1. Introduction
1.1. Overview of RNA Viruses
RNA viruses are a group of widespread pathogens of a wide range of infectious diseases in humans, animals, and plants[
1,
2,
3]. A universal structural feature of RNA viruses is the presence of untranslated regions (UTRs) around one or more open reading frames (ORFs) at the 5 'and 3' ends, often containing sequences or conserved structures required for replication regulation[
4,
5]. Due to physical and structural limitations in RNA genome stability and RNA polymerase fidelity limitations, RNA virus genome sizes differ within an order of magnitude[
6,
7], with the larger genomes consisting of all class I dsDNA viruses[
3]. SARS-CoV-2 and the coronavirus family are among the longest extant RNA viruses in terms of genome size[
8,
9]. RNA viruses are distinguished by their ability to evolve rapidly, largely due to their reliance on RNA-dependent RNA polymerases (RdRPs) for replication. These polymerases lack proof reading mechanisms, resulting in frequent errors during replication[
10]. RNA instability and high replication error rates allow RNA viruses to exhibit high genetic diversity and environmental adaptability, which enables them to rapidly adapt to new hosts and rapidly introduce genetic changes[
11,
12]. Alterations in viral genetic material can result in changes in viral phenotype and population dynamics, potentially leading to changes in virulence or host tropism, resistance to antiviral treatments and evasion of immune detection, facilitating the emergence of novel pathogens[
13,
14]. The continuous emergence of new strains and variants poses a major challenge to public health. This class of viruses includes those causing some of the most severe human health challenges, such as SARS-CoV-2 (the causative agent of COVID-19), HIV, which causes AIDS, and influenza viruses that cause seasonal influenza epidemics and pandemics.
In addition to their impact on human health, RNA viruses can also affect animals. Porcine reproductive and respiratory syndrome virus (PRRSV) is a major pathogen in pigs[
1], causing huge economic losses to the livestock industry. Among the various potential pathogens capable of cross-species transmission, RNA viruses warrant particular concern. For example, human infection with avian influenza viruses (AIVs) is an ongoing public health threat. The main risk factor for human infection with AIVs is direct contact with infected poultry, mainly in Asia and Egypt where live poultry markets are common[
15]. It is very likely that a recent ancestor of the SARS-CoV-2 virus infected bats and possibly other intermediate species[
16]. Notably, RNA viruses have emerged as significant zoonotic pathogens originating from wildlife. Recent studies have consistently identified RNA viruses as the predominant agents of emerging diseases in humans, accounting for approximately 44% of all emerging infectious diseases, with reported ranges between 25% and 44% across different studies. In comparison, bacteria contribute to 10%–49% of these diseases, surpassing other parasitic groups such as fungi (7%–9%), protozoa (11%–25%), and helminths (3%–6%). Therefore, it is crucial to focus on and understand the dynamics of animal health and the interactions between humans and animals to effectively address the challenges posed by these pathogens[
17,
18,
19,
20].
1.2. Current Antiviral Challenges
Antiviral vaccines and therapeutics constitute the primary strategies for the prevention of pandemic viral infections. In recent years, these vaccines and drugs have been extensively employed in the management and prevention of major infections caused by influenza viruses and SARS-CoV-2. Nevertheless, challenges persist as influenza viruses frequently develop resistance to antiviral agents such as oseltamivir, and variants of SARS-CoV-2, including Delta and Omicron, exhibit reduced sensitivity to existing vaccines. Similarly, the development of resistance to antiretroviral therapy (ART) complicates the long-term management of HIV. These issues underscore the significant challenges posed by RNA viruses, attributable to their unique biological characteristics. The reason is that existing vaccines and drugs mainly target specific viral proteins to induce limited immune responses. However, the structure and life cycle of viruses are relatively complex, the replication mechanism of some viruses is still unclear, and the high mutation rate of RNA viruses often causes changes in viral proteins due to viral mutations and the emergence of potential new variants, resulting in a continuous decline in the protection and treatment effects of existing vaccines and drugs, which need to be continuously updated to cope with new viral variants.
1.3. Emergence of CRISPR/Cas13
Clustered regularly interspaced short palindromic repeats (CRISPR) were initially identified as part of an adaptive immune system in prokaryotes, such as bacteria and archaea, which provides defense against viral infections. The term "short palindromic repeats" refers to repetitive DNA sequences found within prokaryotic genomes. The CRISPR system was first discovered in Escherichia coli by the Japanese scientist Yoshizumi Ishino and his team in 1987, although its functional role was not understood at the time[
21]. In 2002, a team led by Ruud Jansen identified and formally named CRISPR/associated proteins (Cas)[
22]. A typical CRISPR/Cas system is composed of Cas genes, leader sequences, repeats, spacers, and tracrRNA, each playing a crucial role in the mechanism of adaptive immunity.
The CRISPR/Cas system is characterized by Cas proteins that are organized into two primary classes, each further divided into three subtypes. The first class is comprised of multi-protein effector complexes, which consist of several distinct Cas proteins, each tasked with specific functions. This class encompasses types I, III, and IV. In contrast, the second class consists of single-protein effector complexes, which are capable of performing their functions through a single Cas protein. This class includes type II (Cas9), type V (Cas12), and type VI (Cas13) (
Table 1). Significantly, the second class of systems is distinguished by its simplicity, efficiency, and operational ease, establishing it as the leading choice for gene editing applications[
23,
24,
25]. Cas9, in particular, is the most widely recognized member of this class, known for its ability to target DNA for precise editing. In 2013, it was first applied to gene editing in mammalian cells[
24], and in 2020, gene editing therapy was first carried out directly in human body. In the same year, the Nobel Prize in Chemistry was awarded to Emmanuelle Charpentier and Jennifer Doudna for their outstanding contributions in CRISPR/Cas9 research[
26]. However, Feng Zhang’s team was the first to identify the distinctive capability of the Cas13 protein family to specifically target and cleave RNA rather than DNA[
27,
28] (
Table 1). Building on this unique feature, they developed a nucleic acid molecular diagnostic technology utilizing the CRISPR/Cas13 system[
29]. Over the past decade, the CRISPR/Cas system has distinguished itself among various gene editing technologies, demonstrating significant potential for application in viral genome editing. It is anticipated to emerge as a novel therapeutic approach for the treatment of viral infectious diseases. In particular, Cas13 functions by recognizing specific RNA sequences through its guide RNA (crRNA) and cleaving them with high specificity. This property makes Cas13 particularly suitable for targeting RNA viruses, whose genomes are composed entirely of RNA. The programmability of Cas13 allows for rapid re-design of crRNAs to target newly emerging viral strains, offering a flexible solution to the challenge of viral mutations. This review explores Cas13's potential as an antiviral tool, focusing on its mechanism of action, applications in RNA viral research, and the challenges it faces before clinical implementation.
