Submitted:
18 June 2025
Posted:
19 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Kinetic Analysis of OsPXG9-Catalyzed Inter- and Intramolecular Oxygen Transfer
2.2. Products of Intermolecular Oxygen Transfer Using Newly Validated Substrates
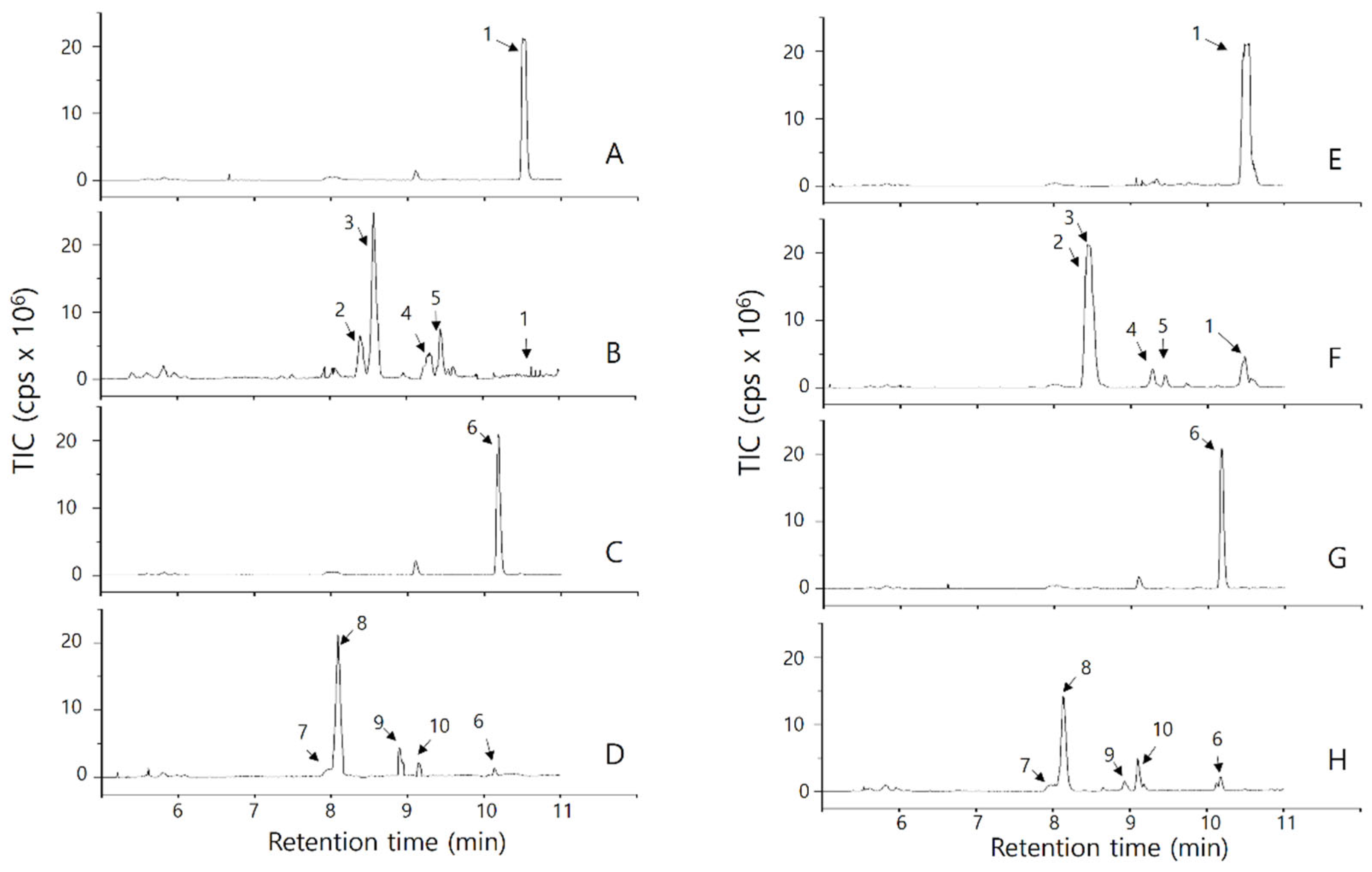
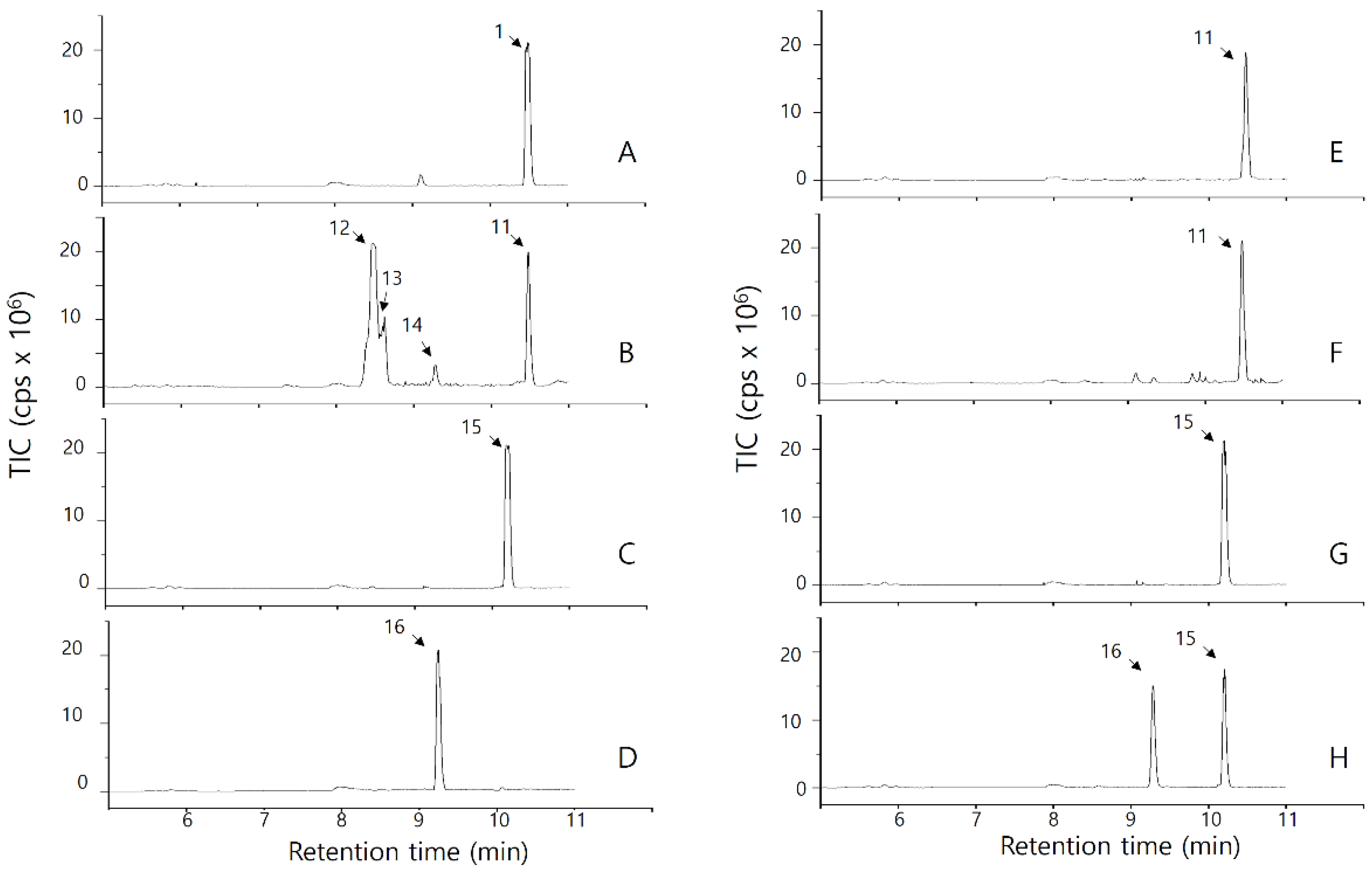
2.2.1. Analysis of OsPXG9 Primary Reaction Products by TLC
2.2.2. Analysis of OsPXG9 Reaction Products in the 9-PXG Pathway
2.2.3. Analysis of OsPXG9 Reaction Products in the 13-PXG Pathway
2.2.4. Relative Abundance of Products from 9- and 13-PXG Pathway Catalyzed by OsPXG9
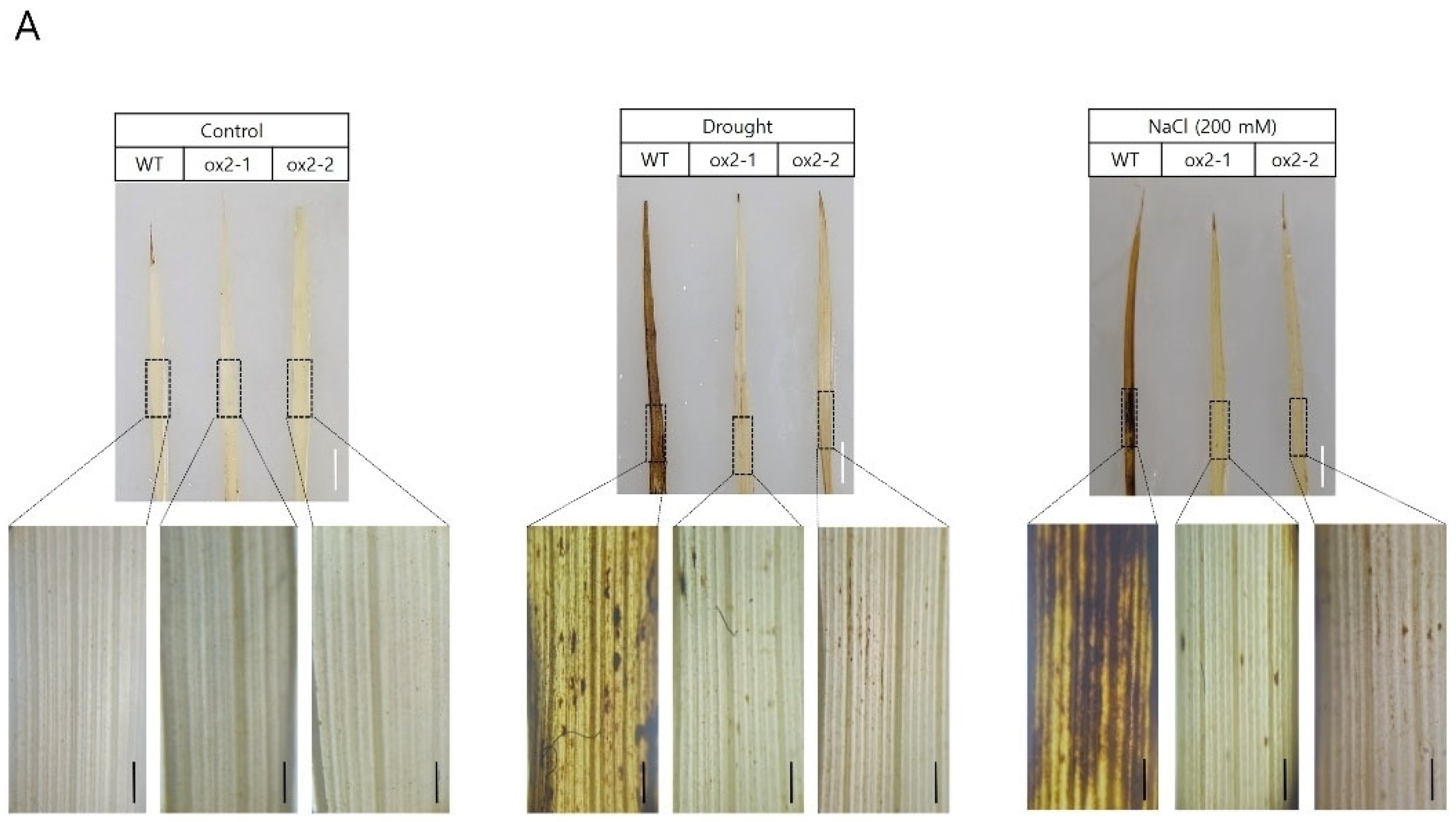
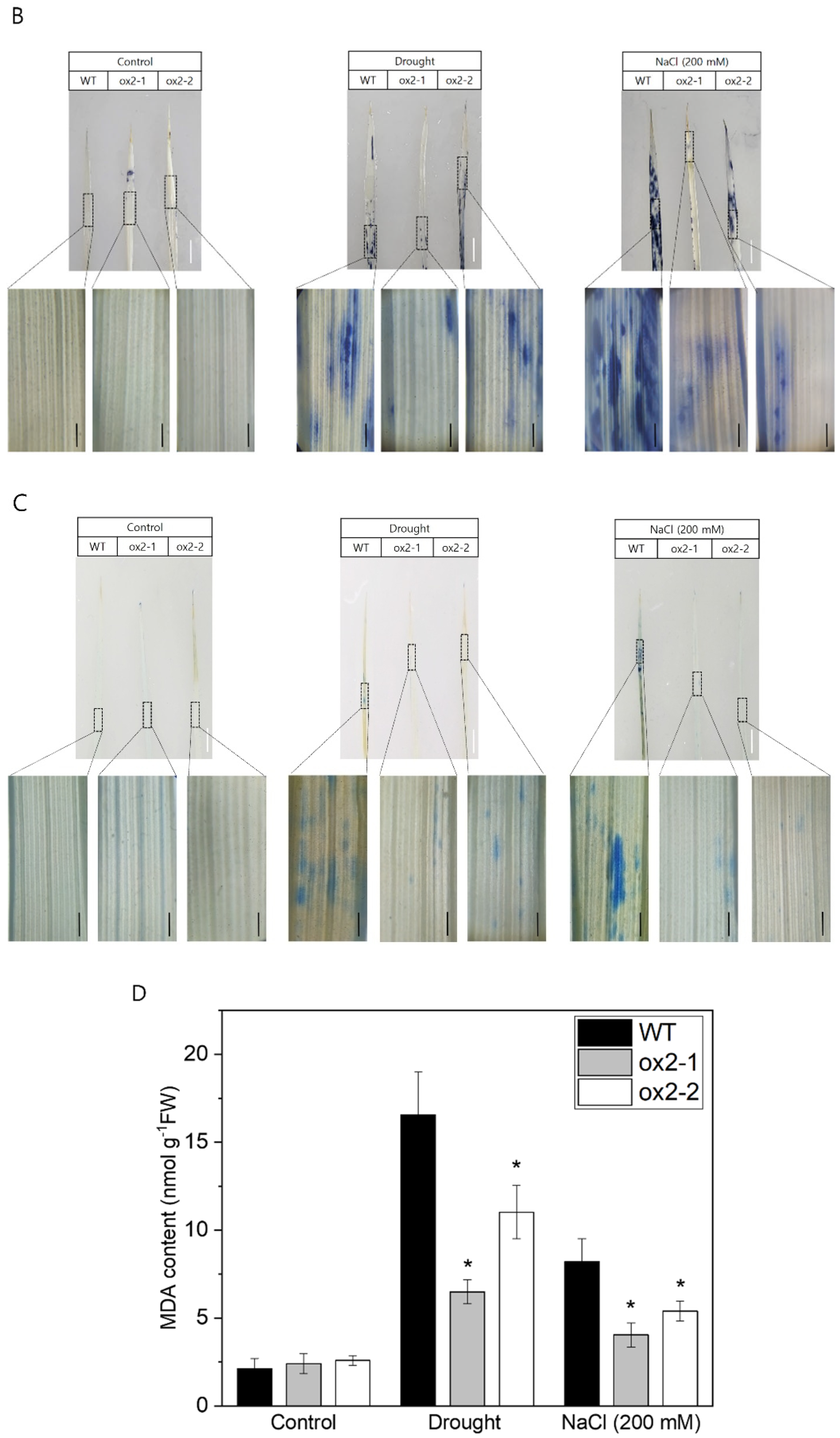
2.3. Overexpression of OsPXG9 Reduces ROS Abundance and Lowers the Frequency of Apoptotic Cell Death after Exposure to Abiotic Stress
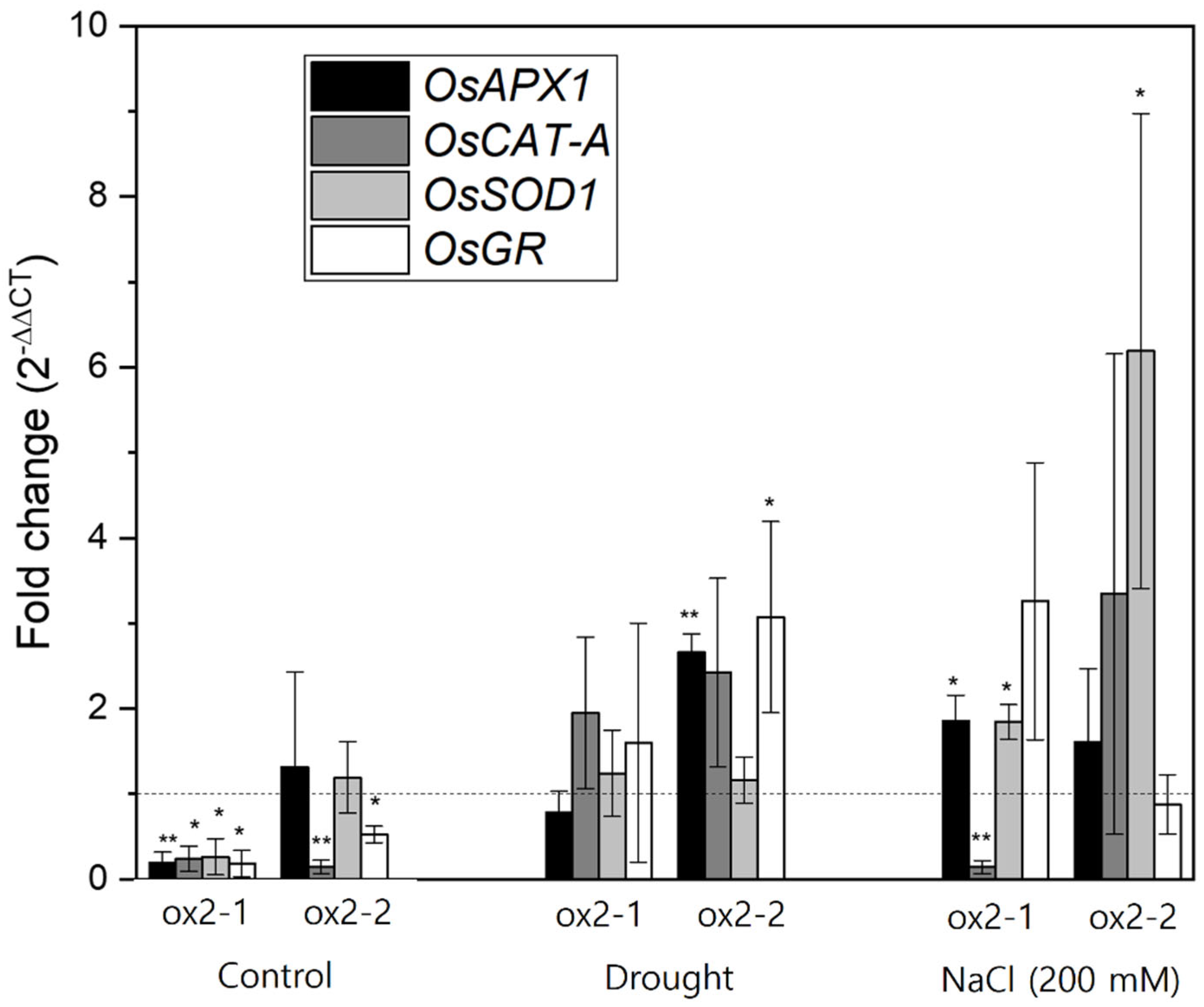
2.4. Overexpression of OsPXG9 Correlates with Reduced Expression of Antioxidant Pathways in Unstressed Plants
3. Discussion
3.1. In vitro Studies of OsPXG9-Dependent Hydroperoxide Scavenging Activity
3.2. OsPXG9 Contributes to Regulate ROS Level and Reduces Cell Death upon Drought and Salt Stress
3.3. Overexpression of OsPXG9 Results in Down-Regulation of Other Antioxidant-Related Enzymes for the Cellular Redox State Balance
4. Materials and Methods
4.1. Vector Construction
4.2. Heterologous Expression and Purification of OsPXG9
4.3. Substrate Preparation, Enzyme Assay, and Kinetic Analysis
4.4. Extraction and Thin Layer Chromatography (TLC) of OsPXG9 Reaction Mixture
4.5. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) and Nuclear Magnetic Resonance Spectroscopy (NMR)
4.6. Agrobacterium-Mediated Transformation
4.7. Plant Materials and Stress Treatments
4.8. Histochemical Staining and MDA Content Quantification
4.9. RNA Extraction and Real-Time Quantitative Reverse Transcription PCR
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 9(S)-10,11-EHOE | 9(S)-10,11-epoxy-9-hydroxy-octadecenoic acid |
