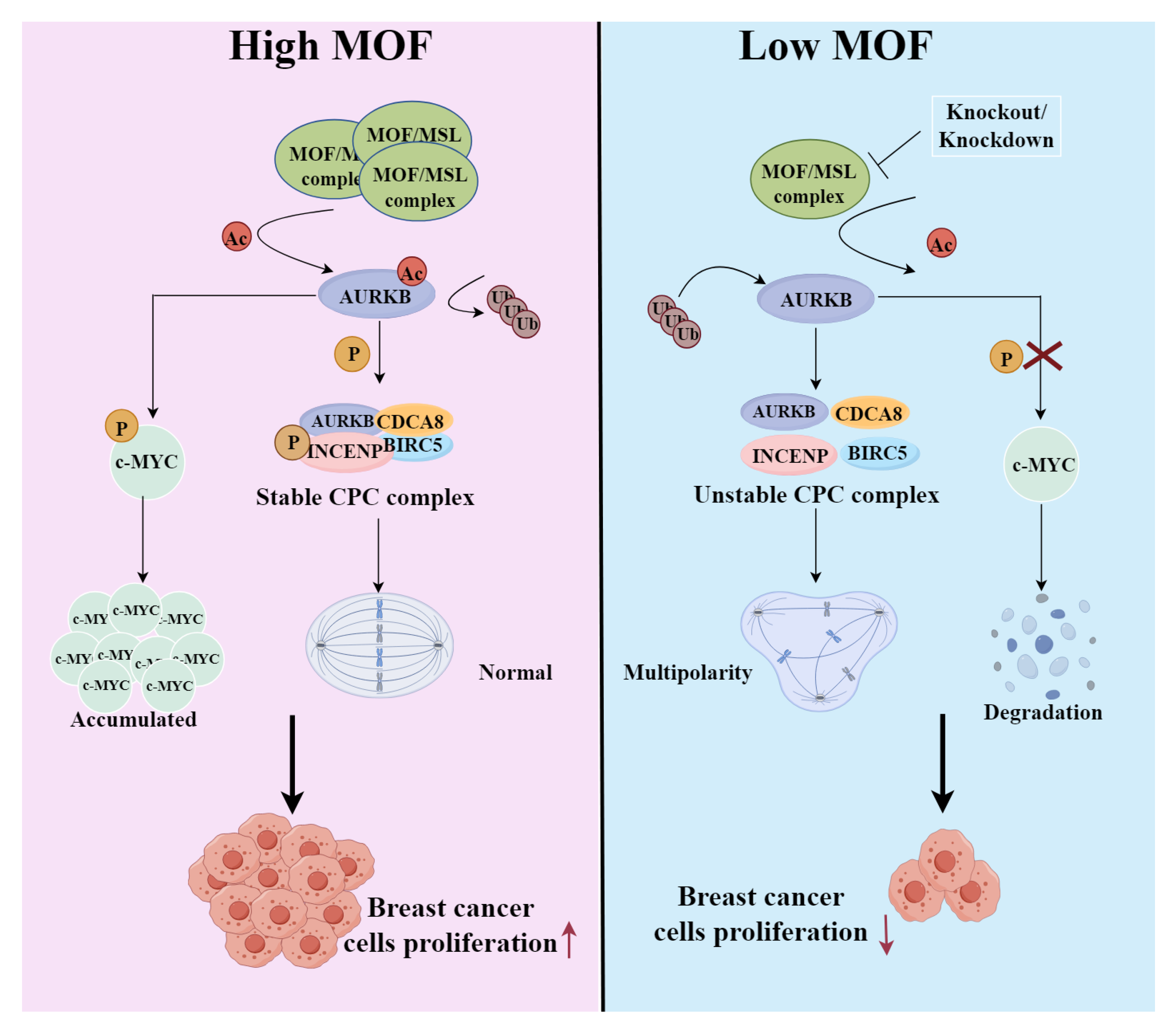
1. Introduction
The Mitosis is a fundamental biological process orchestrated by a complex network of proteins, among which Aurora kinase B (AURKB) plays a pivotal role in mitotic regulation and cell cycle progression [
1]. Aurora kinases, a family of three highly homologous serine/threonine kinases—Aurora A, Aurora B, and Aurora C—execute distinct yet interconnected functions in mitotic control. AURKB, also known by various aliases, including AIK2, AIM1, ARK2, AurB, IPL1, and STK5, was first identified as a cell cycle-dependent kinase in NIH3T3 cells, highlighting its role in cell proliferation [
2].
AURKB is a core component of the chromosome passenger complex (CPC) [
3], functioning in concert with the scaffolding protein inner centromere protein (INCENP) and the non-enzymatic subunits Survivin (BIRC5) and Borealin (CDCA8). The CPC ensures faithful chromosome segregation by dynamically localizing to specific mitotic structures at different stages [
4,
5]. Precise regulation of AURKB-mediated substrate phosphorylation is essential for mitotic progression [
6], particularly during early mitosis, where AURKB phosphorylates kinetochore substrates to correct erroneous microtubule attachments [
7,
8]. As mitosis progresses, the CPC targets additional substrates, coordinating distinct mitotic events. AURKB translocates from metaphase kinetochores to the central spindle during anaphase and subsequently to the contractile ring during telophase [
9,
10,
11], ensuring successful cytokinesis. Aberrant AURKB expression is frequently observed in various cancers, where it is associated with chromosomal instability and heightened malignancy [
12]. Dysregulated AURKB is thought to confer a proliferative advantage to cancer cells [
13].
MOF (males absent on the first), a histone acetyltransferase (HAT) of the MYST family, was initially identified as part of the Drosophila dosage compensation complex [
14,
15]. In humans, MOF shares structural features with its Drosophila homolog dMOF, including a MYST catalytic domain, a chromatin domain, and a C2HC-type zinc finger [
16]. MOF forms two distinct complexes: the male-specific lethal (MSL) complex, which primary acetylates histone H4K16, and the non-specific lethal (NSL) complex, which acetylates histones H4K5, H4K8, and H4K16 [
17,
18]. Beyond histone modification, human MOF plays critical roles in transcriptional regulation, chromatin dynamics, cell proliferation, differentiation, and DNA damage repair [
19,
20,
21]. Increasing evidence suggests that MOF is implicated in tumorigenesis, influencing cancer cell proliferation, apoptosis, and stemness [
22]. For instance, MOF overexpression in non-small cell lung cancer (NSCLC) promotes tumor progression by acetylating Nrf2, thereby contributing to poor prognosis and therapeutic resistance [
23]. Additionally, MOF-mediated acetylation of MDM2 has been linked to cisplatin resistance in ovarian cancer [
24].
Post-translational modifications (PTMs) fine-tune AURKB activity and localization during mitosis, with ubiquitin-mediated degradation being a well-established regulatory mechanism [
25,
26]. Emerging studies suggest that HATs and histone deacetylases (HDACs) modulate AURKB in cancer. In oesophageal cancer, BRD4, a histone acetylation reader, is recruited to the promoters of AURKA and AURKB, while its inhibition by JQ1 induces senescence [
27]. In lymphoma cells, AURKB and HDACs cooperatively regulate proliferation, with inhibition of either trigging cell cycle arrest and apoptosis [
28].
However, direct acetylation of AURKB remains poorly understood. A 2016 study reported TIP60 (KAT5)-dependent AURKB acetylation enhances its kinase activity, ensuring proper mitotic procession and genomic stability [
29]. Similarly, the MOF-containing MSL complex modulates YY1 stability and transcriptional activity via acetylation [
30]. Whether MOF directly acetylates AURKB and how this modification influences AURKB stability and cancer progression remain unknown. Here, we demonstrate that MOF-mediated acetylation of AURKB enhances its stability and activity. Specifically, MOF-driven AURKB acetylation promotes c-MYC accumulation, thereby facilitating malignant proliferation in breast cancer cells.
