Submitted:
03 July 2025
Posted:
04 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
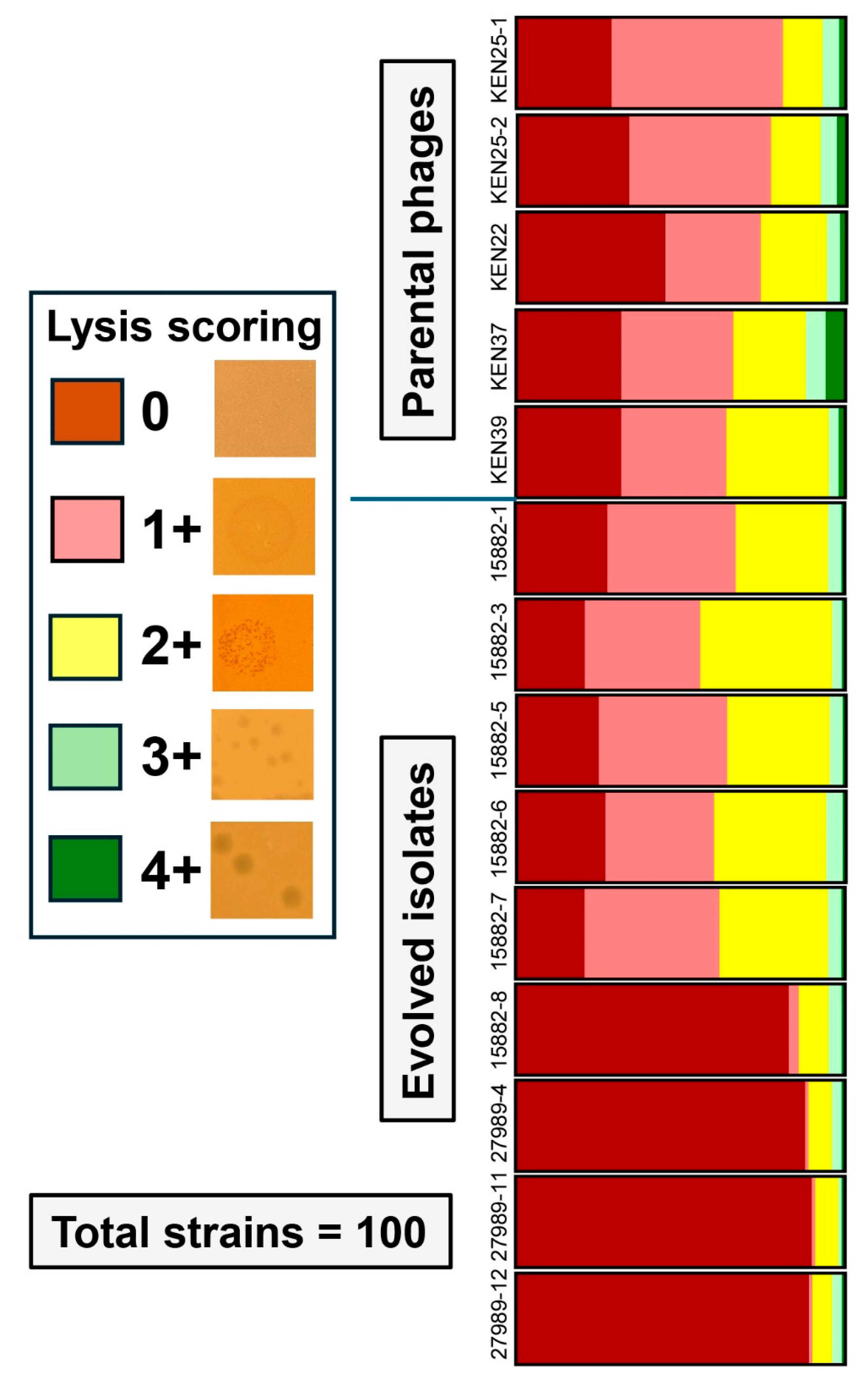
2.1. Training Led to Phage Variants with Altered Lytic Spectra
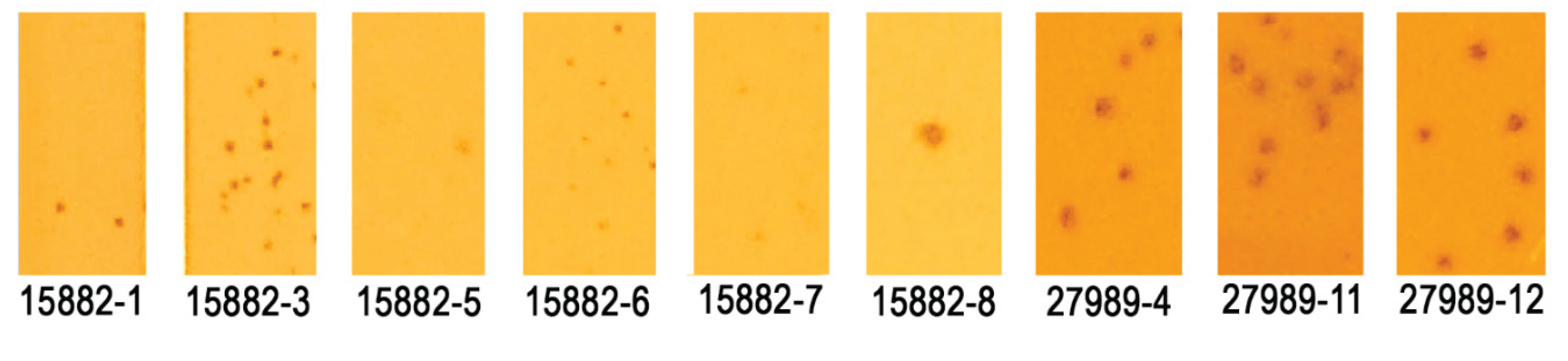
2.2. Serial Propagation on a Phage-susceptible K. pneumoniae Strain Resulted in Host Range Changes
| Score | Observation |
|---|---|
 |
No activity, no lysis (negative result). |
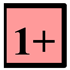 |
Lysis from without: very faint, turbid spots or clear spots in first dilutions, no plaque formation and no negative dynamics of lysis: lysis, lysis, then nothing (negative result). |
 |
Clear or turbid spots, tiny plaques, countable or uncountable, or lack of visible isolated plaques but clear negative dynamics of lysis intensity from lower to higher dilution (slightly positive result). |
 |
Clear spots, clear plaques of medium or small size (strictly positive result). |
 |
Totally clear spots, there are isolated large clear plaques in the highest phage dilutions (highly positive result). |
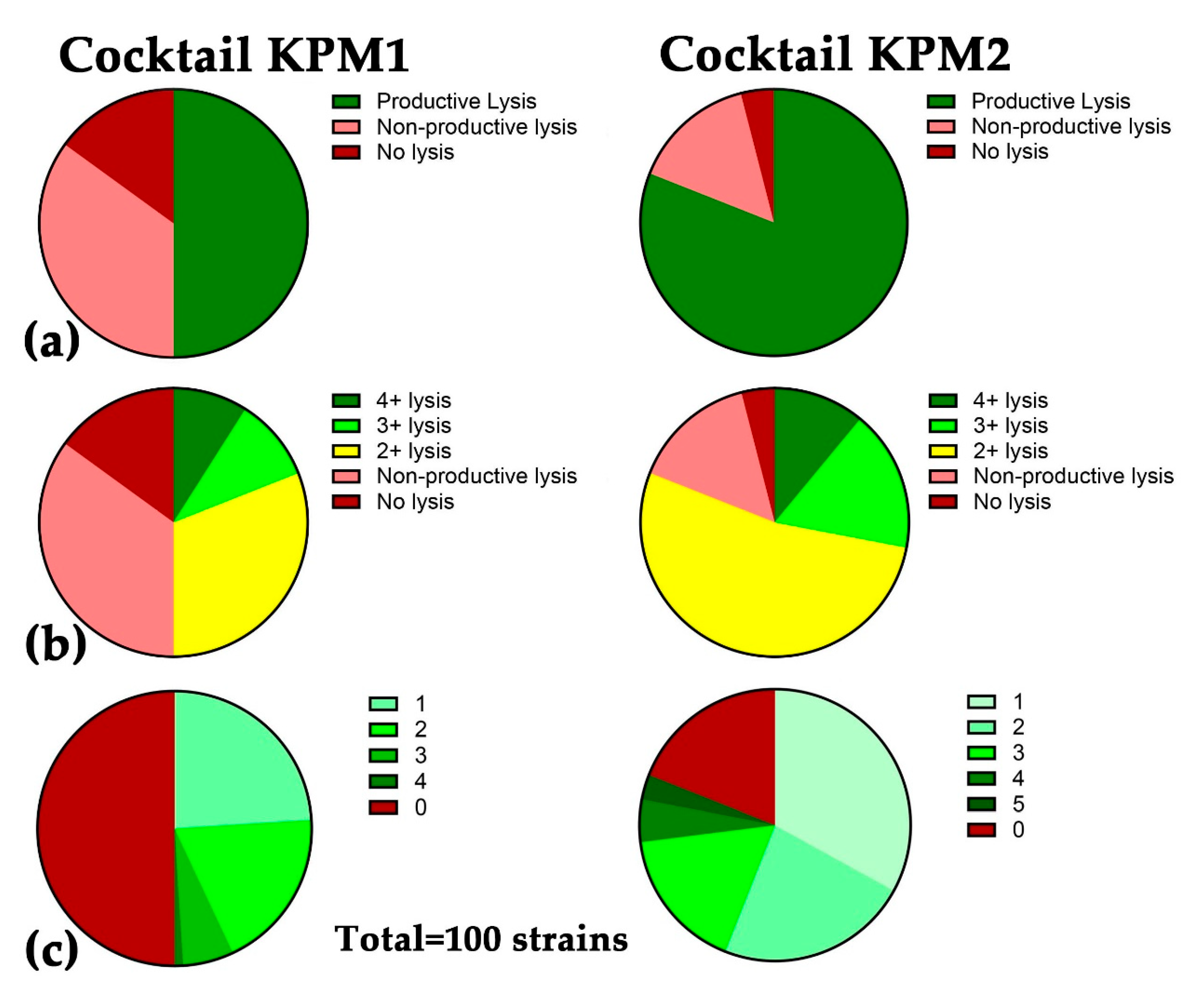
2.3. A Broad-host-range Cocktail Containing an Evolved Phage
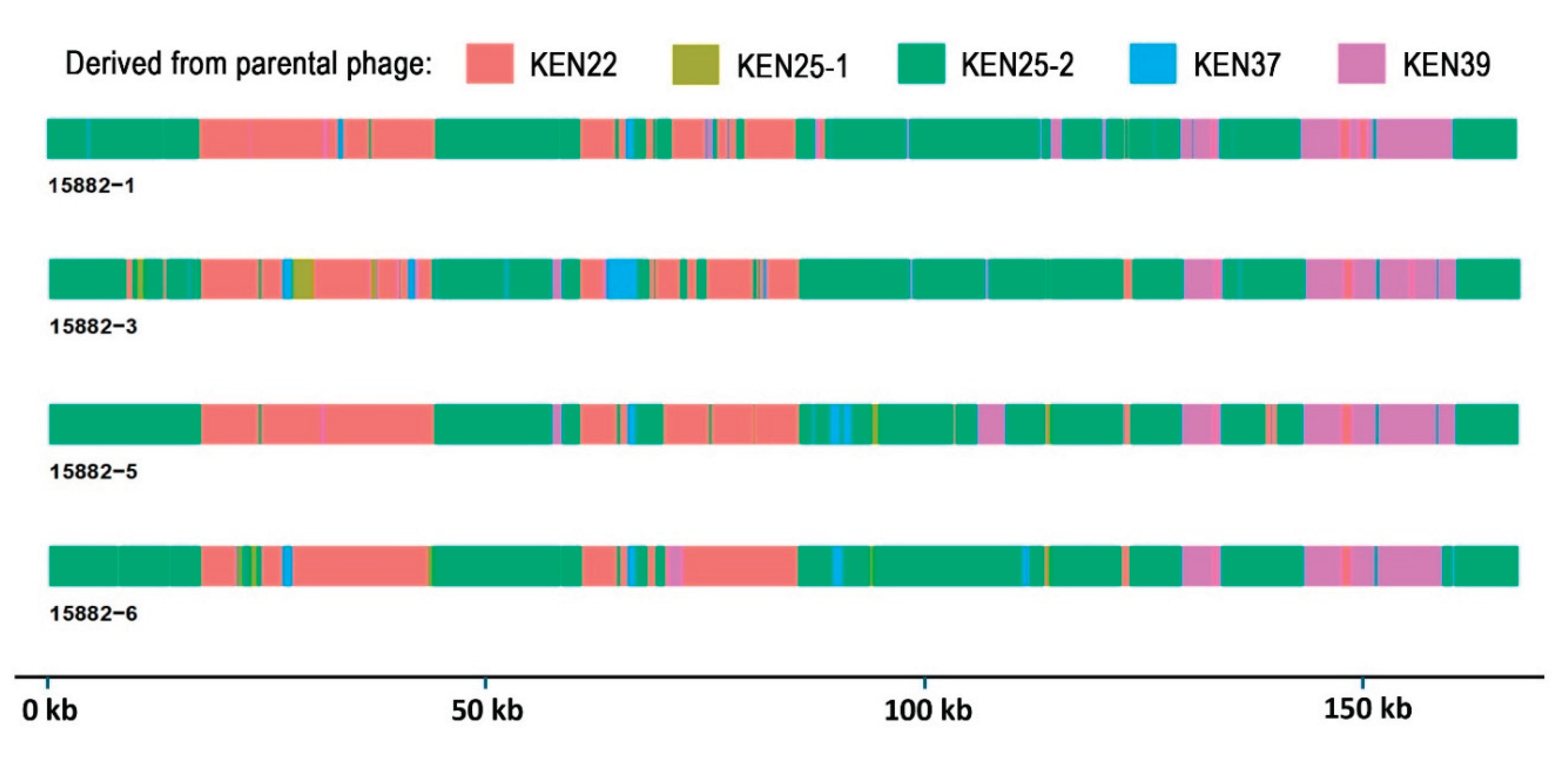
2.4. Sequence Analysis of Trained Phages Reveals Multiple Recombination Events and Accumulation of Mutations
2.5. Recombinant Events Resulted in Few Nonsynonymous Mutations Compared to Parental Phages
| KEN22 | KEN25-1 | KEN25-2 | KEN37 | KEN39 | 15882-1 | 15882-3 | 15882-5 | 15882-6 | |
|---|---|---|---|---|---|---|---|---|---|
| KEN22 | 92.94 | 90.04 | 94.03 | 90.72 | 94.38 | 94.21 | 94.77 | 94.40 | |
| KEN25-1 | 92.94 | 87.70 | 92.12 | 89.44 | 90.17 | 90.23 | 90.52 | 90.19 | |
| KEN25-2 | 90.04 | 87.70 | 91.72 | 88.94 | 94.43 | 94.18 | 93.89 | 94.44 | |
| KEN37 | 94.03 | 92.12 | 91.72 | 91.33 | 92.94 | 93.07 | 92.54 | 92.97 | |
| KEN39 | 90.72 | 89.44 | 88.94 | 91.33 | 89.77 | 89.92 | 90.21 | 89.71 | |
| 15882-1 | 94.38 | 90.17 | 94.43 | 92.94 | 89.77 | 99.34 | 99.15 | 99.65 | |
| 15882-3 | 94.21 | 90.23 | 94.18 | 93.07 | 89.92 | 99.34 | 99.15 | 99.45 | |
| 15882-5 | 94.77 | 90.52 | 93.89 | 92.54 | 90.21 | 99.15 | 99.15 | 99.27 | |
| 15882-6 | 94.40 | 90.19 | 94.44 | 92.97 | 89.71 | 99.65 | 99.45 | 99.27 |
3. Discussion
4. Materials and Methods
4.1. Bacterial strains, Phages, Growth and Storage Conditions
4.2. The Appelmans Training
4.3. Purification of Collected Trained Phage Variants
4.4. Host Range Determination
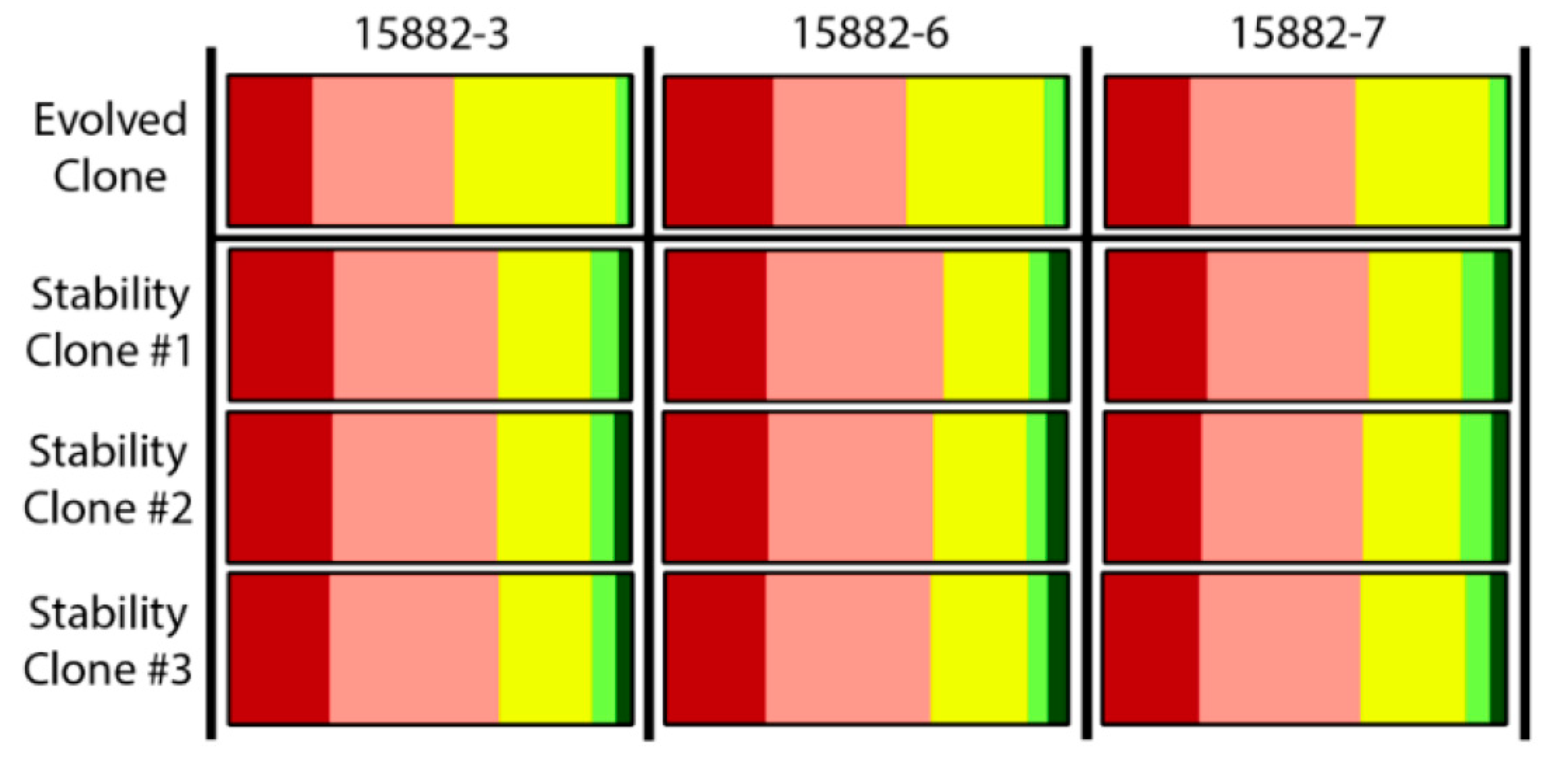
4.5. Assessment of Stability of Host Range Expansion
4.6. DNA Isolation, Library Preparation, Sequencing and Genome Assembly
4.7. Genome Annotation
4.8. Genome Variation Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDS | Protein coding sequence |
| HIB | Heart Infusion Broth with calcium chloride and magnesium sulfate |
| HIB-CM | Heart Infusion Broth with |
| KPM | Klebsiella phage mix |
| LPS | Lipopolysaccharide |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MDR | Multidrug-resistant |
| MIC | Minimum inhibitory concentration |
| MLST | Multilocus sequence typing |
| MRSN | Multidrug-Resistant Organism Repository and Surveillance Network |
| PPR | Pan-phage-resistant |
| PS | Phage-susceptible |
| SNP | Single nucleotide polymorphism |
| ST | Sequence type |
| UTI | Urinary tract infections |
| WRAIR | Walter Reed Army Institute of Research |