| 9(S)-12,13-EHOE | 9(S)-12,13-epoxy-9-hydroxy-octadecenoic acid |
| 9(S)-9,12,13-THO(D)E | 9(S)-9,12,13-trihydroxy-octadec(adi)enoic acid |
| 9(S)-HOD(T)E | 9(S)-hydroxy-octadecadi(tri)enoic acid |
| 9(S)-HPOD(T)E | 9(S)-hydroperoxy-octadecadi(tri)enoic acid |
| 13(S)-9,10-EHOE | 13(S)-9,10-epoxy-13-hydroxy-octadecenoic acid |
| 13(S)-9,10,13-THOE | 13(S)-9,10,13-trihydroxy-octadecenoic acid |
| 13(S)-15,16-EHODE | 13(S)-15,16-epoxy-13-hydroxy-octadecadienoic acid |
| 13(S)-HOD(T)E | 13(S)-hydroxy-octadecadi(tri)enoic acid |
| 13(S)-HPOD(T)E | 13(S)-hydroperoxy-octadecadi(tri)enoic acid |
| APX | ascorbate peroxidase |
| CAT | catalase |
| CLO/PXG | caleosin/peroxygenase |
| CuOOH | cumene hydroperoxide |
| DAB | 3,3'-diaminobenzidine |
| GR | glutathione reductase |
| HOOH | hydrogen peroxide |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| ko | knockout |
| LA | linoleic acid |
| LnA | linolenic acid |
| LOX | lipoxygenase |
| LPO | lipid peroxidation |
| MDA | malondialdehyde |
| NBT | nitroblue tetrazolium |
| OA | oleic acid |
| ox | overexpression |
| PUFAs | polyunsaturated fatty acid |
| ROS | reactive oxygen species |
| SPS | sucrose phosphate synthase |
References
- Hanano, A.; Blee, E.; Murphy, D.J. Caleosin/peroxygenases: multifunctional proteins in plants. Ann Bot 2023, 131, 387–409. [Google Scholar] [CrossRef]
- Frandsen, G.; et al. Novel plant Ca(2+)-binding protein expressed in response to abscisic acid and osmotic stress. J Biol Chem 1996, 271, 343–348. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; et al. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequenceanalysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant and Cell Physiology 1992, 33, 217–224. [Google Scholar] [CrossRef]
