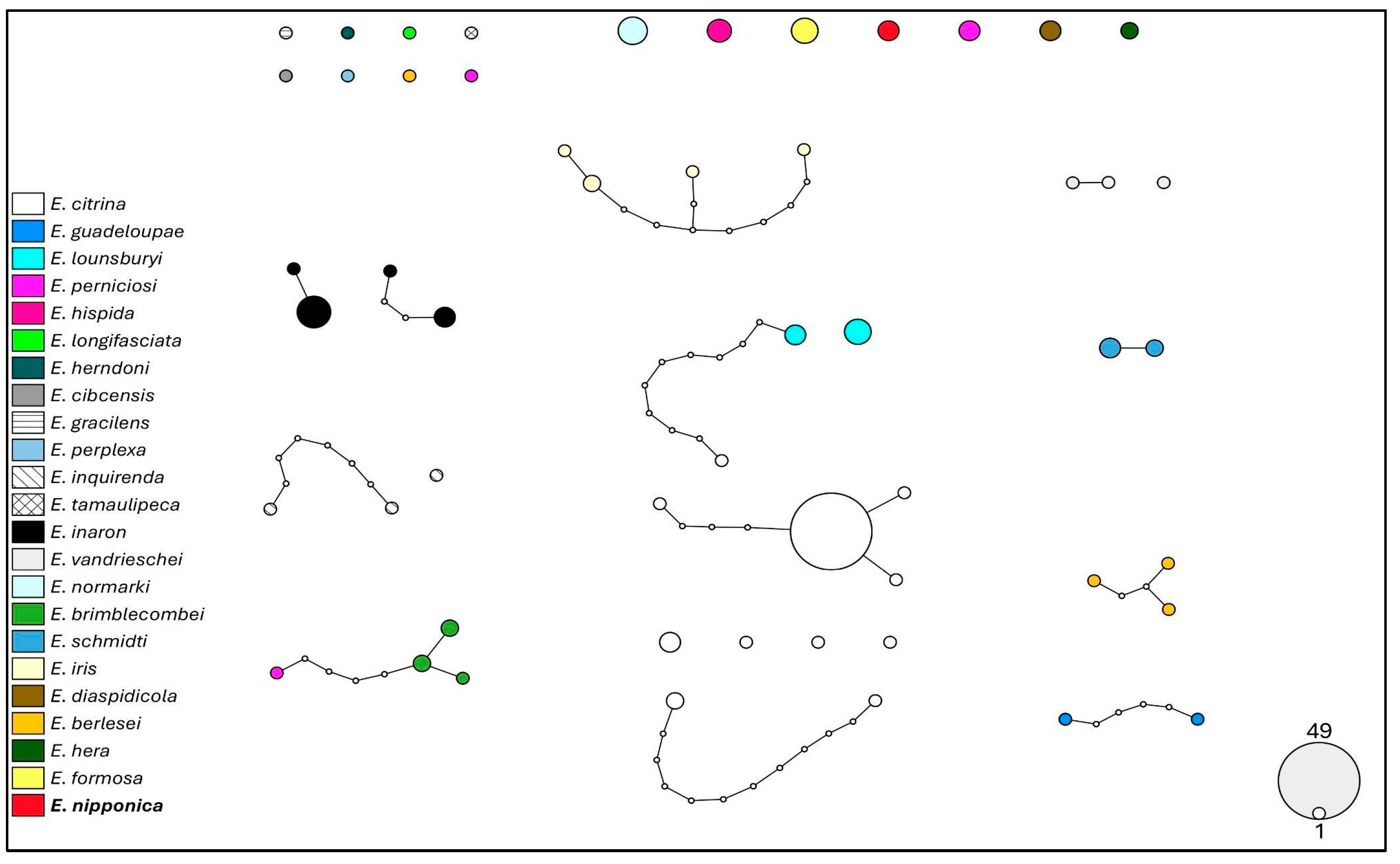
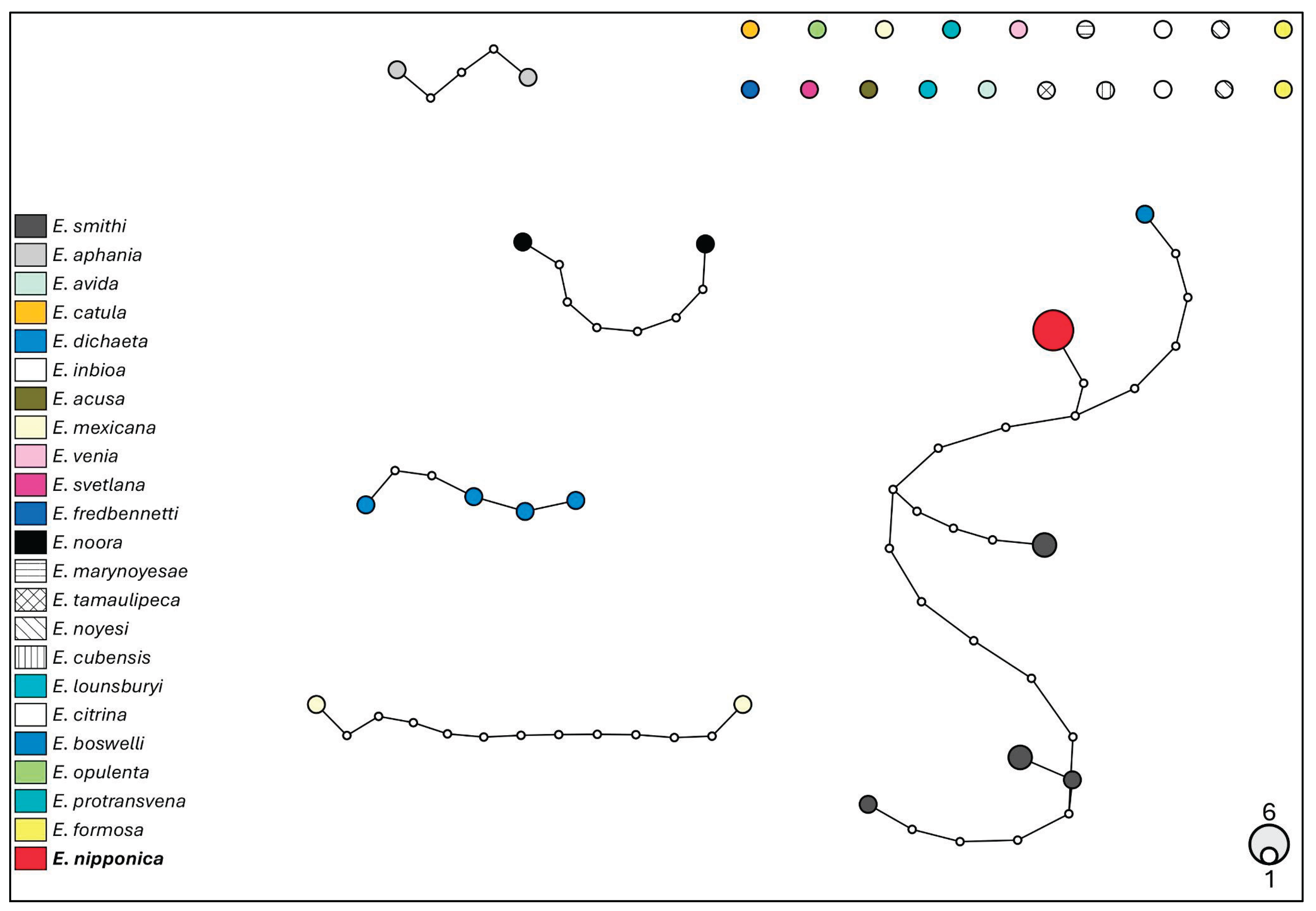
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, J.H. An identification guide to common whitefly pest species of the world (Homoptera: Aleyrodidae). Trop. Pest Manag. 1987, 33, 298–322. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health. Evaluation of a paper by Guarnaccia et al. (2017) on the first report of Phyllosticta citricarpa in Europe. EFSA J. 2018, 16, 1–48. [Google Scholar] [CrossRef]
- Van den Berg, M.A.; De Beer, M.S. Natural enemies of the spiny blackfly, Aleurocanthus spiniferus (Hem. : Aleyrodidae), in Mpumalanga, South Africa. In Proceedings of the International Society of Citriculture, Sun City Resort, South Africa, 12–17 May 1996; pp. 667–669. [Google Scholar]
- Nguyen, R.; Fasulo, T.R. A Citrus blackfly parasitoid, Encarsia perplexa Huang & Polaszek (Insecta: Hymenoptera: Aphelinidae). EDIS 2010. [Google Scholar] [CrossRef]
- Gillespie, P.S. A review of the whitefly genus Aleurocanthus Quaintance & Baker (Hemiptera: Aleyrodidae) in Australia. Zootaxa 2012, 3252, 1–61. [Google Scholar] [CrossRef]
- EPPO. Aleurocanthus spiniferus. Available online: https://gd.eppo.int/taxon/ALEUSI (accessed on 15 July 2024).