| XDR | Extensively drug-resistant |
| XPR | Extensively phage-resistant |
References
- Chang, D.; Sharma, L.; Dela Cruz, C.S.; Zhang, D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef]
- Kochan, T.J.; Nozick, S.H.; Medernach, R.L.; Cheung, B.H.; Gatesy, S.W.M.; Lebrun-Corbin, M.; Mitra, S.D.; Khalatyan, N.; Krapp, F.; Qi, C.; Ozer, E.A.; Hauser, A.R. Genomic surveillance for multidrug-resistant or hypervirulent Klebsiella pneumoniae among United States bloodstream isolates. BMC Infect. Dis. 2022, 22, 603. [Google Scholar] [CrossRef]
- Ullah, S.R.; Jamal, M.; Rahman, A.; Andleeb, S. Comprehensive insights into Klebsiella pneumoniae: unravelling clinical impact, epidemiological trends and antibiotic-resistance challenges. J. Antimicrob. Chemother. 2024, 79, 1484–1492. [Google Scholar] [CrossRef]
- Shoma, S.; Kamruzzaman, M.; Ginn, A.N.; Iredell, J.R.; Partridge, S.R. Characterization of multidrug-resistant Klebsiella pneumoniae from Australia carrying blaNDM-1. Diagn. Microbiol. Infect. Dis. 2014, 78, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Long, S.W.; Olsen, R.J.; Eagar, T.N.; Beres, S.B.; Zhao, P.; Davis, J.J.; Brettin, T.; Xia, F.; Musser, J.M. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: unexpected abundance of clonal group 307. mBio 2017, 8, e00489–17. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.; Macesic, N.; Tolari, G.; Guzman, A.; Uhlemann, A.-C. Multidrug-resistant Klebsiella pneumoniae ST307 in traveler returning from Puerto Rico to Dominican Republic. Emerg. Infect. Dis. 2019, 25, 1583–1585. [Google Scholar] [CrossRef]
- Heiden, S.E.; Hübner, N.O.; Bohnert, J.A.; Heidecke, C.-D.; Kramer, A.; Balau, V.; Gierer, W.; Schaefer, S.; Eckmanns, T.; Gatermann, S.; et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020, 12, 113. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Feldman, C.; Sekyere, J.O.; Maningi, N.E.; Modipane, L.; Essack, S.Y. Pathogenomics and evolutionary epidemiology of multi-drug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci. Rep. 2020, 10, 1232. [Google Scholar] [CrossRef]
- Martin, M.J.; Corey, B.W.; Sannio, F.; Hall, L.R.; MacDonald, U.; Jones, B.T.; Mills, E.G.; Harless, C.; Stam, J.; Maybank, R.; et al. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc. Natl. Acad. Sci. USA 2021, 118, e2110227118. [Google Scholar] [CrossRef]
- Salazar, C.; Antelo, V.; Vieytes, M.; Dávila, C.; Grill, F.; Galiana, A.; Iraola, G. First detection and origin of multi-drug resistant Klebsiella pneumoniae ST15 harboring OXA-48 in South America. J. Glob. Antimicrob. Resist. 2022, 30, 480–484. [Google Scholar] [CrossRef]
- Pham, M.H.; Hoi, L.T.; Beale, M.A.; Khokhar, F.A.; Hoa, N.T.; Musicha, P.; Blackwell, G.A.; Long, H.B.; Huong, D.T.; Binh, N.G.; et al. Evidence of widespread endemic populations of highly multidrug resistant Klebsiella pneumoniae in hospital settings in Hanoi, Vietnam: a prospective cohort study. Lancet Microbe 2023, 4, e255–e263. [Google Scholar] [CrossRef]
- Zarkotou, O.; Pournaras, S.; Tselioti, P.; Dragoumanos, V.; Pitiriga, V.; Ranellou, K.; Prekates, A.; Themeli-Digalaki, K.; Tsakris, A. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin. Microbiol. Infect. 2011, 17, 1798–1803. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Sun, X.; Ma, X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 18. [Google Scholar] [CrossRef]