- Murphy, D.J. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 2012, 249, 541–585. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, A.; Yamazaki, I. Hydroperoxide-dependent hydroxylation involving "H2O2-reducible hemoprotein" in microsomes of pea seeds. A new type enzyme acting on hydroperoxide and a physiological role of seed lipoxygenase. J Biol Chem 1977, 252, 6118–6124. [Google Scholar] [CrossRef] [PubMed]
- Blee, E.; Durst, F. Oxidation of an organosulfur xenobiotic by microsomes from soybean cotyledons. Biochem Biophys Res Commun 1986, 135, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Blee, E.; Schuber, F. Stereochemistry of the epoxidation of fatty acids catalyzed by soybean peroxygenase. Biochem Biophys Res Commun 1990, 173, 1354–1360. [Google Scholar] [CrossRef]
- Blée, E.; et al. The reductase activity of the Arabidopsis caleosin RESPONSIVE TO DESSICATION20 mediates gibberellin-dependent flowering time, abscisic acid sensitivity, and tolerance to oxidative stress. Plant Physiology 2014, 166, 109–124. [Google Scholar] [CrossRef]
- Hanano, A.; et al. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. Journal of Biological Chemistry 2006, 281, 33140–33151. [Google Scholar] [CrossRef]
- Blee, E.; et al. Mechanism of reaction of fatty acid hydroperoxides with soybean peroxygenase. J Biol Chem, 1993, 268, 1708–1715. [Google Scholar] [CrossRef]
- Hamberg, M.; Hamberg, G. Peroxygenase-Catalyzed Fatty Acid Epoxidation in Cereal Seeds (Sequential Oxidation of Linoleic Acid into 9(S),12(S),13(S)-Trihydroxy-10(E)-Octadecenoic Acid). Plant Physiol 1996, 110, 807–815. [Google Scholar] [CrossRef]
- Chehab, E.W.; et al. Oxylipin pathway in rice and Arabidopsis. Journal of integrative plant biology, 2007, 49, 43–51. [Google Scholar] [CrossRef]
- Izquierdo, Y.; et al. Oxylipins From Different Pathways Trigger Mitochondrial Stress Signaling Through Respiratory Complex III. Front Plant Sci 2021, 12, 705373. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; et al. Oxygenated Fatty Acids with Anti-rice Blast Fungus Activity in Rice Plants. Biosci Biotechnol Biochem 1993, 57, 283–287. [Google Scholar] [CrossRef]