2. CRISPR/Cas13: Mechanism and Potential in Targeting RNA Viruses
2.1. Cas13 Discovery and Classification
Cas13 belongs to the Class 2, Type VI CRISPR system, and is characterized by a single protein that performs both RNA recognition and cleavage, making it a unique and versatile tool for RNA manipulation. Cas13 was first discovered in 2015 as part of the expanding CRISPR family of proteins[
28]. Feng Zhang's team employed computational biology methodologies to identify the Cas13a system (also referred to as C2c2) within microbial metagenomic databases[
27,
28]. Their findings, which indicated that the presence of Cas1 and Cas2 genes in the CRISPR/Cas loci is not essential, inspired the development of a novel computational approach. This advancement led to the discovery of the Cas13b system, classified as the type VI-B system in 2017[
30,
31]. By continuously refining these computational methods, Zhang's team expanded their investigation to include smaller effectors in 2018, resulting in the discovery of the Cas13d system[
32]. At approximately 930 amino acids in length, Cas13d is the smallest type VI CRISPR/Cas effector identified to date and is the most widely utilized subtype of Cas13. In 2021, Yang Hui's research group introduced the Cas13X system, the smallest RNA editing tool known, consisting of only 775 amino acids, thereby enhancing its suitability for in vivo delivery[
33]. Building on this, Yang’s team developed a high-fidelity Cas13X protein variant, hfCas13X, which exhibits high gene editing activity with exceptionally low off-target effects, demonstrating significant potential for applications in RNA editing-based in vivo gene therapy[
34]. Each Cas13 isoform has different properties and is suitable for different applications. Cas13's specificity is determined by its crRNA, a small RNA sequence complementary to the target viral RNA. Upon recognition of the target RNA, Cas13 undergoes a conformational change that activates its HEPN (Higher Eukaryotes and Prokaryotes Nucleotide-binding) domains, which are responsible for RNA cleavage. This activation results in precise cutting of the viral RNA, effectively halting viral replication. The features of the Cas13 members identified so far are summarized (
Table 2).
2.2. Mechanism of Action
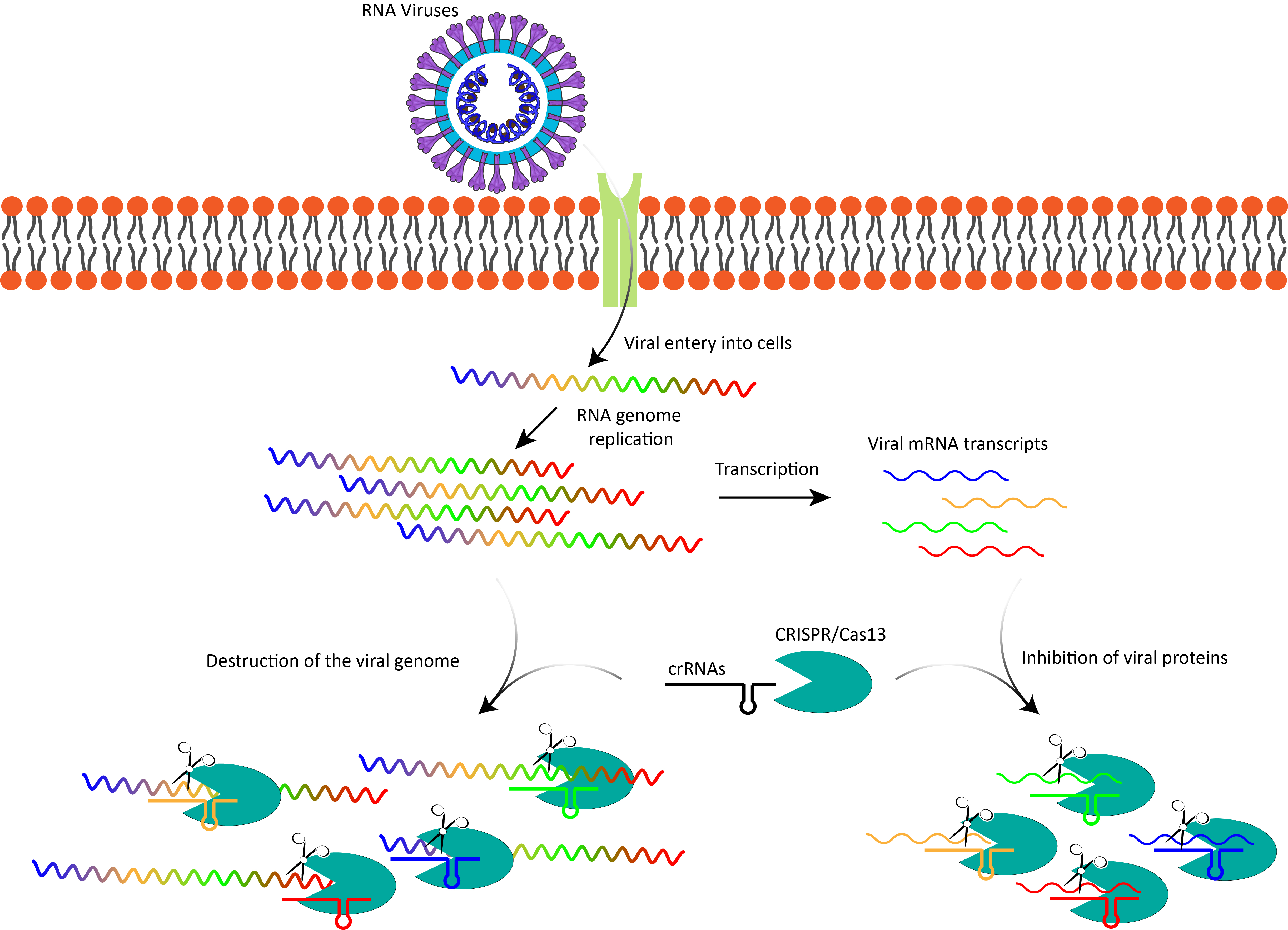
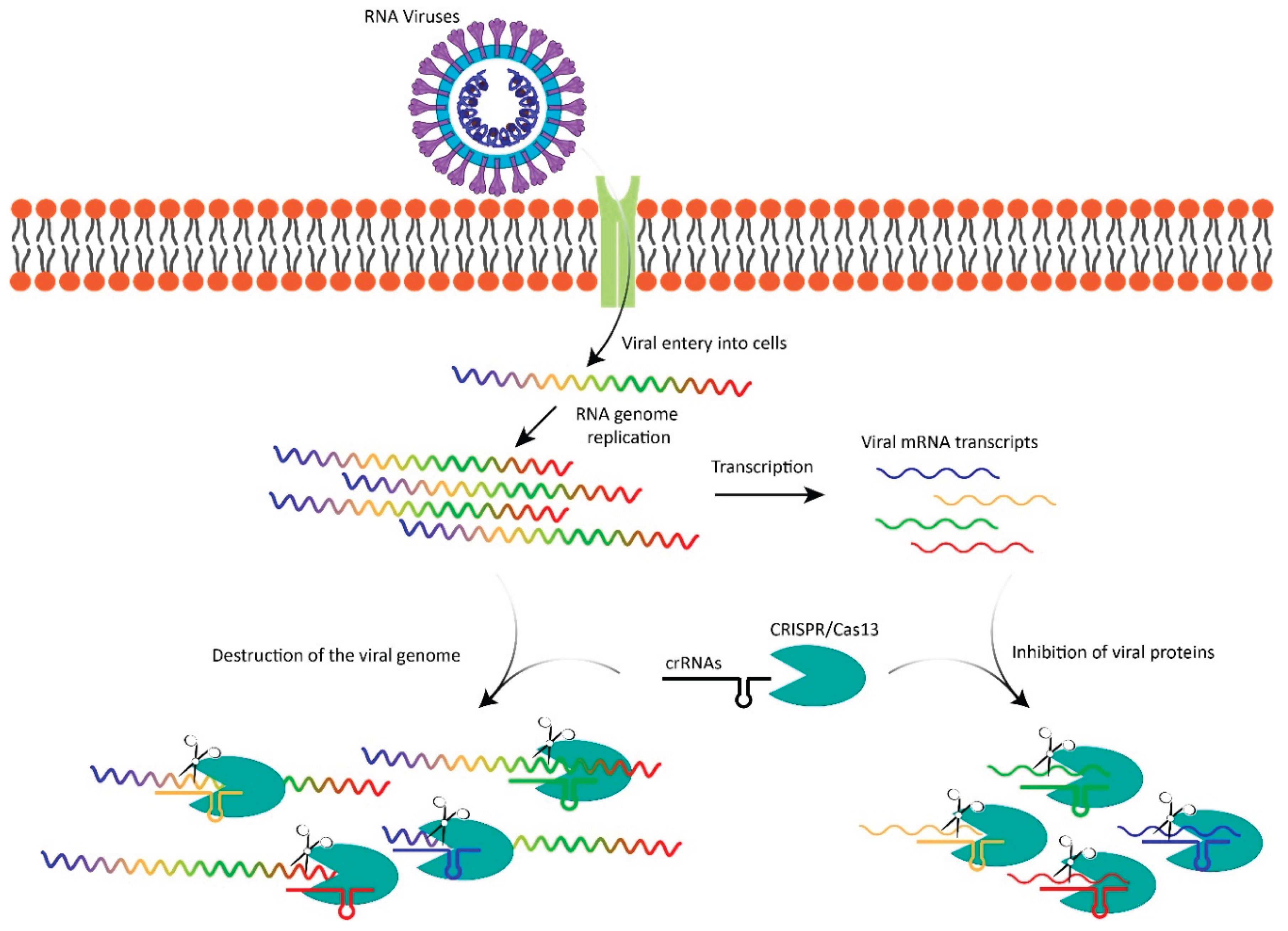
Cas13's mechanism of action involves a three-step process that enables it to target and degrade viral RNA. The first step, recognition and binding, is facilitated by crRNAs that are complementary to specific regions of the viral RNA genome. These crRNAs guide the Cas13 protein to bind precisely to its target within the viral RNA. Upon successful binding, Cas13 undergoes a conformational change, activating its HEPN