- Karlsson, M.; Stanton, R.A.; Ansari, U.; McAllister, G.; Chan, M.Y.; Grass, S.E.; Duffy, N.; Anacker, M.L.; Witwer, M.L.; Rasheed, J.K.; et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob. Agents Chemother. 2019, 63, e00519–19. [Google Scholar] [CrossRef]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ding, Y.; Xu, Y.; Li, Z.; Zeng, Z.; Liu, J. An outbreak of extensively drug-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a teaching hospital in Southwest China. Front. Cell. Infect. Microbiol. 2022, 12, 979219. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Liao, X. Hypervirulent Klebsiella pneumoniae. Infect. Drug Resist. 2023, 16, 5243–5249. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, S.; Ruth, M.M.; Mientjes, M.; de Sévaux, R.G.L.; van Ingen, J. A Dutch case report of successful treatment of chronic relapsing urinary tract infection with bacteriophages in a renal transplant patient. Antimicrob. Agents Chemother. 2019, 64, e01281–19. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J.; et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef]
- Cano, E.J.; Caflisch, K.M.; Bollyky, P.L.; Van Belleghem, J.D.; Patel, R.; Fackler, J.; Brownstein, M.J.; Horne, B.; Biswas, B.; Henry, M.; et al. Phage therapy for limb-threatening prosthetic knee Klebsiella pneumoniae infection: case report and in vitro characterization of anti-biofilm activity. Clin. Infect. Dis. 2021, 73, e144–e151. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Fu, H.; Tian, Z.; Cui, J.; Yan, C.; Xue, G.; Fan, Z.; Du, B.; Feng, J.; Zhao, H.; et al. Bacteriophage effectively rescues pneumonia caused by prevalent multidrug-resistant Klebsiella pneumoniae in the early stage. Microbiol. Spectr. 2022, 10, e0235822. [Google Scholar] [CrossRef]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Ichikawa, M.; Nakamoto, N.; Kredo-Russo, S.; Weinstock, E.; Weiner, I.N.; Khabra, E.; Ben-Ishai, N.; Inbar, D.; Kowalsman, N.; Mordoch, R.; et al. Bacteriophage therapy against pathological Klebsiella pneumoniae ameliorates the course of primary sclerosing cholangitis. Nat. Commun. 2023, 14, 3261. [Google Scholar] [CrossRef]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016, 24, 944–956. [Google Scholar] [CrossRef]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Lusiak-Szelachowska, M.; Zaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 100. [Google Scholar] [CrossRef]
- Lin, T.-L.; Hsieh, P.-F.; Huang, Y.-T.; Lee, W.-C.; Tsai, Y.-T.; Su, P.-A.; Pan, Y.-J.; Hsu, C.-R.; Wu, M.-C.; Wang, J.-T. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 2014, 210, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, J.; Guo, C.; Ge, C.; Wang, X.; Wei, D.; Li, X.; Si, H.; Hu, C. Identification and complete genome of lytic "Kp34likevirus" phage vB_KpnP_Bp5 and therapeutic potency in the treatment of lethal Klebsiella pneumoniae infections in mice. Virus Res. 2021, 297, 198348. [Google Scholar] [CrossRef]
- Ferriol-González, C.; Concha-Eloko, R.; Bernabéu-Gimeno, M.; Fernández-Cuenca, F.; Cañada-García, J.E.; García-Cobos, S.; Sanjuán, R.; Domingo-Calap, P. Targeted phage hunting to specific Klebsiella pneumoniae clinical isolates is an efficient antibiotic resistance and infection control strategy. Microbiol. Spectr. 2024, 12, e00254–24. [Google Scholar] [CrossRef]