- Agrawal, G.K; et al. Rice octadecanoid pathway. Biochemical and biophysical research communications 2004, 317, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu Rev Plant Biol 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Hanano, A.; et al. Involvement of the caleosin/peroxygenase RD20 in the control of cell death during Arabidopsis responses to pathogens. Plant Signal Behav 2015, 10, e991574. [Google Scholar] [CrossRef]
- Hanano, A.; et al. Identification of a dioxin-responsive oxylipin signature in roots of date palm: involvement of a 9-hydroperoxide fatty acid reductase, caleosin/peroxygenase PdPXG2. Sci Rep 2018, 8, 13181. [Google Scholar] [CrossRef]
- Tran, A.D.; Cho, K.; Han, O. Rice peroxygenase catalyzes lipoxygenase-dependent regiospecific epoxidation of lipid peroxides in the response to abiotic stressors. Bioorg Chem 2023, 131, 106285. [Google Scholar] [CrossRef]
- Prost, I.; et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 2005, 139, 1902–1913. [Google Scholar] [CrossRef]
- Shimada, T.L.; et al. Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol 2014, 164, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; et al. Identification, duplication, evolution and expression analyses of caleosins in Brassica plants and Arabidopsis subspecies. Mol Genet Genomics 2016, 291, 971–988. [Google Scholar] [CrossRef]
- Brocard, L.; et al. Proteomic Analysis of Lipid Droplets from Arabidopsis Aging Leaves Brings New Insight into Their Biogenesis and Functions. Front Plant Sci 2017, 8, 894. [Google Scholar] [CrossRef]
- Du, C.; et al. Proteomic identification of lipid-bodies-associated proteins in maize seeds. Acta Physiologiae Plantarum 2019, 41, 70. [Google Scholar] [CrossRef]