2. Materials and Methods
2.1. Antibodies and Reagents
The following antibodies were used in this study: Anti-MSL1 (mouse monoclonal, 24373-1-AP) from Proteintech (Wuhan, China); anti-MOF (rabbit polyclonal, A3390) from ABclonal Technology (Wuhan, China); anti-MSL2 (rabbit polyclonal, ab83911) from Abcam (Shanghai, China); anti-AURKB (rabbit monoclonal, AF1930) from Beyotime (Shanghai, China); anti-c-MYC (9E10), anti-INCENP (sc-376514), anti-CDCA8 (sc-376635) and anti-BIRC5 (sc-17779), anti-Akt (sc-81434), anti-mTOR (sc-517464), anti-ERK (sc-135900), anti-C-jun (sc-166540), anti-MAPK (sc-7972) (mouse monoclonal antibodies), as well as anti-CyclinB1 (sc-752), anti-E-Cadherin (sc-59778) and anti-N-Cadherin (sc-393933) (rabbit polyclonal antibodies), all from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-HA (RLM3003) and anti-H3 (RLM3038) mouse monoclonal antibodies were obtained from Ruiying Biological (Suzhou, China). Anti-H3S10Ph (mouse monoclonal, #26436) was sourced from Upstate (New York USA). Anti-Flag (M2) (A2220), anti-Myc (M2)-agarose (A7470), anti-Flag M2 (mouse monoclonal, F3165), and anti-H4K16ac (H9164) (rabbit polyclonal) antibodies were obtained from Sigma (St. Louis, MO, USA). Anti-c-MYC-T58Ph (rabbit polyclonal) was from Bioss (Beijing, China). Pan-acetylation (Pan-ac, PTM0105RM) (rabbit polyclonal) antibody was from Jingjie Biotechnology (Hangzhou, China). Anti-β-Tubulin (M30109), anti-PanPh (M210030F) (mouse monoclonal), and anti-Ki67 (TW0001F) (rabbit monoclonal) antibodies were purchased from Abmart (Shanghai China). Anti-His (GB151251-100) (mouse monoclonal) antibody was from Servicebio (Wuhan, China). Anti-MSL3L1, and anti-GAPDH (rabbit polyclonal) antibodies were raised against bacterially expressed proteins at Jilin University.
The following reagents were used: cycloheximide (CHX, DH466-1) from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China); the histone acetyltraferase (HAT) inhibitor MG149 (S7476) from Selleck Chemicals (Shanghai, China); the AURKB inhibitor AZD1152-HQPA from Abmole Bioscience (Beijing, China); and hydroxyurea (HU, H8267), Nocodazole (M1404), and MG132 (Z-Leu-Leu-al) from Sigma (St. Louis, MO, USA).
2.2. Cell Culture
HEK293T and HeLa cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The breast cancer lines MCF-7 and MDA-MB-231 were purchased from the Shanghai Biotechnology Co., Ltd. (Shanghai, China). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Meilunbio®, Dalian, China) supplemented with 10% fetal bovine serum ((FBS, Procell, Wuhan, China) and 1% penicillin-streptomycin (P/S, Thermo Fisher Scientific, Waltham, MA, USA). Cells were maintained at 37◦C in a humidified incubator with 5% CO2. Each cell line was authenticated by short tandem repeat (STR) profiling within the past three years. All experiments were conducted using mycoplasma-free cells.
2.3. Plasmid Construction and Transfection
The coding region of full-length AURKB (NM_001313950.2), MSL1 (NM_001012241), MOF (NM_032188), INCENP (NM_020238.3), and MYC (AH002906.2), along with various truncations—including MOF (1-157aa, 1-216aa and 158-458aa)—were subcloned into a pcDNA3.1(–) vectors with Flag, Myc, or HA tags. Additionally, point mutants, including AURKB (K215Q, K215R) and MOF (G327E), were generated. Plasmids were transiently transfected into cells using polyethyleneimine (PEI, 23966, PolySciences, Beijing, China) according to the manufacturer’s instructions.
2.4. Expression of Recombinant Proteins in Escherichia Coli
Full-length AURKB and INCENP were subcloned into pET41a vector. His-GST-tagged AURKB and INCENP proteins that were expressed from pET41a vector in BL21 (DE3) Codon Plus Escherichia coli. Cells.
2.5. siRNA/shRNA Knockdown
293T, MCF-7, and MDA-MB-231 breast cancer cells were transfected with non-targeting (NT) siRNA (D-001206), AURKB siRNA (#1, 5’-CCUGCGUCUCUACAACUAUtt-3’, #2, 5’-UCGUCAAGGUGGACCUAAAtt-3’), and an siRNA SMART pool (Dharmacon, Shanghai, China), using Lipofectamine RNAi MAX (13778150, Invitrogen) according to the manufacturer’s instructions. Seventy-two hours post-transfection, cells were subjected to subsequent experiments. For stable knockdown, the pLVX-shRNA system was used to express shRNA targeting AURKB, MSL1, and MOF in 293T, MCF-7, and MDA-MB-231 cells. The specific shRNA target sequences were: shAURKB (CCUGCGUCUCUACAACUAU), shMSL1 (GCACCGGACGTGTAGGAAAT), and shMOF (CGAAATTGATGCCTGGTAT).
2.6. Immunoprecipitation (IP)
MCF7, MDA-MB-231, and HEK293T cells were cultured in 10 cm tissue culture plates and transiently transfected with Flag- or Myc-tagged plasmids. Forty-eight hours post-transfection, cells were collected and lysed using RIPA buffer containing: 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl, 10% glycerol, 1 mM dithiothreitol (DTT), and a complete protease inhibitor cocktail. Whole-cell lysates were incubated overnight at 4 ◦C with anti-Flag (M2) or anti-Myc-agarose beads. The immunoprecipitated proteins were then eluted using 4 × SDS loading buffer and analyzed by western blot with anti-Flag or anti-Myc antibodies.
2.7. Immunofluorescence Staining
Hela and MCF7 cells were cultured in 24-well plates containing coverslips (Nest, 8D1007) and grown to approximately 30% confluence. Cells were then transfected with plasmids and incubated for 48 hours. Cells were then fixed and immunostained with primary antibodies, followed by FITC/TRITC-conjugated secondary antibodies (1:300, Santa Cruz sc-2012). Nuclei were counterstained with Vectashield containing DAPI (H-1200, Vector Laboratories, Inc., Burlingame, CA, USA). Fluorescent images were acquired using an Olympus BX40F microscope equipped with a 40× silicon immersion objective (Olympus Corporation, Miyazaki, Japan).
2.8. Reverse Transcription PCR
Total RNA was extracted using RNAiso Plus (9109; Takara,Tokyo, Japan). 1 μg of total RNA from each sample was reverse transcribed into cDNA using the PrimeScript 1st Strand synthesis Kit (6110A, Takara, Tokyo, Japan). Relative mRNA levels were quantified using TB Green® Premix Ex Taq™ II (RR820A, Takara, Tokyo, Japan) on the Eco Real-Time PCR System (Illumina, Gene Company Limited, Hong Kong, China). The qPCR primers were as follows: AURKB, 5ʹ-CAGTGGGACACCCGACATC -3ʹ (forward) and 5ʹ-GTACACGTTTCCAAACTTGCC -3ʹ (reverse); MSL1, 5ʹ- CAAGACTCTCCACTCCCCAAAA -3ʹ (forward) and 5ʹ- CCTCCAAGAAGGAATTGCTACAG -3ʹ (reverse); MOF, 5ʹ- CCCAAACCAGTCAGACCAGC -3ʹ (forward) and 5ʹ- GGGCCACCAGAACTGACTTT -3ʹ (reverse); GAPDH, 5ʹ-ATCACTGCCACCCAGAAGAC-3ʹ (forward) and 5ʹ-ATGAGGTCCACCACCCTGTT-3ʹ (reverse).
2.9. In Vitro KAT Assay
Mix the following reactants in a total volume of 20 μl: cold acetyl coenzyme A (12.5 μM), recombinant AURKB proteins expressed in Escherichia coli,anti-Flag MOF beads,HAT reaction buffer to reach 20 μl. The HAT reaction buffer contains 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5% (v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium butyrate. Incubate the reaction mixture at 30°C for 30 to 60 min. Finally, add 4× SDS loading buffer to terminate the reaction, and heat the sample at 95°C for western blot analysis.
2.10. Flow Cytometry Analysis
MCF7 and MDA-MB-231 cells were cultured in DMEM supplemented with 10% FBS. For fixation, cells were harvested and resuspended as single-cell in 70% ethanol at 20ºC for at least 4 hours. Following ethanol fixation, cells were centrifuged at 300×g for 5 min, and the supernatant was discarded. The cell pellets were washed and resuspended in 300 μL PBS containing 0.1% (v/v) Triton X-100 (Sigma, Cat. T8787), 0.3 mg/mL DNase-free RNase A (Sigma, Cat. R5500), and 50 μg/mL propidium iodide (CF0031, Beijing Dingguo, China), followed by incubation at 37 C for 1 hour. Flow cytometry was performed using an EPICS XL™ cytometers (Beckman Coulter), and data were analyzed with ModFit LT software (Verity Software House, USA).