domains. These domains are crucial for the second step, as they cleave the target RNA at specific sites, effectively silencing the viral genome and preventing it from translating into viral proteins. The third step, collateral cleavage, occurs once Cas13 is activated. In this phase, the protein indiscriminately cleaves nearby RNAs in addition to the target. While collateral cleavage can be advantageous in bacterial systems by ensuring comprehensive degradation of viral RNA, it poses a potential challenge in human therapeutic applications, as it risks degrading essential host RNAs and causing unintended side effects. Addressing this issue is critical for safely harnessing Cas13's antiviral potential.
2.3. Advantages of Cas13 over Traditional Antiviral Strategies
The CRISPR/Cas13 system offers several notable advantages over traditional antiviral approaches, positioning it as a promising tool in combating RNA viruses (
Figure 1). One of its key strengths lies in its high specificity, as Cas13 can be programmed with crRNAs to precisely target and degrade RNA sequences unique to specific viruses. This precision minimizes off-target effects, making it well-suited for therapies requiring accurate targeting of viral RNA. Additionally, Cas13 exhibits broad-spectrum potential due to its versatility; it can be rapidly reprogrammed to address a wide range of RNA viruses. This capability enables the quick design of new crRNAs to counter emerging strains, offering a significant advantage over traditional antivirals that often require years to develop. Furthermore, Cas13's programmability makes it adaptable for tackling emerging viral threats, including rapidly mutating viruses like novel variants of SARS-CoV-2, highlighting its flexibility and responsiveness in addressing evolving infectious diseases.
3. Applications of CRISPR/Cas13 in Targeting RNA Viruses
Viral nucleic acids serve as the primary targets for both virus detection and the development of antiviral therapeutics. Various gene editing technologies have the capability to specifically target and modify nucleic acid sequences, thereby facilitating viral nucleic acid detection and the creation of antiviral drugs. The CRISPR/Cas13 system stands out due to its remarkable sensitivity, efficiency, and safety. In recent years, the CRISPR/Cas system has found extensive application in virus detection, and there has been significant exploration into its potential for developing antiviral vaccines and drugs. This section primarily addresses the utilization of Cas13 in treatment as well as detection of a range of human as well as non-human pathogenic viruses (treatment summarized in
Table 3), with the aim of providing a comprehensive overview of the current advancements and serving as a reference for future research.