- Hanano, A.; et al. Specific Caleosin/Peroxygenase and Lipoxygenase Activities Are Tissue-Differentially Expressed in Date Palm (Phoenix dactylifera L.) Seedlings and Are Further Induced Following Exposure to the Toxin 2,3,7,8-tetrachlorodibenzo-p-dioxin. Front Plant Sci 2016, 7, 2025. [Google Scholar] [CrossRef]
- Jiang, P.L.; et al. A unique caleosin in oil bodies of lily pollen. Plant Cell Physiol 2008, 49, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, M.L.; Thomas, T.L. ATS1 and ATS3: two novel embryo-specific genes in Arabidopsis thaliana. Plant Mol Biol 1999, 39, 1153–1163. [Google Scholar] [CrossRef]
- Rejon, J.D.; et al. The Pollen Coat Proteome: At the Cutting Edge of Plant Reproduction. Proteomes 2016, 4. [Google Scholar] [CrossRef]
- Pasaribu, B.; et al. Identification of caleosin and oleosin in oil bodies of pine pollen. Plant Physiol Biochem 2017, 111, 20–29. [Google Scholar] [CrossRef]
- Aubert, Y.; et al. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 2010, 51, 1975–1987. [Google Scholar] [CrossRef]
- Blee, E.; Schuber, F. Efficient epoxidation of unsaturated fatty acids by a hydroperoxide-dependent oxygenase. J Biol Chem 1990, 265, 12887–12894. [Google Scholar] [CrossRef] [PubMed]
- Blee, E.; Schuber, F. Properties of plant peroxygenase. Biochem Soc Trans 1992, 20, 223S. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S. and B.K. Hwang. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol 2010, 152, 948–967. [Google Scholar] [CrossRef]
- Rackova, L.; et al. Free radical scavenging activity and lipoxygenase inhibition of Mahonia aquifolium extract and isoquinoline alkaloids. J Inflamm (Lond) 2007, 4, 15. [Google Scholar] [CrossRef]
- Golldack, D.; et al. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front Plant Sci 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front Plant Sci 2015, 6, 1092. [Google Scholar] [CrossRef]
- Chen, L.H.; et al. Effects of Nitrogen Deficiency on the Photosynthesis, Chlorophyll a Fluorescence, Antioxidant System, and Sulfur Compounds in Oryza sativa. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef]