2.11. EdU Assay
The EdU incorporation assay was performed using the BeyoCleck™ EdU Cell Proliferation Kit and the Alexa Fluor 488 in vitro Imaging Kit (Beyotime, C0071s, Shanghai, China). MCF7 and MDA-MB-231 cells were incubated with 10 µM EdU (5-ethyl-2´-deoxyuridine) at 37◦C for 2 hours. Cells were then fixed with 4% paraformaldehyde for 15 minutes and washed with PBS containing 0.5% Triton-X-100. Nuclei were counterstained with Hoechst 33342 (GC10939, GLPBIO). The proliferation rate was determined according to the manufacturer's instructions. Fluorescent images were aquired using a fluorescence microscope, with three randomly selected fields captured per group.
2.12. Cell Viability Assay
MCF7 and MDA-MB-231 cells (1,000 cells/well) were seeded in 96-well plates, and cell viability was assessed using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay Kit (G3580, Promega Corporation, Madison, WI, USA) at 24, 48, 72, and 96 hours. Absorbance was measured at 490 nm using a microplate reader (Infinite F200 Pro, TECAN, Shanghai, China).
2.13. Colony Formation Assay
Stably transferred MCF7 and MDA-MB-231 cell lines (3000-4000 cells/well) were seeded into 6-well plates and cultured for 10–14 days at 37 C. Colonies were then fixed with 4% paraformaldehyde for 15 minutes and stained with 0.1% crystal violet for 20 min. The number and size of colonies were recorded for comparison and imaged using a digital camera.
2.14. In Vivo Tumor Metastasis Experiments
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Jilin University. Male Balb/c mice (7–8 weeks old, weighing18–20 g) were purchased from Vital River Biotechnology Company (Beijing, China). The animal experiment protocol was approved under the ethics review number (2024) YNPZSY No. (0512). Mice were housed under a 12-hour light/dark cycle with ad libitum access to food and water. Randomization was performed based on body weight, and sample sizes were determined according to the “Resource Equation” method. MDA-MB-231 cells infected with lentiviral-mediated pLVX-Flag-AURKB-WT (n=5) and pLVX-Flag-AURKB K215R (n=5) were subcutaneously injected into mice. Thirth days post-injection, mice were euthanized, and tumor tissues were collected for further analysis.
2.15. Statistical Analysis
Statistical analyses were conducted using data from at least three independent experiments. Data were processed with SPSS software, version 26 (IBM Corp., Armonk, NY, USA). Results are presented as mean ± SD. Differences between two groups were evaluated using an unpaired Student´s t-test, while comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA). A P-value < 0.05 was considered statistically significant.
4. Discussion
RNAi screening identified MOF as a critical regulator of cancer cell survival. MOF has been previously implicated in the regulation of the G2/M cell cycle checkpoint, underscoring its essential role in cancer biology and highlighting its potential as a therapeutic target [
35]. Another key oncogene, AURKB, is widely recognized as a target in multiple cancer types, with AURKB inhibitors showing enhanced efficacy in combination with osimertinib for non-small-cell lung cancer (NSCLC) [
36]. Our study provides novel insights into the molecular interplay between the MOF/MSL complex and the chromosomal passenger complex (CPC). We demonstrate, for the first time, that MOF directly acetylates AURKB, reducing its ubiquitination and enhancing its kinase activity. Furthermore, the MSL1 subunit strengthens the interaction between MOF and AURKB, stabilizing the complex during the G2/M phase. Notably, acetylation of AURKB at K215 is crucial for mitotic progression. In breast cancer cells, MOF-mediated AURKB acetylation enhances AURKB protein levels and promotes c-MYC phosphorylation, leading to its accumulation and subsequent tumor cell proliferation.
The CPC plays a crucial role in mitotic regulation, with AURKB serving as its key catalytic component. AURKB is essential for assembling functional kinetochores, aligning spindle microtubules, and correcting improper microtubule-kinetochore attachments [37, 38]. Our proteomic analysis of MSL1-deficient cells identified differential expression of NUF2, a subunit of the NDC80 (nuclear division cycle 80) complex, which is a well-established AURKB substrate [
39]. AURKB modulates the microtubules-binding affinity of the NDC80 complex by phosphorylating NDC80 at multiple sites, thereby influencing kinetochore-microtubule interactions [
40]. Given that AURKB activity is dynamically regulated by post-translational modifications, our findings suggest that MOF-mediated acetylation serves as an additional regulatory layer that modulates both AURKB stability and kinase activity. Interestingly, we observed co-localization of the MOF/MSL complex and the CPC at the equatorial plate during mid-mitosis, raising the possibility that MOF/MSL-mediated acetylation dynamically regulates AURKB function during chromosome segregation. Further structural studies, including high-resolution cryo-electron microscopy [
41], will be necessary to delineate the precise molecular mechanisms governing MOF/MSL-AURKB interactions during mitosis.
AURKB has garnered significant attention in cancer research due to its elevated expression in multiple malignancies, including colorectal adenocarcinoma, thyroid follicular carcinoma, laryngeal carcinoma, and lung cancer [
42]. In breast cancer, AURKB expression is markedly upregulated and correlates with increased tumor cell proliferation and resistance to therapy [
43]. Furthermore, AURKB overexpression has been implicated in paclitaxel resistance in NSCLC [
44] and is associated with poor prognosis in hematological malignancies, such as acute lymphoblastic leukemia and acute myeloid leukemia [
45]. Similarly, in hepatocellular carcinoma, AURKB mRNA levels are significantly elevated in tumor tissues and serve as an independent prognostic marker for disease aggressiveness [
46]. The c-MYC oncogenes family (MYC, MYCN, MYCL) plays pivotal role in tumorigenesis, particularly in cancers characterized by MYC overexpression or amplification [
47]. Deregulated MYC activity is a hallmark of various malignancies, including breast cancer, where it is linked to aggressive disease progression [
48]. Clinical evidence indicates that c-MYC is frequently overexpressed in breast tumors and contributes to oncogenic transformation and tumor maintenance [49, 50].
Figure 1.
Crosstalk between the MOF/MSL complex and CPC in 293T cells. (A) Summary of MSL1-KO-based mass spectrometry data. (B) Volcano plot of the MSL1-KO protein expression profile, highlighting the downregulation of AURKB, INCENP, and BIRC5. (C) MOF and MSL1 Overexpression increases endogenous AURKB protein levels and kinase activity. 293T cells were transfected with Flag-tagged MOF or MSL1 for 48 hours, and AURKB levels and H3S10 phosphorylation (H3S10p) were detected by Western blot. (D) MOF Knockdown reduces AURKB protein and H3S10p levels. (E,F) Relative AURKB mRNA levels, measured by RT-qPCR, remain unchanged following MOF/MSL1 overexpression or knockdown. GAPDH was used for normalization. (G) MSL1 overexpression restores AURKB, BIRC5, CDCA8, and INCENP protein levels in MSL1-KO cells. (H–K) AURKB protein Half-life assessment. 293T cells transfected with shNT, shMOF, or shMSL1 were treated with 20 μg/mL CHX, and AURKB protein levels were analyzed at 0, 1, 2, 3, 4, and 5 hours. GAPDH and H3 were used as internal controls. (L) In vitro phosphorylation assay: Flag IP was performed in cells overexpressing Flag-AURKB, followed by an in vitro phosphorylation assay using Flag IP eluates, E. coli-expressed/purified His-INCENP, and ATP. Phosphorylated INCENP was detected using an anti-Pan-phospho antibody. (M) Myc-MSL1 overexpression enhances INCENP phosphorylation. GAPDH, β-tubulin, and H3 were used as internal controls throughout.
Figure 1.