3.1. SARS-CoV-2
The global COVID-19 pandemic has driven significant research into Cas13's potential to combat SARS-CoV-2. The Abbott’s team has successfully developed a CRISPR/Cas13-based strategy known as PAC-MAN (Prophylactic Antiviral CRISPR in Human Cells). This approach specifically targets highly conserved regions of the SARS-CoV-2 genome, facilitating the degradation of SARS-CoV-2 RNA within a human lung epithelial cell model[
35]. Blanchard tested Cas13a in a hamster model, where Cas13a was delivered via a nasal nebulizer. Treated hamsters showed significantly reduced viral loads in the lungs and less severe symptoms, illustrating the potential for Cas13-based treatments in respiratory infections like COVID-19[
37]. Zeng’s team used Cas13d to target multiple strains of SARS-CoV-2, including the Alpha, Beta, and Omicron variants, which have shown significant resistance to vaccines and antiviral drugs. In Vero E6 cells, Cas13d reduced viral replication by over 95%, demonstrating its efficacy in neutralizing the virus[
36]. Considering the RNA genome of SARS-CoV-2 and the emergence of new variants, frequent mutations in the virus may lead to recognition failures by CRISPR/Cas13 systems. To address this, Wang’s group employed a comprehensive suite of bioinformatics techniques, including sequence alignment, structural comparison, and molecular docking, to identify conserved regions within the SARS-CoV-2 genome that are targetable by Cas13[
38]. Furthermore, Fareh et al. developed a CRISPR/Cas13b-based system specifically designed to inhibit the replication of various SARS-CoV-2 variants, including the variant of interest B.1.1.7, in infected mammalian cells[
52]. Another studied by Liu et al. used a reprogrammed Cas13d effectors targeting NSP13, NSP14, and nucleocapsid transcripts of SARS-CoV-2 genome and achieved >99% silencing efficiency in human cells including four different variants such as B.1, B.1.1.7(Alpha), D614G B.1.351 (Beta), and B.1.617 (Delta). Based on bioinformatics data predictions, the designed crRNAs could almost 100% target the Omicron variant as well[
41].
The inhibition efficiency can vary significantly and is largely dependent on the crRNA target region within the viral genome. Yu utilized mRNA-encoded Cas13b to target the pseudoknot site upstream of ORF1b. This approach was tested in Vero E6 cells and hACE2 transgenic mice. The results showed a 99% reduction in spike protein expression and a marked attenuation of viral replication[
39]. Cas13 offers a promising approach to inhibit viral replication by targeting conserved regions of the viral genome.
Early diagnosis of viral infections and timely antiviral treatment are crucial in minimizing the risk of disease transmission. Nucleic acid testing is considered the gold standard for confirming viral infections, with fluorescent quantitative PCR being the most widely employed method for viral nucleic acid detection[
53]. However, nucleic acid testing necessitates stringent requirements for sample preservation, nucleic acid extraction conditions, operator proficiency, and the operating environment. These demands pose challenges to large-scale early screening and diagnosis efforts[
54]. In contrast, large-scale screening often relies on antigen detection methods based on colloidal gold, which are quick and easy to perform but suffer from low sensitivity and a tendency for false-negative results. Consequently, the development of a more efficient and user-friendly CRISPR-based platform presents a promising alternative for viral detection. Utilizing SHERLOCK technology for SARS-CoV-2 detection allows for the identification of novel coronavirus strains and their variants within one hour[
55], with a minimum detection threshold of 10 to 100 copies/μL and a 97% concordance rate with clinical positive samples. This method significantly enhances sensitivity and specificity compared to traditional detection technologies. At the same time, Wilson's group has designed a method called CREST (Cas13-Based, Rugged, Equitable, Scalable Testing) to further reduce the cost of using Cas13 for large-scale testing[
56], and Arizti-Sanz's team also innovatively integrated the Cas13 system, fluorescence monitoring and smartphone applications into the SHINE detection system, achieving effective nucleic acid detection of SARS-CoV-2[
57]. It is anticipated that with ongoing optimization, the CRISPR/Cas13 system will evolve into a novel nucleic acid diagnostic tool.
3.2. HIV
According to data from the World Health Organization (WHO), by the end of 2022, approximately 39 million individuals globally were living with human immunodeficiency virus (HIV)[
58]. Highly active antiretroviral therapy remains the primary treatment strategy for patients with HIV-1[
59]. However, HIV presents significant challenges due to its ability to integrate into the host genome and establish latent reservoirs that are resistant to current antiretroviral therapies. Highly active antiretroviral therapy(HAART) is ineffective in eradicating these latent viral reservoirs, rendering HIV a chronic, incurable condition. The CRISPR/Cas system offers a novel approach for HIV treatment by targeting the HIV-1 genome to reduce infection or eliminate the virus. In 2013, the CRISPR/Cas9 system was first utilized for HIV treatment. The Ebina team designed gRNAs (guide RNAs) to target specific HIV sequences in integrated proviral DNA and demonstrated that CRISPR/Cas9 could successfully inhibit HIV-1 genome expression, disrupt viral gene expression and reduce viral replication in Jurkat cell lines, showing that targeting multiple sites in the genome simultaneously could enhance excision efficiency[
60]. Unlike Cas9, Cas13 does not alter the DNA sequence, potentially allowing cells to tolerate and recover from off-target effects without permanent DNA damage. The CRISPR/Cas13 system offers a promising approach by targeting HIV RNA in both active and latent infections. Yin showed that Cas13a could substantially reduce HIV RNA levels in HEK293T cells[
61]. By targeting viral RNA, Cas13a effectively suppressed viral replication, highlighting its potential to diminish latent viral reservoirs, which remain a major obstacle in HIV cure research. The ability of Cas13 to specifically target RNA offers the possibility of disrupting the transcription of latent HIV, providing hope for reducing or even eradicating these viral reservoirs. This approach could lead to functional cures, effectively eliminating HIV from the body and reducing the dependence on lifelong antiretroviral therapy.
More recent advancements in Cas13-based HIV research further explored its potential in therapy and viral detection. In 2021, RfxCas13d (CasRx) combined with HIV-specific gRNAs effectively inhibited HIV-1 replication in cell models, primary CD4⁺ T cells, and reactivated latent HIV, demonstrating its therapeutic potential[
62] Later, a membrane-based digital Cas13a system enabled absolute quantification of HIV-1 viral particles without amplification, optimizing crRNA design for enhanced detection sensitivity[
63]. In 2023, a point-of-care Cas13a-based method provided a stable, simple, and highly sensitive tool for early HIV diagnosis and treatment monitoring, reinforcing Cas13’s role in next-generation HIV management[
64].