- Meesapyodsuk, D.; Qiu, X. A peroxygenase pathway involved in the biosynthesis of epoxy fatty acids in oat. Plant Physiology 2011, 157, 454–463. [Google Scholar] [CrossRef]
- Blée, E.; et al. A non-canonical caleosin from A rabidopsis efficiently epoxidizes physiological unsaturated fatty acids with complete stereoselectivity. The FEBS journal 2012, 279, 3981–3995. [Google Scholar] [CrossRef]
- Jing, P.; et al. OsClo5 functions as a transcriptional co-repressor by interacting with OsDi19-5 to negatively affect salt stress tolerance in rice seedlings. Plant J 2021, 105, 800–815. [Google Scholar] [CrossRef]
- Zeng, X.; et al. Rice OsClo5, a caleosin protein, negatively regulates cold tolerance through the jasmonate signalling pathway. Plant Biol (Stuttg) 2022, 24, 52–61. [Google Scholar] [CrossRef]
- Li, H.; Poulos, T.L. Structural variation in heme enzymes: a comparative analysis of peroxidase and P450 crystal structures. Structure 1994, 2, 461–464. [Google Scholar] [CrossRef]
- Ashraf, M.A.; et al. Abiotic stress-induced oxidative stress in rice, in Advances in Rice Research for Abiotic Stress Tolerance. 2019, Elsevier. p. 489-504.
- Shanker, R.; Sharma, P. Drought induces oxidative stress and enhances the activities of antioxidant enzymes in growing rice seedling. Journal of Plant and Growth Regulation 2005, 46, 209–221. [Google Scholar]
- Jia, Y.; et al. Rice OsANN9 Enhances Drought Tolerance through Modulating ROS Scavenging Systems. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front Plant Sci 2014, 5, 685. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; et al. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet 2018, 14, e1007662. [Google Scholar] [CrossRef]
- De la Riva, G.A.; et al. Oxidative Stress (OS) in Plants, Beneficial Interactions with Their Microbiome and Practical Implications for Agricultural Biotechnology. 2024.