Crosstalk between the MOF/MSL complex and CPC in 293T cells. (A) Summary of MSL1-KO-based mass spectrometry data. (B) Volcano plot of the MSL1-KO protein expression profile, highlighting the downregulation of AURKB, INCENP, and BIRC5. (C) MOF and MSL1 Overexpression increases endogenous AURKB protein levels and kinase activity. 293T cells were transfected with Flag-tagged MOF or MSL1 for 48 hours, and AURKB levels and H3S10 phosphorylation (H3S10p) were detected by Western blot. (D) MOF Knockdown reduces AURKB protein and H3S10p levels. (E,F) Relative AURKB mRNA levels, measured by RT-qPCR, remain unchanged following MOF/MSL1 overexpression or knockdown. GAPDH was used for normalization. (G) MSL1 overexpression restores AURKB, BIRC5, CDCA8, and INCENP protein levels in MSL1-KO cells. (H–K) AURKB protein Half-life assessment. 293T cells transfected with shNT, shMOF, or shMSL1 were treated with 20 μg/mL CHX, and AURKB protein levels were analyzed at 0, 1, 2, 3, 4, and 5 hours. GAPDH and H3 were used as internal controls. (L) In vitro phosphorylation assay: Flag IP was performed in cells overexpressing Flag-AURKB, followed by an in vitro phosphorylation assay using Flag IP eluates, E. coli-expressed/purified His-INCENP, and ATP. Phosphorylated INCENP was detected using an anti-Pan-phospho antibody. (M) Myc-MSL1 overexpression enhances INCENP phosphorylation. GAPDH, β-tubulin, and H3 were used as internal controls throughout.
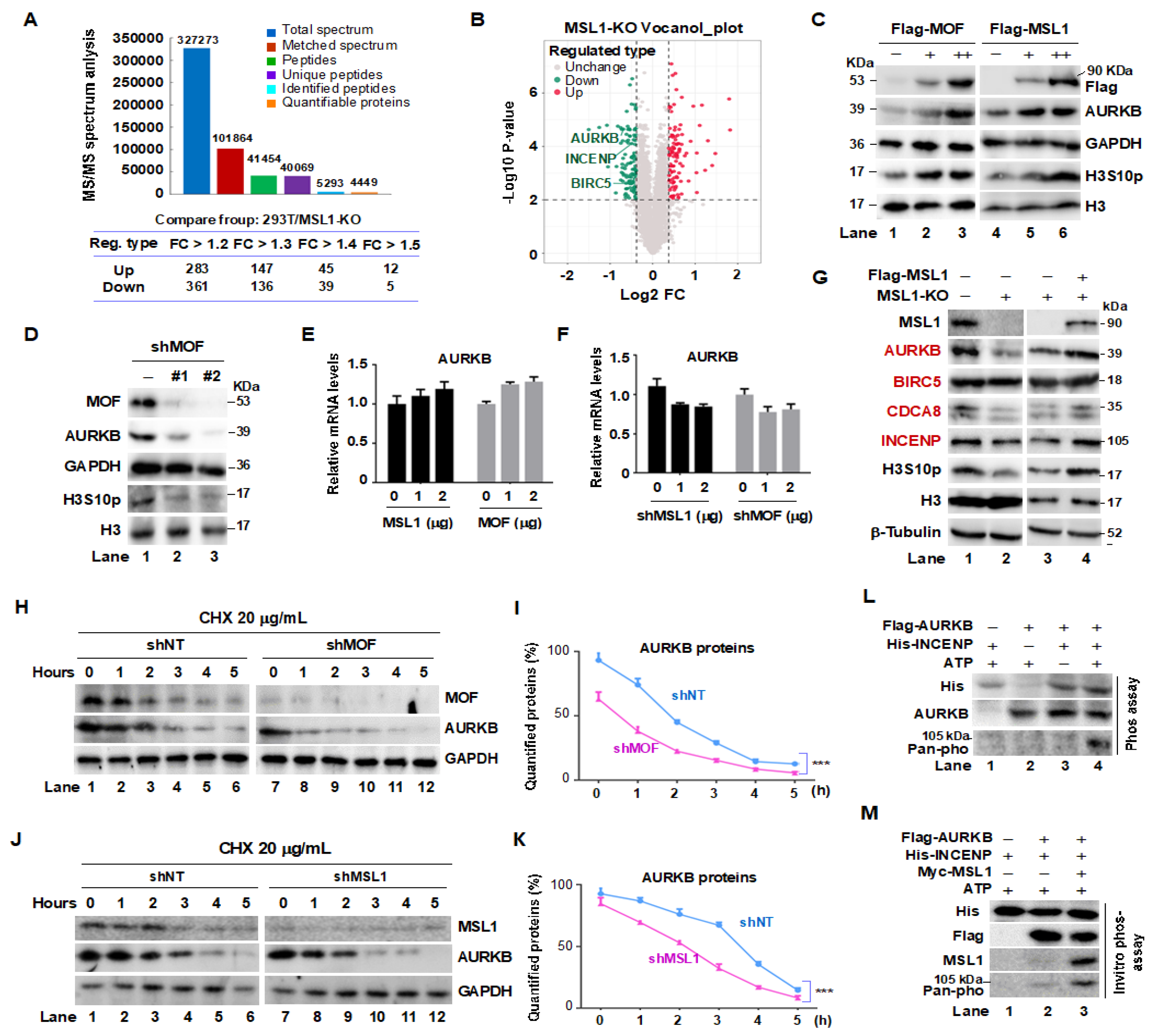
Figure 2.
Interaction and co-distribution of MOF/MSL1 and AURKB during mitosis in HeLa cells. (A–C) Interaction between the MOF/MSL complex and AURKB. Endogenous MOF, MSL1, and AURKB proteins bound to Flag-AURKB, Flag-MOF, or Flag-MSL1 were identified through Flag IP in 293T cells. (D, E) Co-transfection and Co-IP experiments further validate the interaction between AURKB and the MOF/MSL complex in 293T cells. (F, G) Domain mapping analysis. MOF deletion mutants were generated based on distinct structural domains (upper panel). Full-length Flag-AURKB and MOF truncations were transfected into 293T cells. After 48 hours, Flag IP was conducted, and bound proteins were analyzed by Western blot. (H–J) IF staining showing the subcellular localization of AURKB, MOF, and MSL1 (green) at different mitotic phases. β-tubulin (red) marks the mitotic phases, and DAPI (blue) stains nuclei. Scale bar: 20 μm. Images were captured with a 40× objective. (K,L) Co-IP assays confirm interactions between the MOF/MSL1 complex and the CPC. 293T cells were co-transfected with Flag-INCENP and Myc-tagged MOF or MSL1. 48 hours later, Flag IP was performed, and bound proteins, including phospho-INCENP, were detected by Western blot. (M) MSL1 Knockdown destabilizes the CPC. Flag IP was carried out in cells overexpressing Flag-INCENP with or without shMSL1. Input and IP eluates were analyzed for Western blot for the indicated proteins.
Figure 2.
Interaction and co-distribution of MOF/MSL1 and AURKB during mitosis in HeLa cells. (A–C) Interaction between the MOF/MSL complex and AURKB. Endogenous MOF, MSL1, and AURKB proteins bound to Flag-AURKB, Flag-MOF, or Flag-MSL1 were identified through Flag IP in 293T cells. (D, E) Co-transfection and Co-IP experiments further validate the interaction between AURKB and the MOF/MSL complex in 293T cells. (F, G) Domain mapping analysis. MOF deletion mutants were generated based on distinct structural domains (upper panel). Full-length Flag-AURKB and MOF truncations were transfected into 293T cells. After 48 hours, Flag IP was conducted, and bound proteins were analyzed by Western blot. (H–J) IF staining showing the subcellular localization of AURKB, MOF, and MSL1 (green) at different mitotic phases. β-tubulin (red) marks the mitotic phases, and DAPI (blue) stains nuclei. Scale bar: 20 μm. Images were captured with a 40× objective. (K,L) Co-IP assays confirm interactions between the MOF/MSL1 complex and the CPC. 293T cells were co-transfected with Flag-INCENP and Myc-tagged MOF or MSL1. 48 hours later, Flag IP was performed, and bound proteins, including phospho-INCENP, were detected by Western blot. (M) MSL1 Knockdown destabilizes the CPC. Flag IP was carried out in cells overexpressing Flag-INCENP with or without shMSL1. Input and IP eluates were analyzed for Western blot for the indicated proteins.