- Martin, M.J.; Stribling, W.; Ong, A.C.; Maybank, R.; Kwak, Y.I.; Rosado-Mendez, J.A.; Preston, L.N.; Lane, K.F.; Julius, M.; Jones, A.R.; et al. A panel of diverse Klebsiella pneumoniae clinical isolates for research and development. Microb. Genom. 2023, 9, mgen000967. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Pirnay, J.P.; De Vos, D. Guidelines to compose an ideal bacteriophage cocktail. Methods Mol. Biol. 2018, 1693, 99–110. [Google Scholar] [CrossRef]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in vitro evolution of therapeutic bacteriophages: the Appelmans protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef]
- Peters, T.L.; Urick, C.D.; Georges, M.; Burke, K.A.; Kirillina, O.A.; Mzhavia, N.; Musila, L.; Filippov, A.A.; Nikolich, M.P. Genome sequences of five Klebsiella bacteriophages that belong to the genus Jiaodavirus. Microbiol. Resour. Announc. 2024, 13, e0105624. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Gannon, L.; Muscatt, G.; Dunstan, R.; Michniewski, S.; Sapkota, H.; Kiljunen, S.J.; Kolsi, A.; Skurnik, M.; et al. Isolation and characterization of Klebsiella phages for phage therapy. Phage (New Rochelle) 2021, 2, 26–42. [Google Scholar] [CrossRef]
- Bird, J.T.; Burke, K.A.; Urick, C.D.; Braverman, J.L.; Mzhavia, N.; Ellison, D.W.; Nikolich, M.P.; Filippov, A.A. Genome sequence of the Klebsiella quasipneumoniae bacteriophage EKq1 with activity against Klebsiella pneumoniae. Microbiol. Resour. Announc. 2024, 13, e0095423. [Google Scholar] [CrossRef]
- Raaijmakers, H.; Törö, I.; Birkenbihl, R.; Kemper, B.; Suck, D. Conformational flexibility in T4 endonuclease VII revealed by crystallography: implications for substrate binding and cleavage. J. Mol. Biol. 2001, 302, 2. [Google Scholar] [CrossRef]
- Granel,l M. ; Namura, M.; Alvira, S.; Kanamaru, S.; Van Raaij, M.J. Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fiber protein Gp34. Viruses 2017, 9, 168. [Google Scholar] [CrossRef]
- Ayala, R.; Moiseenko, A.V.; Chen, T.H.; Eugene, E.E.; Golomidova, A.K.; Orekhov, P.S.; Street, M.A.; Sokolova, O.S.; Letarov, A.V.; Wolf, M. Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Nat. Commun. 2023, 14, 8205. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Bittrich, S.; Segura, J.; Duarte, J.M.; Burley, S.K.; Rose, Y. RCSB protein Data Bank: exploring protein 3D similarities via comprehensive structural alignments. Bioinformatics 2024, 40, btae370. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, W.; Xiao, H.; Yang, F.; Song, J; Cheng, L. ; Liu, H. Asymmetric structure of podophage GP4 reveals a novel architecture of three types of tail fibers. J. Mol. Biol. 2023, 435, 168258. [Google Scholar] [CrossRef]
- Garcia-Doval, C.; Castón, J.R.; Luque, D.; Granell, M.; Otero, J.M.; Llamas-Saiz, A.L.; Renouard, M.; Boulanger, P.; Van Raaij, M.J. Structure of the receptor-binding carboxy-terminal domain of the bacteriophage T5 L-shaped tail fibre with and without its intra-molecular chaperone. Viruses 2015, 7, 6424–6440. [Google Scholar] [CrossRef]