3.3. Dengue, Influenza and Other RNA Viruses
Dengue virus and other flaviviruses, such as Zika and West Nile virus, are responsible for millions of infections annually, particularly in tropical regions. Cas13 has shown potential in inhibiting these viruses by directly targeting their RNA genomes. Li’s group applied Cas13a to target the NS3 gene of the dengue virus, which is crucial for viral replication[
43]. By introducing Cas13a into infected Vero cells, they observed a 95% reduction in viral RNA levels within two days, demonstrating Cas13's effectiveness in disrupting dengue virus replication. Santangelo’s research team was the first to report the use of lipid nanoparticles (LNPs) for delivering mRNA encoding Cas13a and guide RNA targeting the dengue virus (DENV) type 2 and 3 genomes. This treatment was administered one day after infection with a lethal viral dose. The approach successfully reduced DENV RNA levels in a mouse model and protected the mice from mortality[
45]. The team is now collaborating with TFF Pharmaceuticals to explore the feasibility of producing dry powder inhalation formulations, advancing the potential for practical and efficient therapeutic applications.
The team led by Chiara Zurla and Philip J. Santangelo at Emory University pioneered a method for delivering mRNA encoding Cas13a and a replicase targeting the highly conserved regions of the influenza virus PB1/PB2 and SARS-CoV-2 via PBAE polymers in 2021[
37]. This strategy utilizes CRISPR RNA (crRNA) specific to the nucleocapsid gene, effectively degrading viral RNA in lung tissue following aerosol inhalation. This approach has demonstrated efficacy in mitigating symptoms of viral infections and offers a therapeutic avenue for influenza A and SARS-CoV-2. Building on this, the researchers employed PBAE polymers to encapsulate the mRNA for delivery through a human aerosol inhalation device. Notably, even when administered 24 hours post-infection in mice, the viral RNA in lung tissue, as quantified by qPCR, was reduced by 90% over three days. Moreover, cytopathic effect (CPE) indicators were significantly diminished, underscoring the effectiveness of this strategy in inhibiting viral replication and providing therapeutic benefits post-infection. mRNA-encoded LbuCas13a, along with two crRNAs targeting H1N1 and H3N2 strains, was tested in A549 cells and hamsters. Recently, another group developed a similar strategy utilizing Cas13 to inhibit influenza viruses[
47]. They employed mRNA-encoded LbuCas13a combined with two crRNAs designed to target H1N1 and H3N2 strains. This approach was tested in both A549 cells and hamsters. In vitro studies demonstrated effective RNA degradation when the treatment was administered 24 hours post-infection. Furthermore, in vivo experiments in hamsters revealed a significant 1–2 log reduction in viral titers, underscoring the potential of this method for combating influenza infections. In a subsequent development in 2024, The highly mutagenic nature of influenza A virus (IAV) makes it a difficult target for vaccines and antivirals. Cas13 offers an alternative by targeting the RNA of various strains of influenza, potentially preventing viral replication before new mutations can arise. In cell culture models, Cas13a was shown to reduce IAV replication by fourfold, offering a potential therapeutic pathway for seasonal and pandemic influenza[
46].
Similarly, Zika virus is a single-stranded RNA (ssRNA) virus. Hao Pei's team successfully used Cas13 to cleave Zika virus ssRNA in mammalian cells and screened several effective targets on the Zika virus genome, such as gE-2, GNS1-1, GNS1-2, GNS3-1, and gNS4B-1. These targets were designed for the highly conserved regions in the Zika virus genome. Multiple Zika virus mutants can be targeted, thereby preventing the escape of the virus mutants[
65].
Hepatitis C Virus (HCV), another RNA virus, has also been targeted with Cas13, particularly at its internal ribosome entry site (IRES). Early studies have demonstrated that Cas13 can significantly reduce HCV replication, paving the way for its use in treating chronic infections like hepatitis C[
49]. Additionally, Cas13a-based diagnostic technology has demonstrated a high sensitivity and specificity (81.9% as compared to 66.7% for conventional qPCR detection method) in detecting the Hepatitis delta virus (HDV). This technology shows promise as a potential tool for monitoring HDV infection progression and evaluating therapeutic efficacy[
66].
Borna disease virus (BoDV-1) also contains an RNA genome, which infects the central nervous system of various animals, including humans, causing fatal encephalitis and persistent neuronal infections with neurobehavioral effects. Sasaki et al. showed that the CRISPR/Cas13 system effectively reduces viral mRNAs and genomic RNA in persistently infected cells, demonstrating its potential to suppress BoDV-1 in both acute and persistent infections[
51].
4. Challenges and Limitations of Cas13-Based Antiviral Therapies
4.1. Collateral Cleavage and Off-Target Effects
While Cas13’s specificity is a key advantage, its collateral cleavage activity poses a significant challenge. Once activated, Cas13 can degrade other non-target RNAs in the vicinity, raising the risk of unintended degradation of host transcripts and cell toxicity. In human cells, such off-target RNA degradation could disrupt essential cellular functions, leading to cytotoxicity or unintended side effects. Another way to increase the cleavage specificity requires strict design of guide RNAs.
4.2. Delivery Challenges
Efficiently delivering Cas13 and its associated crRNAs into target cells remains a major obstacle. Viral vectors, such as adeno-associated viruses (AAVs), are commonly used to deliver CRISPR systems but face limitations, including immune responses and limited capacity to deliver large cargo. Alternatively, non-viral delivery systems, such as LNPs, have shown promise, particularly for delivering mRNA-based therapeutics like Cas13, but achieving tissue-specific targeting remains a key challenge.
4.3. Immunogenicity
Another limitation of Cas13-based therapies is the potential for immunogenicity. Cas13 proteins, especially when delivered via viral vectors, may be recognized by the host immune system, triggering an immune response that reduces the effectiveness of the treatment. This is particularly concerning for chronic infections that require repeated administrations, as the immune system may neutralize the Cas13 proteins or delivery vectors over time.
4.4. Limited in Vivo Data
Although Cas13 has shown great promise in in vitro studies and some in vivo animal models, there is still limited data on its long-term safety and efficacy in humans. Most Cas13 antiviral studies have been conducted in cell lines rather than in live animal models. Few studies have tested Cas13 in mice or non-human primates, making it difficult to assess immune response, pharmacokinetics, and long-term safety. The complexity of viral infections in living organisms, such as tissue tropism and immune evasion, requires rigorous in vivo validation. Until now, Cas13 antiviral therapies have not yet entered advanced preclinical or clinical trials, limiting real-world validation.