- Sahu, P.K.; et al. ROS generated from biotic stress: Effects on plants and alleviation by endophytic microbes. Front Plant Sci 2022, 13, 1042936. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V. Mechanisms of oxidative stress in plants: from classical chemistry to cell biology. Environmental and experimental botany 2015, 109, 212–228. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; et al. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants (Basel) 2020, 9. [Google Scholar] [CrossRef]
- Qamer, Z.; et al. Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. Journal of Cotton Research 2021, 4, 9. [Google Scholar] [CrossRef]
- Huang, L.; et al. The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem Biophys Res Commun 2018, 495, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Jantasuriyarat, C.; et al. Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol 2005, 138, 105–115. [Google Scholar] [CrossRef]
- Kaminaka, H.; et al. Gene cloning and expression of cytosolic glutathione reductase in rice (Oryza sativa L.). Plant Cell Physiol 1998, 39, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Ohsuga, H.; Tanaka, K. Nucleotide sequences of two cDNA clones encoding different Cu/Zn-superoxide dismutases expressed in developing rice seed (Oryza sativa L.). Plant Mol Biol 1992, 19, 323–327. [Google Scholar] [CrossRef]
- Tsai, Y.C.; et al. Expression of ascorbate peroxidase and glutathione reductase in roots of rice seedlings in response to NaCl and H2O2. J Plant Physiol 2005, 162, 291–299. [Google Scholar] [CrossRef]
- Goyal, M.M.; Basak, A. Human catalase: looking for complete identity. Protein Cell 2010, 1, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Alamu, O.; et al. Differential Sensitivity of Two Endothelial Cell Lines to Hydrogen Peroxide Toxicity: Relevance for In Vitro Studies of the Blood-Brain Barrier. Cells 2020, 9. [Google Scholar] [CrossRef]
- Fridovich, I. The trail to superoxide dismutase. Protein Sci 1998, 7, 2688–2690. [Google Scholar] [CrossRef]
- Kelly, G.J.; Latzko, E. Soluble ascorbate peroxidase: detection in plants and use in vitamim C estimation. Naturwissenschaften. 1979, 66, 617–619. [Google Scholar]
- Asada, K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annual review of plant biology 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Mittler, R.; et al. Reactive oxygen gene network of plants. Trends Plant Sci 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Chelikani, P.; I. Fita, and P.C. Loewen. Diversity of structures and properties among catalases. Cell Mol Life Sci 2004, 61, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.A.; et al. Compensatory Modulation of Seed Storage Protein Synthesis and Alteration of Starch Accumulation by Selective Editing of 13 kDa Prolamin Genes by CRISPR-Cas9 in Rice. Int J Mol Sci 2024, 25. [Google Scholar] [CrossRef]
- Patti, G.J.; Tautenhahn, R.; Siuzdak, G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nature protocols 2012, 7, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; et al. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Weng, H.; et al. Estimating number of transgene copies in transgenic rapeseed by real-time PCR assay with HMG I/Y as an endogenous reference gene. Plant Molecular Biology Reporter 2004, 22, 289–300. [Google Scholar] [CrossRef]
| Substrateb | Oxygen acceptor |
9(S)-HODE | 9(S)-HOTE | 13(S)-HODE | ||||||||
| Oxygen donor |
HOOH | CuOOH | HOOH | CuOOH | HOOH | CuOOH | ||||||
| kcat (s⁻¹) | 20.1 ± | 1.6 | 175.8 ± | 11.1 | 8.3 ± | 1.6 | 297.6 ± | 122.1 | 96.1 ± | 30.2 | N/Rc | |
| 5.8 ± | 1.2 | 24.7 ± | 5.1 | 7.8 ± | 0.6 | 25.5 ± | 2.7 | 0.6 ± | 0.2 | N/R | ||
| Km (μM) | 23.9 ± | 4.8 | 84.8 ± | 16.9 | 7.4 ± | 1.8 | 231.4 ± | 95.0 | 224.5 ± | 35.0 | N/R | |
| 131.6 ± | 56.7 | 99.2 ± | 38.9 | 224.6 ± | 27.8 | 57.6 ± | 15.9 | 269.1 ± | 128.6 | N/R | ||
| kcat/Km (s⁻¹μM⁻¹) | 0.9 ± | 0.1 | 2.1 ± | 0.3 | 1.1 ± | 0.3 | 1.3 ± | 0.1 | 0.4 ± | 0.1 | N/R | |
| 46.5 × 10⁻³ ± 9.6 × 10⁻³ |
260.8 × 10⁻³ ± 50.3 × 10⁻³ |
34.7 × 10⁻³ ± 2.6 × 10⁻³ |
454.9 × 10⁻³ ± 69.7 × 10⁻³ |
2.4 × 10⁻³ ± 0.4 × 10⁻³ |
N/R | |||||||
| Peak number |