- Granell, M.; Namura, M.; Alvira, S.; Kanamaru, S.; Van Raaij, M.J. Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fiber protein gp34. Viruses 2017, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Kanamaru, S.; Ishiwata, Y.; Suzuki, T.; Rossmann, M.G.; Arisaka, F. Control of bacteriophage T4 tail lysozyme activity during the infection process. J. Mol. Biol. 2005, 346, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
- Follador, R.; Heinz, E.; Wyres, K.L.; Ellington, M.J.; Kowarik, M.; Holt, K.E.; Thomson, N.R. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016, 2, e000073. [Google Scholar] [CrossRef]
- Hesse, S.; Rajaure, M.; Wall, E.; Johnson, J.; Bliskovsky, V.; Gottesman, S.; Adhya, S. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. mBio 2020, 11, e02530–19. [Google Scholar] [CrossRef]
- Beamud, B.; García-González, N.; Gómez-Ortega, M.; González-Candelas, F.; Domingo-Calap, P.; Sanjuan, R. Genetic determinants of host tropism in Klebsiella phages. Cell Rep. 2023, 42, 112048. [Google Scholar] [CrossRef]
- Domingo-Calap, P.; Beamud, B.; Mora-Quilis, L.; González-Candelas, F.; Sanjuán, R. Isolation and characterization of two Klebsiella pneumoniae phages encoding divergent depolymerases. Int. J. Mol. Sci. 2020, 21, 3160. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Latka, A.; Berisio, R.; Squeglia, F.; Maciejewska, B.; Briers, Y.; Drulis-Kawa, Z. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front. Microbiol. 2018, 9, 2517. [Google Scholar] [CrossRef]
- Wu, J.W.; Wang, J.-T.; Lin, T.-L.; Liu, Y.-Z.; Wu, L.-T.; Pan, Y.-J. Identification of three capsule depolymerases in a bacteriophage infecting Klebsiella pneumoniae capsular types K7, K20, and K27 and therapeutic application. J. Biomed. Sci. 2023, 30, 31. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Chen, L.; Guo, G.; Li, P.; Ma, J.; Chen, R.; Du, H.; Liu, Y.; Zhang, W. Identification of a phage-derived depolymerase specific for KL47 capsule of Klebsiella pneumoniae and its therapeutic potential in mice. Virol. Sin. 2022, m37, 538–546. [Google Scholar] [CrossRef]
- Pan, Y.-J.; Lin, T.-L.; Chen, C.-C.; Tsai, Y.-T.; Cheng, Y.-H.; Chen, Y.-Y.; Hsieh, P.-F.; Lin, Y.-T.; Wang, J.-T. Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 2017, 91, e02457–16. [Google Scholar] [CrossRef]
- Lourenco, M.; Osbelt, L.; Passet, V.; Gravey, F.; Megrian, D.; Strowig, T.; Rodrigues, C.; Brisse, S. Phages against noncapsulated Klebsiella pneumoniae: broader host range, slower resistance. Microbiol. Spectr. 2023, 11, e0481222. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Mackiewicz, P.; Kęsik-Szeloch, A.; Maciaszczyk-Dziubinska, E.; Weber-Dąbrowska, B.; Dorotkiewicz-Jach, A.; Augustyniak, D.; Majkowska-Skrobek, G.; Bocer, T.; Empel, J.; et al. Isolation and characterisation of KP34 – a novel ɸKMV-like bacteriophage for Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2011, 90, 1333–1345. [Google Scholar] [CrossRef]
- Wu, L.-T.; Chang, S.-Y.; Yen, M.-R.; Yang, T.-C.; Tseng, Y.-H. Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl. Environ. Microbiol. 2007, 73, 2532–2540. [Google Scholar] [CrossRef]
- Karumidze, N.; Kusradze, I.; Rigvava, S.; Goderdzishvili, M.; Rajakumar, K.; Alavidze, Z. Isolation and characterisation of lytic bacteriophages of Klebsiella pneumoniae and Klebsiella oxytoca. Curr. Microbiol. 2013, 66, 251–258. [Google Scholar] [CrossRef]