4.5. Regulatory and Clinical Approval Hurdles:
While Cas13-based antiviral therapies hold great potential, their translation from research to clinical use faces several regulatory and approval challenges. These hurdles include 1. Delivery optimization needs demonstrating stable, targeted delivery in animal models before advancing to human trials. 2. Efficacy validation in Preclinical and Clinical Trials requires Phase I–III clinical trials to establish safety, dosing, efficacy, and long-term effects. 3. Safety concerns to conduct preclinical animal studies to assess toxicity, immunogenicity, and long-term effects before moving to human trials. 4. Regulatory frameworks that must be addressed with compliance with bioethical guidelines and international regulations for gene-editing therapies before Cas13-based therapies can be approved for human use.
5. Future Directions and Ethical Considerations
5.1. Research Advancements
Advancements in Cas13-based therapies are being driven by ongoing efforts to enhance specificity, optimize delivery mechanisms, and expand antiviral applications. Research is actively refining crRNA design and employing protein engineering to improve Cas13 specificity, aiming to reduce off-target effects and minimize collateral cleavage while preserving high on-target efficiency. Beyond traditional considerations for selecting the optimal target region and refining crRNA length and composition, significant advancements in computational tools for off-target prediction and crRNA selection are essential. Perturb-Seq, which integrates pooled CRISPR-Cas13 crRNA screening with scRNA-seq readouts, offers a powerful approach to generating large-scale data. This wealth of information can foster collaboration with bioinformaticians, enabling the development of machine-learning models to refine crRNA design. Ultimately, these efforts could lead to a universal computational tool capable of precisely selecting crRNAs for targeting RNA-based viruses with high specificity and efficiency. Modifying gRNA chemically or structurally by introducing Locked Nucleic Acids (LNAs) or optimizing spacer sequence may also increase specificity and prevent off-target activation.
Simultaneously, innovative delivery methods, such as tissue-specific AAV serotypes, are being developed to improve the targeting of infected tissues both in vivo and in vitro. These approaches seek to address current challenges associated with systemic delivery by introducing tissue specific promoters or tagging with cell specific peptides for direct and precise expression of Cas13. In the case of using LNPs, engineering organ- or tissue-specific LNPs by modifying lipid composition or adding targeting ligands(e.g., transferrin for brain targeting) will be worthy to explore. Further efforts with bioengineering smaller Cas13 proteins or modifying non-essential Cas13 regions while maintaining RNA cleavage activity are also needed to overcome the challenges with AAV deliver capacity. Both modification on AAV and Cas13 may also help for solving immunogenicity.
Furthermore, Cas13 holds significant potential as a broad-spectrum antiviral platform. By targeting conserved RNA sequences shared across various RNA viruses, it could be engineered to combat multiple viral threats with minimal modifications, offering a versatile solution to emerging infectious diseases.
Each subtype of the Cas13 family exhibits unique properties in terms of protein size, crRNA requirements, and targeting efficiency and cleavage activity against different viruses. Different members of the Cas13 family are used to combat viruses in different cell lines, there numerous factors, including Cas13 orthologs, target regions within viral genomes, crRNA design, and levels of Cas13 protein expression, can contribute to the variability in inhibitory effects on different viruses across various hosts. Therefore, optimizing these elements is essential to enhance the antiviral efficacy of Cas13.
5.2. Ethical Considerations
Cas13-based therapies, though promising, face several challenges that must be addressed to ensure their safety, efficacy, and accessibility in vivo. A key concern is the potential for off-target effects, as Cas13's RNA-targeting mechanism, while temporary, could inadvertently affect the host transcriptome. Rigorous preclinical testing is crucial to mitigate these risks and ensure the safety of such therapies. Another critical issue is the immunogenicity and long-term safety of Cas13 proteins and their delivery systems. The risk of immune reactions must be thoroughly evaluated, prompting the exploration of immune-evasive Cas13 variants or alternative non-viral delivery methods that minimize immune recognition. Additionally, as these advanced therapies progress, ensuring equitable access is imperative. Populations in low- and middle-income countries, which are often most affected by RNA viral outbreaks, must benefit from these innovations. Global collaboration will play a vital role in making these potentially life-saving technologies accessible to all, regardless of socioeconomic barriers.
6. Conclusion
The CRISPR/Cas13 system represents a breakthrough in antiviral research, offering a novel approach to targeting RNA viruses. Its unique ability to degrade viral RNA with high specificity provides a promising alternative to traditional antiviral therapies. Cas13’s programmability and broad-spectrum potential make it a versatile tool in the fight against RNA viruses, especially those that mutate rapidly, such as SARS-CoV-2 and HIV. However, challenges related to off-target effects, delivery, and immunogenicity must be addressed before Cas13 can be widely adopted in clinical practice. With ongoing research and development, Cas13 has the potential to revolutionize antiviral therapies and provide new solutions for combating some of the world’s most pressing viral threats.