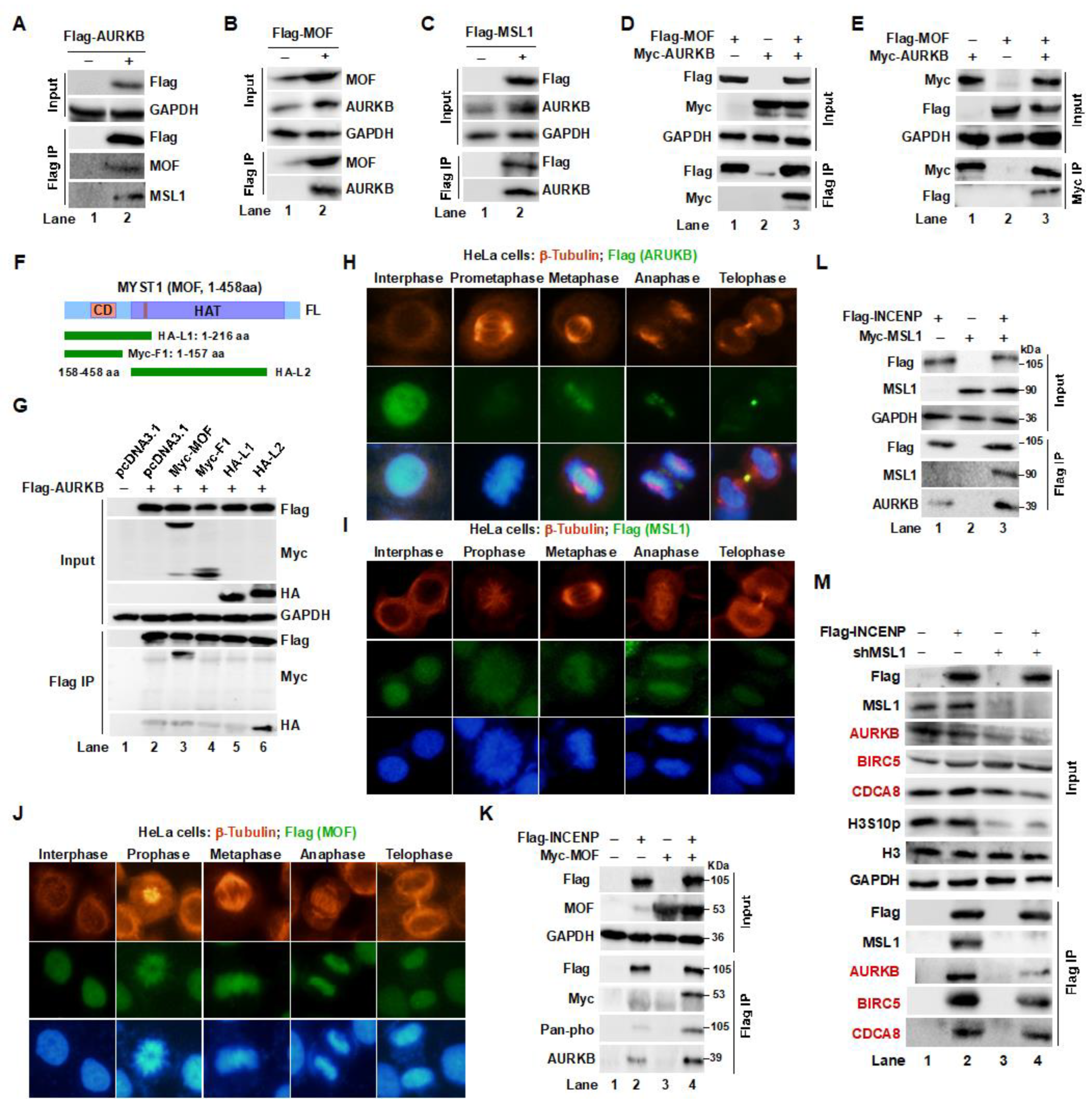
Figure 3.
Acetylation of AURKB by the MOF/MSL1 complex stabilizes the CPC in 293T cells. (A, B) MOF/MSL1 complex stabilizes AURKB. Acetylation and stability of AURKB were assessed following co-transfection of Flag-AURKB with Myc-tagged MOF or MSL1, in the presence of HA-Ubiquitin and the proteasome inhibitor MG132, followed by Flag IP. (C) AURKB instability in 293T cells upon MOF knockdown (shMOF). (D) The MOF G327E mutant, which lacks enzymatic activity, failed to acetylate AURKB. (E) In vitro lysine acetyltransferase (KAT) assay. Recombinant His-AURKB protein was expressed and purified from E. coli. The KAT assay was performed by incubating recombinant His-AURKB with Flag IP eluates from MOF-overexpressing 293T cells and acetyl coenzyme-A (AcCoA) (upper panel). Acetylation of His-AURKB was assessed by Western blot using an anti-Pan-ac antibody (lower panel, lane 4). (F, G) Positive regulation of AURKB by MOF/MSL1. The effects of MOF on Flag-AURKB were analyzed in 293T cells with MSL1 overexpression or knockdown. Protein levels in the input and Flag IP eluates were detected by Western blot using specific antibodies. (H) Role of MSL1 in CPC complex integrity. Myc-AURKB and Flag-INCENP were co-transfected into MSL1-KO 293T cells. The interaction between AURKB and INCENP, as well as the acetylation, phosphorylation, and ubiquitination levels of AURKB and H3S10p, were analyzed by Western blot using specific antibodies.
Figure 3.
Acetylation of AURKB by the MOF/MSL1 complex stabilizes the CPC in 293T cells. (A, B) MOF/MSL1 complex stabilizes AURKB. Acetylation and stability of AURKB were assessed following co-transfection of Flag-AURKB with Myc-tagged MOF or MSL1, in the presence of HA-Ubiquitin and the proteasome inhibitor MG132, followed by Flag IP. (C) AURKB instability in 293T cells upon MOF knockdown (shMOF). (D) The MOF G327E mutant, which lacks enzymatic activity, failed to acetylate AURKB. (E) In vitro lysine acetyltransferase (KAT) assay. Recombinant His-AURKB protein was expressed and purified from E. coli. The KAT assay was performed by incubating recombinant His-AURKB with Flag IP eluates from MOF-overexpressing 293T cells and acetyl coenzyme-A (AcCoA) (upper panel). Acetylation of His-AURKB was assessed by Western blot using an anti-Pan-ac antibody (lower panel, lane 4). (F, G) Positive regulation of AURKB by MOF/MSL1. The effects of MOF on Flag-AURKB were analyzed in 293T cells with MSL1 overexpression or knockdown. Protein levels in the input and Flag IP eluates were detected by Western blot using specific antibodies. (H) Role of MSL1 in CPC complex integrity. Myc-AURKB and Flag-INCENP were co-transfected into MSL1-KO 293T cells. The interaction between AURKB and INCENP, as well as the acetylation, phosphorylation, and ubiquitination levels of AURKB and H3S10p, were analyzed by Western blot using specific antibodies.