- Li, M.; Guo, M.; Chen, L.; Zhu, C.; Xiao, Y.; Li, P.; Guo, H.; Chen, L.; Zhang, W.; Du, H. Isolation and characterization of novel lytic bacteriophages infecting epidemic carbapenem-resistant Klebsiella pneumoniae strains. Front. Microbiol. 2020, 1, 1554. [Google Scholar] [CrossRef]
- Hall, J.P.J.; Harrison, E.; Brockhurst, M.A. Viral host-adaptation: insights from evolution experiments with phages. Curr. Opin. Virol. 2013, 3, 572–577. [Google Scholar] [CrossRef]
- Appelmans, R. Le dosage du bacteriophage. Compt. Rend. Soc. Biol. 1921, 85, 1098–1099. [Google Scholar]
- O'Flaherty, S.; Ross, R.P.; Meaney, W.; Fitzgerald, G.F.; Elbreki, M.F.; Coffey, A. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl. Environ. Microbiol. 2005, 71, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Kvachadze, L.; Balarjishvili, N.; Meskhi, T.; Tevdoradze, E.; Skhirtladze, N.; Pataridze, T.; Adamia, R.; Topuria, T.; Kutter, E.; Rohde, C.; et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb. Biotechnol. 2011, 4, 643–650. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Farlow, J.; Su, W.; Kvachadze, L.; Balarjishvili, N.; Kutateladze, M.; Nikolich, M.P. Correlation of host range expansion of therapeutic bacteriophage Sb-1 with allele state at a hypervariable repeat locus. Appl. Environ. Microbiol. 2019, 85, e01209–19. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, C.; Bonhomme, M.; Medina, M.; Pouilly, M.; Rousseau, C.; Troesch, E.; Martins-Simoes, P.; Stegger, M.; Verhoeven, P.O.; Laumay, F.; et al. Potential of training of anti-Staphylococcus aureus therapeutic phages against Staphylococcus epidermidis multidrug-resistant isolates is restricted by inter- and intra-sequence type specificity. mSystems 2024, 9, e0085024. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B. Analysis of the Appelmans protocol for the generation of therapeutic bacteriophages. Ph.D. thesis, Texas Tech Univeristy Health Sciences Center, Lubbock, Texas, USA, 2011.
- Mapes, A.C.; Trautner, B.W.; Liao, K.S.; Ramig, R.F. Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 2016, 6, e1096995. [Google Scholar] [CrossRef]
- Lossouarn, J.; Beurrier, E.; Bouteau, A.; Moncaut, E.; Sir Silmane, M.; Portalier, H.; Zouari, A.; Cattoir, V.; Serror, P.; Petit, M.-A. The virtue of training: extending phage host spectra against vancomycin-resistant Enterococcus faecium strains using the Appelmans method. Antimicrob. Agents Chemother. 2024, 68, e0143923. [Google Scholar] [CrossRef]
- Blasco, L.; Bleriot, I.; González de Aledo, M.; Fernández-García, L.; Pacios, O.; Oliveira, H.; López, M.; Ortiz-Cartagena, C.; Fernández-Cuenca, F.; Pascual, Á.; et al. Development of an anti-Acinetobacter baumannii biofilm phage cocktail: genomic adaptation to the host. Antimicrob. Agents Chemother. 2022, 66, e0192321. [Google Scholar] [CrossRef]
- Peters, T.L.; Song, Y.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Mutant and recombinant phages selected from in vitro coevolution conditions overcome phage-resistant Listeria monocytogenes. Appl. Environ. Microbiol. 2020, 86, e02138–20. [Google Scholar] [CrossRef]
- Sybesma, W.; Zbinden, R.; Chanishvili, N.; Kutateladze, M.; Chkhotua, A.; Ujmajuridze, A.; Mehnert, U.; Kessler, T.M. Bacteriophages as potential treatment for urinary tract infections. Front. Microbiol. 2016, 7, 465. [Google Scholar] [CrossRef]
- Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Leitner, L.; Mehnert, U.; Chkhotua, A.; Kessler, T.M.; Sybesma, W. Adapted bacteriophages for treating urinary tract infections. Front. Microbiol. 2018, 9, 1832. [Google Scholar] [CrossRef]
- Vu, T.N.; Clark, J.R.; Jang, E.; D'Souza, R.; Nguyen, L.P.; Pinto, N.A.; Yoo, S.; Abadie, R.; Maresso, A.W.; Yong, D. Appelmans protocol - a directed in vitro evolution enables induction and recombination of prophages with expanded host range. Virus Res. 2024, 339, 199272. [Google Scholar] [CrossRef]
- Peters, T.L.; Schow, J.; Spencer, E.; Van Leuven, J.T.; Wichman, H.; Miller, C. Directed evolution of bacteriophages: thwarted by prolific prophage. Appl. Environ. Microbiol. 2024, 90, e0088424. [Google Scholar] [CrossRef]
- Jakob, N.; Hammerl, J.A.; Swierczewski, B.E.; Würstle, S.; Bugert, J.J. Appelmans protocol for in vitro Klebsiella pneumoniae phage host range expansion leads to induction of the novel temperate linear plasmid prophage vB_KpnS-KpLi5. Virus Genes 2025, 61, 132–135. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.P.; De Vos, D. Guidelines to compose an ideal bacteriophage cocktail. Methods Mol. Biol. 2024, 2734, 49–66. [Google Scholar] [CrossRef]
- Nikolich, M.P.; Filippov, A.A. Bacteriophage therapy: developments and directions. Antibiotics (Basel) 2020, 9, 135. [Google Scholar] [CrossRef]
- Mencke, J.L.; He, Y.; Filippov, A.A.; Nikolich, M.P.; Belew, A.T.; Fouts, D.E.; McGann, P.T.; Swierczewski, B.E.; Getnet, D.; Ellison, D.W.; et al. Identification and characterization of vB_PreP_EPr2, a lytic bacteriophage of pan-drug resistant Providencia rettgeri. Viruses 2022, 14, 708. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017; 7, 8292. [CrossRef]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef]
- McNair, K.; Zhou, C.; Dinsdale, E.A.; Souza, B.; Edwards, R.A. PHANOTATE: a novel approach to gene identification in phage genomes. Bioinformatics 2019, 35, 4537–4542. [Google Scholar] [CrossRef] [PubMed]
- Terzian, P.; Olo Ndela, E.; Galiez, C.; Lossouarn, J.; Pérez Bucio, R.E.; Mom, R.; Toussaint, A.; Petit, M.-A.; Enault, F. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genomics and Bioinformatics 2021, 3, 1–12. [Google Scholar] [CrossRef]
- Chen, L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.; Cheng, A.A.; Liu, S.; et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Larralde, M.; Zeller, G. PyHMMER: a Python library binding to HMMER for efficient sequence analysis. Bioinformatics 2023, 39, btad214. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 2007, 8, 209. [Google Scholar] [CrossRef]
- Cook, R.; Brown, N.; Redgwell, T.; Rihtman, B.; Barnes, M.; Clokie, M.; Stekel, D.J.; Hobman, J.; Jones, M.A.; Millard, A. INfrastructure for a PHAge REference Database: identification of large-scale biases in the current collection of cultured phage genomes. Phage (New Rochelle) 2021, 2, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Khelik, K.; Lagesen, K.; Sandve, G.K.; Rognes, T.; Nederbragt, A.J. NucDiff: in-depth characterization and annotation of differences between two sets of DNA sequences. BMC Bioinformatics 2017, 18, 338. [Google Scholar] [CrossRef]
- Hackl, T.; Ankenbrand, M.; van Adrichem, B.; Wilkins, D.; Haslinger, K. gggenomes: effective and versatile visualizations for comparative genomics. arXiv:2411.13556. https://thackl.github.io/gggenomes/authors.html.