Author Contributions
Conceptualization, X.T., B.C., G.H. and X.X.; writing—original draft preparation, X.T., J.L., J.W. and X.X.; writing—review and editing, X.T., B.C., K.T., G.H. and X.X.; funding acquisition, G.H. and X.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by German Heart Research Foundation(DSHF), grant number ”F/19/23” and by German Center for Cardiovascular Research(DZHK), grant number ”DZHK B24-022SE”.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| UTR |
Untranslated Regions |
| ORFs |
Open Reading Frames |
| RdRPs |
RNA-Dependent RNA Polymerases |
| PRRSV |
Porcine Reproductive and Respiratory Syndrome Virus |
| AIVs |
Avian Influenza Viruses |
| ART |
Antiretroviral Therapy |
| CRISPR |
Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas |
CRISPR/Associated Proteins |
| HEPN |
Higher Eukaryotes and Prokaryotes Nucleotide-Binding |
| WHO |
World Health Organization |
| HIV |
Human Immunodeficiency Virus |
| HAART |
Highly Active Antiretroviral Therapy |
| LNPs |
Lipid Nanoparticles |
| DENV |
Dengue Virus |
| crRNA |
CRISPR RNA |
| CPE |
Cytopathic Effect |
| IAV |
Influenza A Virus |
| ssRNA |
Single-Stranded RNA |
| HCV |
Hepatitis C Virus |
| IRES |
Internal Ribosome Entry Site |
| HDV |
Hepatitis Delta Virus |
| BoDV-1 |
Borna Disease Virus |
| AAVs |
Adeno-Associated Viruses |
| LNAs |
Locked Nucleic Acids |
References
- Bolduc, B.; Shaughnessy, D.P.; Wolf, Y.I.; Koonin, E.V.; Roberto, F.F.; Young, M. Identification of Novel Positive-Strand RNA Viruses by Metagenomic Analysis of Archaea-Dominated Yellowstone Hot Springs. Journal of Virology 2012, 86, 5562–5573. [Google Scholar] [CrossRef] [PubMed]
- Callanan, J.; Stockdale, S.R.; Adriaenssens, E.M.; Kuhn, J.H.; Rumnieks, J.; Pallen, M.J.; Shkoporov, A.N.; Draper, L.A.; Ross, R.P.; Hill, C. Leviviricetes: expanding and restructuring the taxonomy of bacteria-infecting single-stranded RNA viruses. Microbial Genomics 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Agol, V.I. The Baltimore Classification of Viruses 50 Years Later: How Does It Stand in the Light of Virus Evolution? Microbiology and Molecular Biology Reviews 2021, 85, e00053–e00021. [Google Scholar] [CrossRef] [PubMed]
- Dreher, T.W. F UNCTIONS OF THE 3′-U NTRANSLATED R EGIONS OF P OSITIVE S TRAND RNA V IRAL G ENOMES. Annual Review of Phytopathology 1999, 37, 151–174. [Google Scholar] [CrossRef]
- Strauss, E.G.; Strauss, J.H. RNA viruses: genome structure and evolution. Current Opinion in Genetics & Development 1991, 1, 485–493. [Google Scholar] [CrossRef]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A New Wine Saccharomyces cerevisiae Killer Toxin (Klus), Encoded by a Double-Stranded RNA Virus, with Broad Antifungal Activity Is Evolutionarily Related to a Chromosomal Host Gene. Applied and Environmental Microbiology 2011, 77, 1822–1832. [Google Scholar] [CrossRef]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A planarian nidovirus expands the limits of RNA genome size. PLOS Pathogens 2018, 14, e1007314. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Research 2006, 117, 17–37. [Google Scholar] [CrossRef]
- Nayak, B.; Kumar, S.; Collins, P.L.; Samal, S.K. Molecular characterization and complete genome sequence of avian paramyxovirus type 4 prototype strain duck/Hong Kong/D3/75. Virology Journal 2008, 5, 124. [Google Scholar] [CrossRef]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef]
- Mattenberger, F.; Vila-Nistal, M.; Geller, R. Increased RNA virus population diversity improves adaptability. Scientific Reports 2021, 11, 6824. [Google Scholar] [CrossRef] [PubMed]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cellular and Molecular Life Sciences 2016, 73, 4433–4448. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: patterns and determinants. Nature Reviews. Genetics 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Graham, R.L.; Baric, R.S. Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission. Journal of Virology 2010, 84, 3134–3146. [Google Scholar] [CrossRef]
- Li, Y.-T.; Linster, M.; Mendenhall, I.H.; Su, Y.C.F.; Smith, G.J.D. Avian influenza viruses in humans: lessons from past outbreaks. British Medical Bulletin 2019, 132, 81–95. [Google Scholar] [CrossRef]
- Gray, G.C.; Robie, E.R.; Studstill, C.J.; Nunn, C.L. Mitigating Future Respiratory Virus Pandemics: New Threats and Approaches to Consider. Viruses 2021, 13, 637. [Google Scholar] [CrossRef]
- Binder, S.; Levitt, A.M.; Sacks, J.J.; Hughes, J.M. Emerging infectious diseases: public health issues for the 21st century. Science (New York, N.Y.) 1999, 284, 1311–1313. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242–249. [Google Scholar] [CrossRef]
- Woolhouse, M.E.J.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerging Infectious Diseases 2005, 11, 1842–1847. [Google Scholar] [CrossRef]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. Journal of Bacteriology 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Embden, J.D.A.v.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences 2012, 109. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Ledford, H. CRISPR treatment inserted directly into the body for first time. Nature 2020, 579, 185–185. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Molecular Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e614. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Molecular Cell 2017, 65, 618–630.e617. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Molecular Cell 2018, 70, 327–339.e325. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y.; et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat Methods 2021, 18, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Huang, J.; Xiao, Q.; He, B.; Dong, X.; Liu, Y.; Yang, X.; Han, D.; Wang, Z.; Ying, W.; et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effect. Nature Biotechnology 2021. [Google Scholar] [CrossRef]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876.e812. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, Y.; Nguyenla, X.H.; Abbott, T.R.; Han, M.; Zhu, Y.; Chemparathy, A.; Lin, X.; Chen, X.; Wang, H.; et al. Broad-spectrum CRISPR-mediated inhibition of SARS-CoV-2 variants and endemic coronaviruses in vitro. Nature Communications 2022, 13, 2766. [Google Scholar] [CrossRef]