Retention time (min) |
[M-H]- m/z |
MS/MSa | Chemical formula |
Molecular weight |
Mass bank score |
Identification |
| 1 | 10.52 | 295.2 | [M-H]-: 295; [M-H2O-H]-: 277; [M-CH3(CH2)4(CH)4-H]-: 171 | C18H32O3 | 296.2351 | 0.9555 | 9(S)-hydroxy-10,12-octadecadienoic acid (9(S)-HODE) |
| 2 | 8.40 | 329.3 | [M-H]-: 329; [M-H2O-H]-: 311; [M-H2O-H2O-H]-: 293; [M-CH3(CH2)4CH-OH]-: 229; [M-CH3(CH2)4CH-OH-H2O]-: 211; [M-CH3(CH2)4(CH)3-(OH)2]-: 183; [M-CH3(CH2)4(CH)4-(OH)2]-: 171 | C18H34O5 | 330.2406 | 0.4548 | 9(S)-9,12,13-trihydroxy-10-octadecenoic acid (9(S)-9,12,13-THOE) |
| 3 | 8.57 | 0.4806 | |||||
| 4 | 9.33 | 311.2 | [M-H]-: 311; [M-H2O-H]-: 293; [M-CO2-H2O-H]-: 249; [M-CH3(CH2)4(CH)2]-: 211; [M-CH3(CH2)4(CH)2-H2O]-: 193; [M-CH3(CH2)4(CH)3-O]-: 185; [M-CH3(CH2)4(CH)4-O]-: 171 | C18H32O4 | 312.2301 | N/A | 9(S)-10,11-epoxy-9-hydroxy-12-octadecenoic acid (9(S)-10,11-EHOE) |
| 5 | 9.50 | 311.2 | [M-H]-: 311; [M-H2O-H]-: 293; [M-CO2-H2O-H]-: 249; [M-CH3(CH2)4CH-O]-: 211; [M-CH3(CH2)4CH-O-H2O]-: 193; [M-CH3(CH2)4(CH)4-O]-: 171 | C18H32O4 | 312.2301 | N/A | 9(S)-12,13-epoxy-9-hydroxy-10-octadecenoic acid (9(S)-12,13-EHOE) |
| 6 | 10.19 | 293.2 | [M-H]-: 293; [M-H2O-H]-: 275; [M-CH3(CH2)4(CH)4-H2O-H]-: 171 | C18H30O3 | 294.2195 | 0.9420 | 9(S)-hydroxy-10,12-octadecatrienoic acid (9(S)-HOTE) |
| 7 | 8.13 | 327.2 | [M-H]-: 327; [M-H2O-H]-: 309; [M-H2O-H2O-H]-: 291; [M-CH3(CH2)4CH-OH]-: 229; [M-CH3(CH2)4CH-OH-H2O]-: 211; [M-CH3(CH2)4(CH)3-(OH)2]-: 171; [M-CH3(CH2)4(CH)4-(OH)2]-: 171 | C18H32O5 | 328.2250 | 0.4540 | 9(S)-9,12,13-trihydroxy-10,15-octadecadienoic acid (9(S)-9,12,13-THODE) |
| 8 | 8.18 | 0.4487 | |||||
| 9 | 8.94 | 309.2 | [M-H]-: 309; [M-H2O-H]-: 291; [M-CO2-H2O-H]-: 247; [M-CH3(CH2)4-(CH)2]-: 211; [M-CH3(CH2)4-(CH)2]-H2O]-: 193; [M-CH3(CH2)4(CH)3-O]-: 185; [M-CH3(CH2)4(CH)4-O]-: 171 | C18H30O4 | 310.2101 | N/A | 9(S)-10,11-epoxy-9-hydroxy-12-octadecadienoic acid (9(S)-10,11-EHODE) |
| 10 | 9.18 | 309.2 | [M-H]-: 309; [M-H2O-H]-: 291; [M-CO2-H2O-H]-: 247; [M-CH3(CH2)4CH-O]-: 211; [M-CH3(CH2)4CH-O-H2O]-: 193; [M-CH3(CH2)4(CH)4-O]-: 171 | C18H30O4 | 310.2101 | 0.5902 | 9(S)-12,13-epoxy-9-hydroxy-10-octadecadienoic acid (9(S)-12,13-EHODE) |
| 11 | 10.49 | 295.2 | [M-H]-: 295; [M-H2O-H]-: 277; [M-CH3(CH2)4CH-OH-H]-: 195, | C18H32O3 | 296.2351 | 0.8635 | 13(S)-hydroxy-9,11-octadecadienoic acid (13(S)-HODE) |
| 12 | 8.49 | 329.2 | [M-H]-: 329; [M-H2O-H]-: 311; [M-H2O-H2O-H]-: 293; [M-CH3(CH2)4CH-OH]-: 229; [M-CH3(CH2)4CH-OH-H2O]-: 211; [M-CH3(CH2)4(CH)4-(OH)2]-: 171; [M-CO2-(CH2)7-CH-OH-H 2O]-: 139 | C18H34O5 | 330.2406 | N/A | 13(S)-9,10,13-trihydroxy-11-octadecenoic acid (13(S)-9,10,13-THOE) |
| 13 | 8.54 | N/A | |||||
| 14 | 9.35 | 311.2 | [M-H]-: 311; [M-H2O-H]-: 293; [M-H2O- CO2-H]-: 249; [M-CH3(CH2)4CH-OH-H]-: 211; [M-CH3(CH2)4(CH)3-OH-H]-: 183; [M-CH3(CH2)4(CH)3-OH-CO2-H]-: 139; [M-CH3(CH2)4(CH)4-OH-H]-: 171; | C18H32O4 | 312.2301 | N/A | 13(S)-9,10-epoxy-13-hydroxy-11-octadecenoic acid (13(S)-9,10-EHOE ) |
| 15 | 10.20 | 293.2 | [M-H]-: 293; [M-H2O-H]-: 275; [M-CH3(CH2)2(CH)2-H]-: 223; [M-CH3(CH2)2(CH)3-OH-H]-: 195 | C18H30O3 | 294.2195 | 0.9417 | 13(S)-hydroxy-9,11,15-octadecatrienoic acid (13(S)-HOTE) |
| 16 | 9.29 | 309.2 | [M-H]-: 309; [M-H2O-H]-: 291; [M-CH3(CH2)2(CH)2-O]-: 223; [M-CH3(CH2)2(CH)3-OH]-: 195; [M-CH3(CH2)2(CH)2-O-CO2]-: 179 | C18H30O4 | 310.4292 | N/A | 13(S)-15,16-epoxy-13-hydroxy-9,11-octadecadienoic acid (13(S)-15,16-EHODE) |
| Path | Oxygen acceptor | Oxygen donor | Product | Krel |
| 9-PXG | 9(S)-HODE | HOOH | 9(S)-9,12,13-THOE | 22.6 |
| 9(S)-10,11-EHOE | 4.0 | |||
| 9(S)-12,13-EHOE | 4.7 | |||
| CuOOH | 9(S)-9,12,13-THOE | 5.3 | ||
| 9(S)-10,11-EHOE | 0.4 | |||
| 9(S)-12,13-EHOE | 0.1 | |||
| 9(S)-HOTE | HOOH | 9(S)-9,12,13-THODE | 12.7 | |
| 9(S)-10,11-EHODE | 2.0 | |||
| 9(S)-12,13-EHODE | 0.9 | |||
| CuOOH | 9(S)-9,12,13-THODE | 7.1 | ||
| 9(S)-10,11-EHODE | 0.7 | |||
| 9(S)-12,13-EHODE | 1.8 | |||
| 13-PXG | 13(S)-HODE | HOOH | 13(S)-9,10,13-THOE | 3.0 |
| 13(S)-9,10-EHOE | 0.2 | |||
| 13(S)-HOTE | HOOH | 13(S)-15,16-EHODE | 31.7 | |
| CuOOH | 13(S)-15,16-EHODE | 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