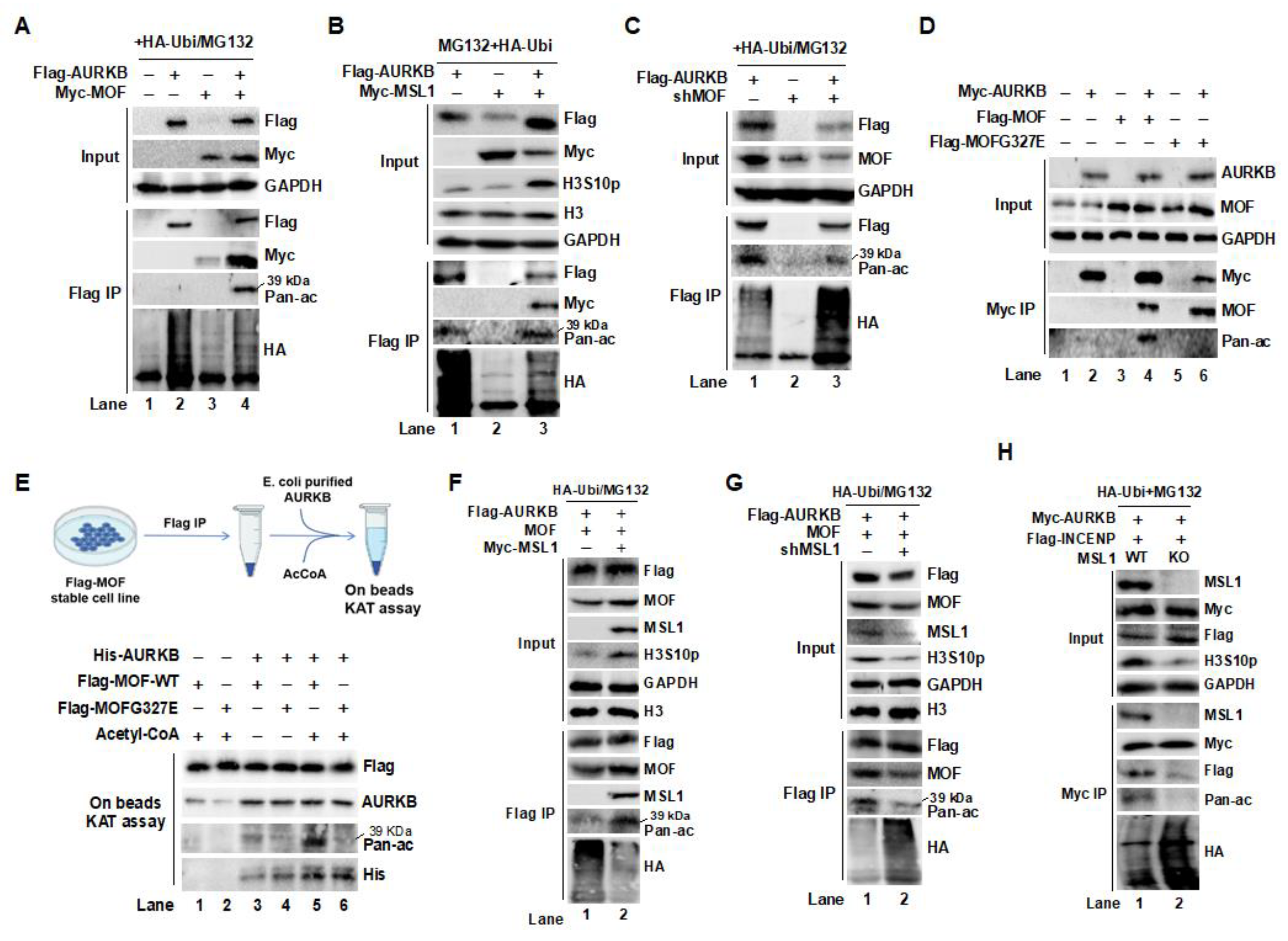
Figure 4.
Lysine 215 is the primary acetylation site targeted by the MOF/MSL complex on AURKB in 293T cells. (A) 3D docking analysis of MOF (pink) and AURKB (green) reveals binding sites, with a zoomed-in view in the right panel. Hydrogen bonds between the two proteins are indicated by yellow dashed lines. (B) Prediction of AURKB K215 acetylation based on the PhosphoSitePlus® database. (C) Flag IP and Western blot analysis confirm AURKB K215 as the primary acetylation site targeted by the MOF/MSL complex. (D) Mutation of the K215 site significantly reduces AURKB protein half-life. (E) Acetylation of AURKB at K215 is essential for maintaining its stability and kinase activity. (F) MOF stabilizes AURKB by acetylating the K215 site, preventing its degradation. (G) The AURKB K215R (non-acetylatable) mutant retains kinase activity on its substrate INCENP, compared to the WT and acetylation-mimic mutant AURKB (K215Q). (H) The K215R mutation disrupts AURKB’s ability to form the CPC and reduces the expression of key complex components, including INCENP, CDCA8, and BIR5. Additionally, the K215R mutation decreases c-MYC expression while increasing phosphorylation of c-MYC at T58, a modification associated with c-MYC degradation. GAPDH and H3 were used as internal controls. (I) The MSL/MOF complex acetylates AURKB at K215, which is crucial for maintaining CPC integrity and AURKB activity.
Figure 4.
Lysine 215 is the primary acetylation site targeted by the MOF/MSL complex on AURKB in 293T cells. (A) 3D docking analysis of MOF (pink) and AURKB (green) reveals binding sites, with a zoomed-in view in the right panel. Hydrogen bonds between the two proteins are indicated by yellow dashed lines. (B) Prediction of AURKB K215 acetylation based on the PhosphoSitePlus® database. (C) Flag IP and Western blot analysis confirm AURKB K215 as the primary acetylation site targeted by the MOF/MSL complex. (D) Mutation of the K215 site significantly reduces AURKB protein half-life. (E) Acetylation of AURKB at K215 is essential for maintaining its stability and kinase activity. (F) MOF stabilizes AURKB by acetylating the K215 site, preventing its degradation. (G) The AURKB K215R (non-acetylatable) mutant retains kinase activity on its substrate INCENP, compared to the WT and acetylation-mimic mutant AURKB (K215Q). (H) The K215R mutation disrupts AURKB’s ability to form the CPC and reduces the expression of key complex components, including INCENP, CDCA8, and BIR5. Additionally, the K215R mutation decreases c-MYC expression while increasing phosphorylation of c-MYC at T58, a modification associated with c-MYC degradation. GAPDH and H3 were used as internal controls. (I) The MSL/MOF complex acetylates AURKB at K215, which is crucial for maintaining CPC integrity and AURKB activity.
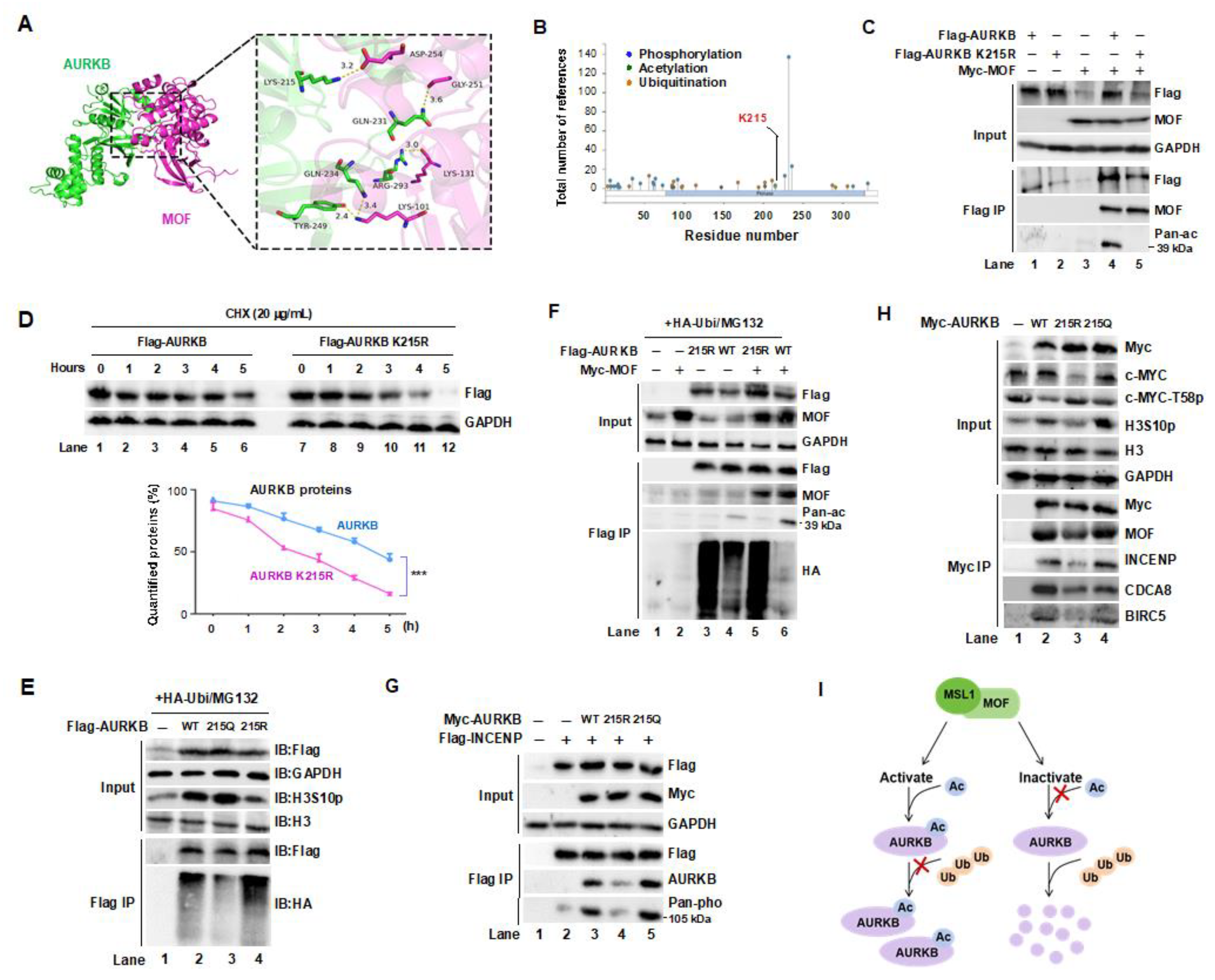
Figure 5.