- Porcelli, F. First record of Aleurocanthus spiniferus (Homoptera: Aleyrodidae) in Apulia, Southern Italy. EPPO Bull. 2008, 38, 516–518. [Google Scholar] [CrossRef]
- Šimala, M.; Pintar, M.; Milek, T.M.; Markotić, V. Results of a two year survey (2015-2016) of quarantine whitefly species from genus Aleurocanthus Quaintance & Baker 1914 on citrus in Croatia. In Proceedings of the 13th Slovenian Conference on Plant Protection, Rimske Toplice, Slovenia, 7–8 March 2017. [Google Scholar]
- Radonjić, S.; Hrnčić, S.; Malumphy, C. First record of Aleurocanthus spiniferus (Quaintance) (Hemiptera Aleyrodidae) in Montenegro. Redia 2014, 97, 141–145. [Google Scholar]
- Kapantaidaki, D.E.; Antonatos, S.; Kontodimas, D.; Milonas, P.; Papachristos, D.P. Presence of the invasive whitefly Aleurocanthus spiniferus (Hemiptera: Aleyrodidae) in Greece. EPPO Bull. 2019, 49, 127–131. [Google Scholar] [CrossRef]
- Nugnes, F.; Laudonia, S.; Jesu, G.; Jansen, M.G.M.; Bernardo, U.; Porcelli, F. Aleurocanthus spiniferus (Hemiptera: Aleyrodidae) in some European countries: Diffusion, hosts, molecular characterization, and natural enemies. Insects 2020, 11, 42. [Google Scholar] [CrossRef]
- Streito, J.C.; Mendes, E.; Sanquer, E.; Strugarek, M.; Ouvrad, D.; Robin-Havret, V.; Poncet, L.; Lannou, C.; Rossi, J.P. Incursion Preparedness, Citizen Science and Early Detection of Invasive Insects: The Case of Aleurocanthus spiniferus (Hemiptera, Aleyrodidae) in France. Insects 2023, 14, 916. [Google Scholar] [CrossRef]
- Bariselli, M.; Bortolotti, P.P.; Nannini, R. Scheda 27 - Aleurocanthus spiniferus. In Schede tecniche per il riconoscimento degli organismi nocivi da quarantena; Regione Emilia-Romagna, Assessorato Agricoltura, Caccia e Pesca, Servizio fitosanitario Emilia-Romagna: Bologna, Italy, 2019; p. 4. [Google Scholar]
- Rapisarda, C.; Longo, S. First report from Sicily (Italy) of the orange spiny whitefly, Aleurocanthus spiniferus (Quaintance) (Hemiptera: Aleyrodidae), and its potential risk for the Italian citrus industry. EPPO Bull. 2021, 51, 329–332. [Google Scholar] [CrossRef]
- Melone, G.; Ascolese, R.; Nugnes, F.; Porcelli, F.; Rapisarda, C.; Farina, A.; Picciotti, U.; Garganese, F.; Laudonia, S. An Eretmocerus species, parasitoid of Aleurocanthus spiniferus, was found in Europe: The secret savior of threatened plants. Sustainability 2024, 16, 2970. [Google Scholar] [CrossRef]
- Gill, R.J. The morphology of whiteflies. In Whiteflies: Their bionomics, pest status and management; Gerling, D., Ed.; Intercept: Andover, Hants, UK, 1990; pp. 13–46. [Google Scholar]
- Mound, L.A.; Halsey, S.H. Whitefly of the World; British Museum (Natural History) and John Wiley & Sons: Chichester, UK, 1978; p. 340. [Google Scholar]
- Cioffi, M.; Cornara, D.; Corrado, I.; Jansen, M.G.M.; Porcelli, F. The status of Aleurocanthus spiniferus from its unwanted introduction in Italy to date. Bull. Insectology 2013, 66, 273–281. [Google Scholar]
- Evans, G. The whiteflies (Hemiptera: Aleyrodidae) of the world and their host plants and natural enemies; USDA/Animal Plant Health Inspection Service (APHIS): Riverdale, MD, USA, 2008. [Google Scholar]