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucl. Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
| Strain | Sample type | Antibiotic susceptibility 1 | Phage susceptibility 2 | Sequence type | KL serotype | OL serotype |
|---|---|---|---|---|---|---|
| MRSN 4759 | Urine | MDR | XPR | ST37 | KL38 | O3b |
| MRSN 6778 | Urine | MDR | XPR | ST1842 | KL3 | O2v2 |
| MRSN 15687 | Urine | MDR | XPR | ST5446 | KL62 | O2v1 |
| MRSN 15882 | Perianal | MDR | XPR | ST1686 | KL113 | O1v1 |
| MRSN 22232 | Respiratory | XDR | XPR | ST405 | KL151 | O4 |
| MRSN 27989 | Wound | MDR | XPR | ST2279 | KL3 | O2v2 |
| MRSN 479404 | Wound | XDR | XPR | ST16 | KL51 | O3b |
| MRSN 511348 | Unknown | XDR | XPR | ST14 | KL2 | O1v1 |
| MRSN 614201 | Environ. | MDR | PPR | ST1838 | KL14 | O3b |
| MRSN 681054 | Urine | XDR | PPR | ST340 | KL15 | O4 |
| MRSN 414780 | Urine | XDR | PS | ST323 | KL21 | O3b |
| Phage | Propagation strain | Genome length, bp | Family | Genus | Host range, % |
|---|---|---|---|---|---|
| KEN22 | Kp. MRSN 11382 | 166,645 | Straboviridae | Jiaodavirus | 26 |
| KEN25-1 | Kp MRSN 529046 | 169,768 | Straboviridae | Jiaodavirus | 19 |
| KEN25-2 | Kp MRSN 529046 | 165,574 | Straboviridae | Jiaodavirus | 23 |
| KEN37 | Kp MRSN 529046 | 166,503 | Straboviridae | Jiaodavirus | 34 |
| KEN39 | Kp MRSN 529046 | 166,254 | Straboviridae | Jiaodavirus | 36 |
| KEN42 | Kp MRSN 3619 | 38,200 | Autographiviridae | Teetrevirus | 18 |
| KEN1821 | Kp MRSN 529046 | 168,619 | Straboviridae | Jiaodavirus | 36 |
| AFR4 | Kp MRSN 3619 | 48,962 | Drexlerviridae | Webervirus | 17 |
| EKq1 | Kq MRSN 829456 | 48,244 | Fmr. Siphoviridae* | Unclass. | 15 |
| EKq2 | Kq MRSN 829456 | 51,496 | Drexlerviridae | Webervirus | 7 |
| Phage | Host range | Expansion? |
|---|---|---|
| KEN22 | 26 | Parental phage |
| KEN25-1 | 19 | Parental phage |
| KEN25-2 | 23 | Parental phage |
| KEN37 | 34 | Parental phage |
| KEN39 | 36 | Parental phage |
| 15882-1 | 33 | No |
| 15882-3 | 44 | Yes |
| 15882-5 | 36 | No |
| 15882-6 | 40 | Yes |
| 15882-7 | 38 | Yes |
| 15882-8 | 14 | No |
| 27989-4 | 11 | No |
| 27989-11 | 9 | No |
| 27989-12 | 10 | No |
| Cocktail | Phage ID | Genome size, bp | Family | Genus | Host Range | Mix host range |
|---|---|---|---|---|---|---|
| KPM1 | AFR4 | 48,962 | Drexlerviridae | Webervirus | 17% | 50% |
| KEN39 | 166,254 | Straboviridae | Jiaodavirus | 36% | ||
| KEN42 | 38,200 | Autographiviridae | Teetrevirus | 18% | ||
| EKq1 | 48,244 | Fmr. Siphoviridae* | Unclass. | 15% | ||
| EKq2 | 51,496 | Drexlerviridae | Webervirus | 7% | ||
| KPM2 | AFR4 | 48,962 | Drexlerviridae | Webervirus | 17% | 81% |
| KEN39 | 166,254 | Straboviridae | Jiaodavirus | 36% | ||
| KEN42 | 38,200 | Autographiviridae | Teetrevirus | 18% | ||
| EKq1 | 48,244 | Fmr. Siphoviridae* | Unclass. | 15% | ||
| EKq2 | 51,496 | Drexlerviridae | Webervirus | 7% | ||
| KEN1821 | 168,619 | Straboviridae | Jiaodavirus | 36% | ||
| 15882-3 | 167,537 | Straboviridae | Jiaodavirus | 44% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