MOF/MSL complex-mediated acetylation of AURKB at K215 regulates G2/M phase progression. (A,B) Immunofluorescence (IF) staining of β-Tubulin (red or green), AURKB (red), H3S10p (green) in HeLa cells treated with shMOF (A) or shMSL1 (B). Nuclei were stained with DAPI (blue). Scale bars: 200 μm. (C) IF staining of β-Tubulin (red) and AURKB (green) in MCF-7 cells treated with shMOF. Nuclei were stained with DAPI (blue). Scale bars:100 μm. (D) IF staining of β-Tubulin (red) in HeLa cells overexpressing wild-type (WT) or K215R mutant AURKB Nuclei were stained with DAPI (blue). Scale bars:20 μm. (E–H) Quantification of spindle multipolar cells corresponding to panels A D, respectively. (I,J) Validation of MOF and MSL1 knockdown efficiency in the experiments shown in panels A C. (K) Transient transfection of the AURKB K215R mutant in HeLa cells delayed cell cycle progression compared to the WT AURKB. Effect of MOF overexpression on AURKB protein levels in MCF7 and MDA-MB-231 cells. (L) Subcellular fractionation of HeLa cells arrested in the S and M phases using hydroxyurea (HU) and nocodazole, respectively. Cytoplasmic and nuclear fractions were separated by centrifugation and analyzed by Western blot. GAPDH was used as a cytoplasmic marker. (M) Flow cytometry analysis of cell cycle progression. MCF-7 and MDA_MB-231 cells were transfected with wild-type or mutant AURKB were analyzed in AURKB knockdown backgrounds. (N) Co-localization of MOF, MSL1, and AURKB across different cell cycle phases. HeLa cells were synchronized at specific phases: (1) G1 phase by serum starvation (24 hours); (2) S phase by HU treatment (1 mM, 24 hours); (3) G2/M phase by nocodazole treatment (500 ng/mL, 16 hours); (4) M phase by release from nocodazole arrest (1 hour).
Figure 5.
MOF/MSL complex-mediated acetylation of AURKB at K215 regulates G2/M phase progression. (A,B) Immunofluorescence (IF) staining of β-Tubulin (red or green), AURKB (red), H3S10p (green) in HeLa cells treated with shMOF (A) or shMSL1 (B). Nuclei were stained with DAPI (blue). Scale bars: 200 μm. (C) IF staining of β-Tubulin (red) and AURKB (green) in MCF-7 cells treated with shMOF. Nuclei were stained with DAPI (blue). Scale bars:100 μm. (D) IF staining of β-Tubulin (red) in HeLa cells overexpressing wild-type (WT) or K215R mutant AURKB Nuclei were stained with DAPI (blue). Scale bars:20 μm. (E–H) Quantification of spindle multipolar cells corresponding to panels A D, respectively. (I,J) Validation of MOF and MSL1 knockdown efficiency in the experiments shown in panels A C. (K) Transient transfection of the AURKB K215R mutant in HeLa cells delayed cell cycle progression compared to the WT AURKB. Effect of MOF overexpression on AURKB protein levels in MCF7 and MDA-MB-231 cells. (L) Subcellular fractionation of HeLa cells arrested in the S and M phases using hydroxyurea (HU) and nocodazole, respectively. Cytoplasmic and nuclear fractions were separated by centrifugation and analyzed by Western blot. GAPDH was used as a cytoplasmic marker. (M) Flow cytometry analysis of cell cycle progression. MCF-7 and MDA_MB-231 cells were transfected with wild-type or mutant AURKB were analyzed in AURKB knockdown backgrounds. (N) Co-localization of MOF, MSL1, and AURKB across different cell cycle phases. HeLa cells were synchronized at specific phases: (1) G1 phase by serum starvation (24 hours); (2) S phase by HU treatment (1 mM, 24 hours); (3) G2/M phase by nocodazole treatment (500 ng/mL, 16 hours); (4) M phase by release from nocodazole arrest (1 hour).
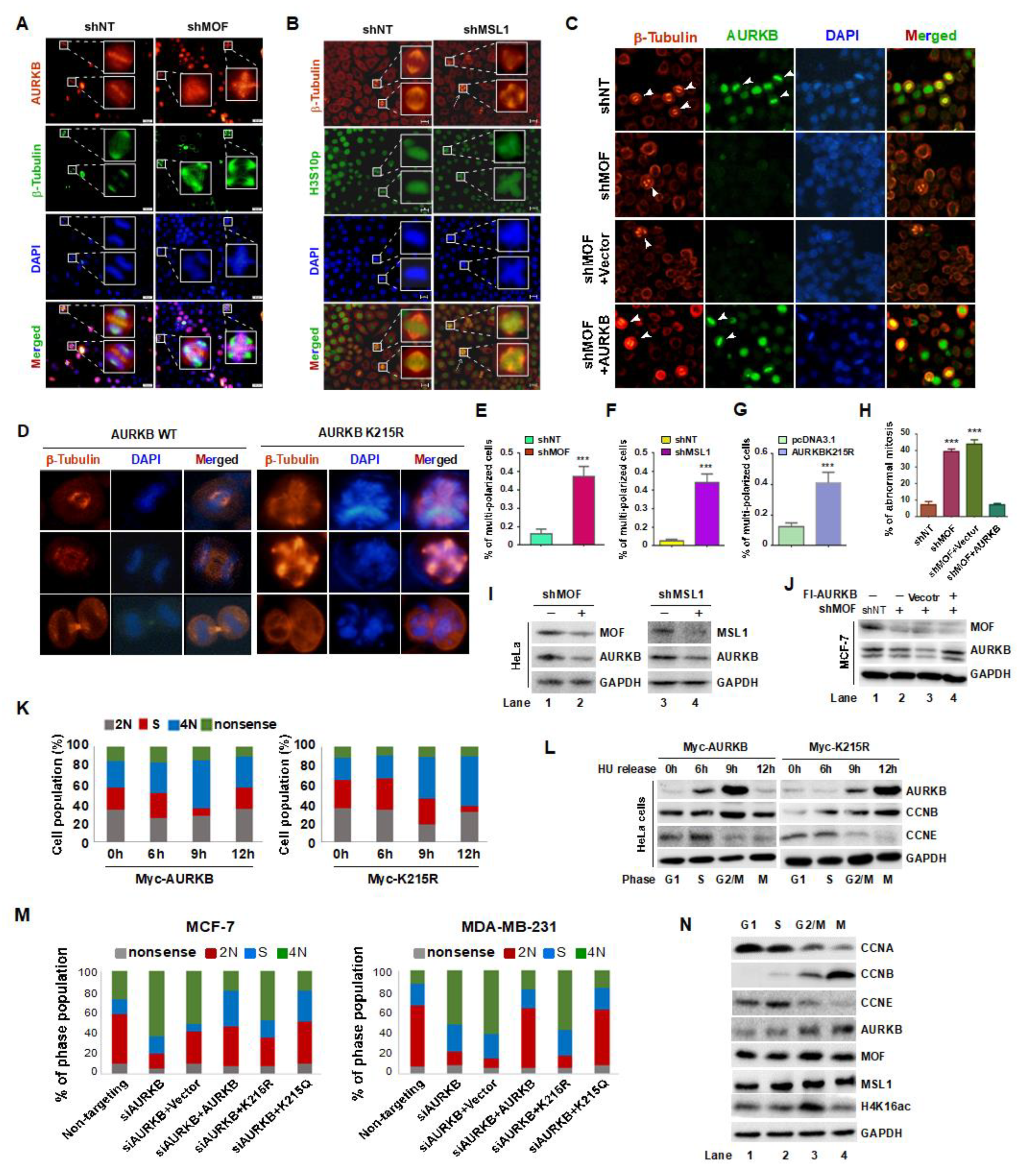
Figure 6.