- Blanchard, E.L.; Vanover, D.; Bawage, S.S.; Tiwari, P.M.; Rotolo, L.; Beyersdorf, J.; Peck, H.E.; Bruno, N.C.; Hincapie, R.; Michel, F.; et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nature Biotechnology 2021, 39, 717–726. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, J.; Wang, Q.; Wang, Y.; Kang, C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics 2021, 11, 649–664. [Google Scholar] [CrossRef]
- Yu, D.; Han, H.J.; Yu, J.; Kim, J.; Lee, G.H.; Yang, J.H.; Song, B.M.; Tark, D.; Choi, B.S.; Kang, S.M.; et al. Pseudoknot-targeting Cas13b combats SARS-CoV-2 infection by suppressing viral replication. Mol Ther 2023, 31, 1675–1687. [Google Scholar] [CrossRef]
- Hussein, M.; Andrade Dos Ramos, Z.; Vink, M.A.; Kroon, P.; Yu, Z.; Enjuanes, L.; Zuniga, S.; Berkhout, B.; Herrera-Carrillo, E. Efficient CRISPR-Cas13d-Based Antiviral Strategy to Combat SARS-CoV-2. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, X.; Kan, C.; Li, L.; Zhang, Y.; Gao, Y.; Zhang, S.; Zhou, L.; Zhao, H.; Li, M.; et al. CRISPR-Cas13d effectively targets SARS-CoV-2 variants, including Delta and Omicron, and inhibits viral infection. MedComm (2020) 2023, 4, e208. [Google Scholar] [CrossRef] [PubMed]
- Keng, C.T.; Yogarajah, T.; Lee, R.C.H.; Muhammad, I.B.H.; Chia, B.S.; Vasandani, S.R.; Lim, D.S.; Guo, K.; Wong, Y.H.; Mok, C.K.; et al. AAV-CRISPR-Cas13 eliminates human enterovirus and prevents death of infected mice. EBioMedicine 2023, 93, 104682. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Dong, X.; Li, Q.; Li, M.; Li, J.; Guo, Y.; Jin, X.; Zhou, Y.; Song, H.; et al. CRISPR-Cas13a Cleavage of Dengue Virus NS3 Gene Efficiently Inhibits Viral Replication. Mol Ther Nucleic Acids 2020, 19, 1460–1469. [Google Scholar] [CrossRef]
- Singsuksawat, E.; Onnome, S.; Posiri, P.; Suphatrakul, A.; Srisuk, N.; Nantachokchawapan, R.; Praneechit, H.; Sae-Kow, C.; Chidpratum, P.; Sa-Ngiamsuntorn, K.; et al. Potent programmable antiviral against dengue virus in primary human cells by Cas13b RNP with short spacer and delivery by VLP. Mol Ther Methods Clin Dev 2021, 21, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Zurla, C.; Auroni, T.T.; Vanover, D.; Chaves, L.C.S.; Sadhwani, H.; Pathak, H.; Basu, R.; Beyersdorf, J.P.; Amuda, O.O.; et al. mRNA-encoded Cas13 can be used to treat dengue infections in mice. Nat Microbiol 2024, 9, 2160–2172. [Google Scholar] [CrossRef]
- Challagulla, A.; Schat, K.A.; Doran, T.J. In Vitro Inhibition of Influenza Virus Using CRISPR/Cas13a in Chicken Cells. Methods and Protocols 2021, 4, 40. [Google Scholar] [CrossRef]
- Chaves, L.C.S.; Orr-Burks, N.; Vanover, D.; Mosur, V.V.; Hosking, S.R.; Kumar, E.K.P.; Jeong, H.; Jung, Y.; Assumpcao, J.A.F.; Peck, H.E.; et al. mRNA-encoded Cas13 treatment of Influenza via site-specific degradation of genomic RNA. PLoS Pathog 2024, 20, e1012345. [Google Scholar] [CrossRef]
- Cui, J.; Techakriengkrai, N.; Nedumpun, T.; Suradhat, S. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci Rep 2020, 10, 9617. [Google Scholar] [CrossRef]
- Ashraf, M.U.; Salman, H.M.; Khalid, M.F.; Khan, M.H.F.; Anwar, S.; Afzal, S.; Idrees, M.; Chaudhary, S.U. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie 2021, 136, 111239. [Google Scholar] [CrossRef]
- Adler, B.A.; Hessler, T.; Cress, B.F.; Lahiri, A.; Mutalik, V.K.; Barrangou, R.; Banfield, J.; Doudna, J.A. Broad-spectrum CRISPR-Cas13a enables efficient phage genome editing. Nat Microbiol 2022, 7, 1967–1979. [Google Scholar] [CrossRef]
- Sasaki, S.; Ogawa, H.; Katoh, H.; Honda, T. Suppression of Borna Disease Virus Replication during Its Persistent Infection Using the CRISPR/Cas13b System. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef] [PubMed]
- Fareh, M.; Zhao, W.; Hu, W.; Casan, J.M.L.; Kumar, A.; Symons, J.; Zerbato, J.M.; Fong, D.; Voskoboinik, I.; Ekert, P.G.; et al. Reprogrammed CRISPR-Cas13b suppresses SARS-CoV-2 replication and circumvents its mutational escape through mismatch tolerance. Nature Communications 2021, 12, 4270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Meng, H.Y.; Liu, H.H.; Ye, Q. Advances in laboratory detection methods and technology application of SARS-CoV-2. J Med Virol 2022, 94, 1357–1365. [Google Scholar] [CrossRef]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.-P.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of quantitative real-time RT-PCR assays for the detection of six honeybee viruses. Journal of Virological Methods 2019, 270, 70–78. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.-l.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. 2020. [CrossRef]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A Scalable, Easy-to-Deploy Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. Journal of Clinical Microbiology 2021, 59, e02402–02420. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.-S.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nature Communications 2020, 11, 5921. [Google Scholar] [CrossRef]
- Scott, G.Y.; Worku, D. HIV vaccination: Navigating the path to a transformative breakthrough—A review of current evidence. Health Science Reports 2024, 7, e70089. [Google Scholar] [CrossRef]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 2013, 3, 2510. [Google Scholar] [CrossRef]
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Molecular Therapy. Nucleic Acids 2020, 21, 147–155. [Google Scholar] [CrossRef]
- Nguyen, H.; Wilson, H.; Jayakumar, S.; Kulkarni, V.; Kulkarni, S. Efficient Inhibition of HIV Using CRISPR/Cas13d Nuclease System. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Jiang, Y.; Politza, A.J.; Liu, T.; Greene, W.H.; Zhu, Y.; Nunez, J.J.; Lian, X.; Guan, W. STAMP-Based Digital CRISPR-Cas13a for Amplification-Free Quantification of HIV-1 Plasma Viral Loads. ACS Nano 2023, 17, 10701–10712. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, B.; Yang, L.; Kou, Z.; Wu, H.; Zhang, T.; Liu, L.; Han, Y.; Niu, M.; Sun, Y.; et al. Highly sensitive and rapid point-of-care testing for HIV-1 infection based on CRISPR-Cas13a system. BMC Infect Dis 2023, 23, 627. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, M.; Chen, Y.; Jing, X.; Zhang, N.; Zhou, X.; Li, X.; Long, G.; Hao, P. Targeted inhibition of Zika virus infection in human cells by CRISPR-Cas13b. Virus Research 2022, 312, 198707. [Google Scholar] [CrossRef]
- Tian, Y.; Fan, Z.; Zhang, X.; Xu, L.; Cao, Y.; Pan, Z.; Mo, Y.; Gao, Y.; Zheng, S.; Huang, J.; et al. CRISPR/Cas13a-Assisted accurate and portable hepatitis D virus RNA detection. Emerg Microbes Infect 2023, 12, 2276337. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).