- Silvestri, F. Contribuzione alla conoscenza degli Aleurodidae (Insecta: Hemiptera) viventi su Citrus in Estremo Oriente e dei loro parassiti. II. Descrizione e notizie biologiche dei parassiti di Aleurodidi viventi su Citrus. Boll. Lab. Zool. Gen. Agrar. R. Ist. Super. Agrar. Portici 1927, 21, 20–60. [Google Scholar]
- Van den Berg, M.A.; Hoppner, G.; Greenland, J. An economic study of the biological control of the spiny blackfly, Aleurocanthus spiniferus (Hemiptera: Aleyrodidae), in a citrus orchard in Swaziland. Biocontrol Sci. Techn. 2000, 10, 27–32. [Google Scholar] [CrossRef]
- Muniappan, R.; Purea, M.; Sengebau, F.; Reddy, G.V.P. Orange spiny whitefly, Aleurocanthus spiniferus (Quaintance) (Homoptera: Aleyrodidae), and its parasitoids in the Republic of Palau. Proc. Hawaiian Entomol. Soc. 2006, 38, 21–25. [Google Scholar]
- Gyeltshen, J.; Hodges, A.; Hodges, G.S. Orange Spiny Whitefly, Aleurocanthus spiniferus Quaintance (Insecta: Hemiptera: Aleyrodidae); University of Florida IFAS Extension: Gainesville, FL, USA, 2019. [Google Scholar]
- Laudonia, S.; Melone, G.; Ascolese, R.; Nugnes, F. Eretmocerus iulii Laudonia et Melone sp.n.: parasitoid associated with Aleurocanthus spiniferus. Bull. Insectology 2024, 77, 263–269. [Google Scholar]
- Colazza, S.; Bin, F. Efficiency of Trissolcus basalis (Hymenoptera: Scelionidae) as an egg parasitoid of Nezara viridula (Heteroptera: Pentatomidae) in Central Italy. Environ. Entomol. 1995, 24, 1703–1707. [Google Scholar] [CrossRef]
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Mächler, M.; Bolker, B.M. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. The R J. 2017, 9, 378–400. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Noyes, J.S. Collecting and preserving chalcid wasps (Hymenoptera: Chalcidoidea). J. Nat. Hist. 1982, 16, 315–334. [Google Scholar] [CrossRef]
- Nugnes, F.; Russo, E.; Viggiani, G.; Bernardo, U. First record of an invasive fruit fly belonging to Bactrocera dorsalis complex (Diptera: Tephritidae) in Europe. Insects 2018, 9, 182. [Google Scholar] [CrossRef]
- Uesugi, R.; Sato, Y.; Han, B.; Huang, Z.D.; Yara, K.; Furuhashi, K. Molecular evidence for multiple phylogenetic groups within two species of invasive spiny whiteflies and their parasitoid wasp. Bull. Entomol. Res. 2016, 106, 328–340. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved Polymerase Chain Reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Park, J.K.; O’Foighil, D. Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Mol. Phylogenetics Evol. 2000, 14, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Nunn, G.B.; Theisen, B.F.; Christensen, B.; Arctander, P. Simplicity correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. J. Mol. Evol. 1997, 42, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Polaszek, A.; Ayshford, T.; Yahya, B.E.; Fusu, L. Wallaceaphytis: an unusual new genus of parasitoid wasp (Hymenoptera: Aphelinidae) from Borneo. J. Nat. Hist. 2014, 48, 1111–1123. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, M.; Kushwaha, M.; Makarana, G.; Yadav, M.R. Integrated use of organic and inorganic nutrient sources influences the nutrient content, uptake and nutrient use efficiencies of fodder oats (Avena sativa). Indian J. Agron. 2021, 66, 466–473. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Borowiec, M.L. Spruceup: fast and flexible identification, visualization, and removal of outliers from large multiple sequence alignments. J. Open-Source Softw. 2019, 4, 1635. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Martin, J.H. The whitefly fauna of Australia (Sternorrhyncha: Aleyrodidae). A taxonomic account and identification guide; CSIRO Entomology Technical Paper 38; CSIRO Entomology: Canberra, Australia, 1999; p. 197. [Google Scholar]
- Dubey, A.K.; Sundararaj, R. Whitefly species of the genus Aleurocanthus Quaintance & Baker (Hemiptera: Aleyrodidae) from India, with descriptions of six new species. Orient. Insects 2005, 39, 295–321. [Google Scholar] [CrossRef]
- Dubey, A.K.; Ko, C.C. Sexual dimorphism among species of Aleurocanthus Quaintance & Baker (Hemiptera: Aleyrodidae) in Taiwan, with one new species and an identification key. Zootaxa 2012, 3177, 1–23. [Google Scholar] [CrossRef]
- Huang, J.; Polaszek, A. A revision of the Chinese species of Encarsia Förster (Hymenoptera: Aphelinidae): parasitoids of whiteflies, scale insects and aphids (Hemiptera: Aleyrodidae, Diaspididae, Aphidoidea). J. Nat. Hist. 1998, 32, 1825–1966. [Google Scholar] [CrossRef]
- Viggiani, G. Le specie italiane del genere Encarsia Foerster (Hymenoptera: Aphelinidae). Boll. Lab. Entomol. Agrar. Filippo Silvestri Portici 1987, 44, 121–179. [Google Scholar]
| Specimen Code | Preliminary Identification | Year | Sampling site | Host Plant/Host |
|---|---|---|---|---|
| As GBP V1 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Vitis vinifera |
| As GBP V2 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Vitis vinifera |
| As GBP V3 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Vitis vinifera |
| As GBP V4 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Vitis vinifera |
| As GBP M1 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Pyrus communis |
| As GBP M2 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Pyrus communis |
| As GBP M3 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Pyrus communis |
| As GBP M4 | Aleurocanthus sp. | 2023 | Giardino Botanico La Pica San Felice sul Panaro |
Pyrus communis |
| As M226 1 | Aleurocanthus sp. | 2023 | Azienda Magnanini Carpi |
Pyrus communis |
| As M226 2 | Aleurocanthus sp. | 2023 | Azienda Magnanini Carpi |
Pyrus communis |
| As M226 3 | Aleurocanthus sp. | 2023 | Azienda Magnanini Carpi |
Pyrus communis |
| As bom 5 | Aleurocanthus sp. | 2018 | Bomporto | Orchard hedge |
| 25a | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| 25b | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| 41° | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| 53° | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| 53b | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| 56a | Encarsia sp. | 2022 | Giardino Botanico La Pica S. Felice sul Panaro |
Aleurocanthus sp. |
| median [IQR] | estimate | z value | P value | |
|---|---|---|---|---|
| Malus domestica | 1[0,2] | - | - | - |
| Pyrus communis | 4[1,8] | 1.45 | 22.21 | <0.001 *** |
| Vitis vinifera | 16[8,27] | 2.891 | 45.88 | <0.001 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