MOF-mediated acetylation of AURKB at K215 regulates the proliferation of MCF-7 and MDA-MB-231 cells. (A) EdU incorporation assay to assess cell proliferation. Cells were incubated with EdU for 2 hours and analyzed using the BeyoCleck™ EdU Cell Proliferation Kit (C0071s). Scale bar: 50 μm. (B) Quantification of EdU-positive cells. (C) CCK-8 assay to evaluate cell viability. (D) Colony formation assays, with colonies stained using crystal violet. (E) Quantification of colony formation efficiency. (F) Generation of stable MDA-MB-231 cell lines expressing Flag-tagged AURKB or the AKRKB K215R mutant. (G) Schematic representation of the animal experiment design. (H) Tumor growth progression over time. (I) Representative images of tumors at the end of the experiment (n = 5 per group). (J) Tumor weights at the experiments termination in mice injected with AURKB or AURKB K215R-expressing cells. ***p < 0.001 compared to the AURKB group. (K) Western blot analysis of protein expression in whole-cell lysates from four tumors per group.
Figure 6.
MOF-mediated acetylation of AURKB at K215 regulates the proliferation of MCF-7 and MDA-MB-231 cells. (A) EdU incorporation assay to assess cell proliferation. Cells were incubated with EdU for 2 hours and analyzed using the BeyoCleck™ EdU Cell Proliferation Kit (C0071s). Scale bar: 50 μm. (B) Quantification of EdU-positive cells. (C) CCK-8 assay to evaluate cell viability. (D) Colony formation assays, with colonies stained using crystal violet. (E) Quantification of colony formation efficiency. (F) Generation of stable MDA-MB-231 cell lines expressing Flag-tagged AURKB or the AKRKB K215R mutant. (G) Schematic representation of the animal experiment design. (H) Tumor growth progression over time. (I) Representative images of tumors at the end of the experiment (n = 5 per group). (J) Tumor weights at the experiments termination in mice injected with AURKB or AURKB K215R-expressing cells. ***p < 0.001 compared to the AURKB group. (K) Western blot analysis of protein expression in whole-cell lysates from four tumors per group.
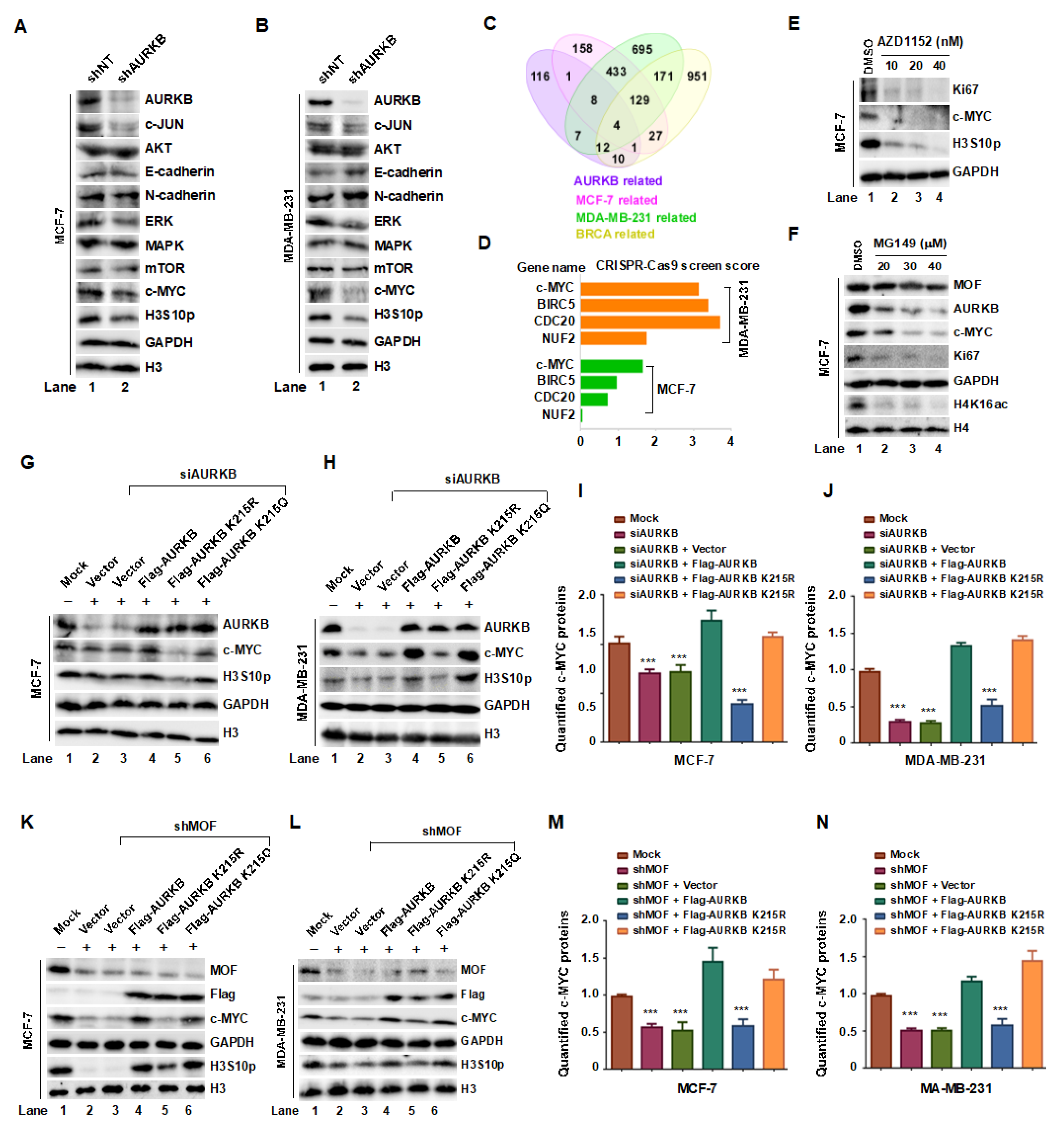
Figure 7.
The MOF-AURKB K215ac axis promotes breast cancer cell proliferation by stabilizing c-MYC protein. (A,B) Western blot analysis of the indicated protein expression levels in MCF-7 and MDA-MB-231 cells transfected with either shNT or shAURKB. (C,D) Venn diagram showing the intersection of data from BioGRID ORCS, STRING and TCGA Databases, identifying AURKB downstream targets involved in breast cancer proliferation. (E) Treatment of MCF-7 cells with the AURKB kinase inhibitor AZD1152 (10, 20, and 40 nM for 24 hours) resulted in decreased in c-MYC- levels. (F) Inhibition of MOF activity using the enzyme inhibitor MG149 reduced the expression of AURKB, c-MYC, and Ki67 in MCF7 cells. (G,H) Effects of overexpressing WT, K215R, and K215Q AURKB in MCF-7 and MDA-MB-231 cells on c-MYC levels following AURKB knockdown. GAPDH and histone H3 were used as internal controls. (I,J) Quantification of c-MYC protein levels. ***P < 0.001 compared to non-targeting siRNA group. (K,L) Effects of overexpressing WT, K215R, and K215Q AURKB on c-MYC, H3S10p, and AURKB levels following MOF knockdown. (M,N) Quantification of c-MYC protein levels. ***P < 0.001 compared to non-targeting shRNA group.
Figure 7.
The MOF-AURKB K215ac axis promotes breast cancer cell proliferation by stabilizing c-MYC protein. (A,B) Western blot analysis of the indicated protein expression levels in MCF-7 and MDA-MB-231 cells transfected with either shNT or shAURKB. (C,D) Venn diagram showing the intersection of data from BioGRID ORCS, STRING and TCGA Databases, identifying AURKB downstream targets involved in breast cancer proliferation. (E) Treatment of MCF-7 cells with the AURKB kinase inhibitor AZD1152 (10, 20, and 40 nM for 24 hours) resulted in decreased in c-MYC- levels. (F) Inhibition of MOF activity using the enzyme inhibitor MG149 reduced the expression of AURKB, c-MYC, and Ki67 in MCF7 cells. (G,H) Effects of overexpressing WT, K215R, and K215Q AURKB in MCF-7 and MDA-MB-231 cells on c-MYC levels following AURKB knockdown. GAPDH and histone H3 were used as internal controls. (I,J) Quantification of c-MYC protein levels. ***P < 0.001 compared to non-targeting siRNA group. (K,L) Effects of overexpressing WT, K215R, and K215Q AURKB on c-MYC, H3S10p, and AURKB levels following MOF knockdown. (M,N) Quantification of c-MYC protein levels. ***P < 0.001 compared to non-targeting shRNA group.