Submitted:
03 June 2025
Posted:
03 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- We employ GNNs to identify VFs leveraging PPI networks. This approach inte-grates both topological information from PPI networks and protein sequence fea-tures. Notably, we pioneer in transforming the VF identification task into a class-imbalanced node classification problem within the graph domain.
- We propose a novel framework for VF identification that combines generative and contrastive self-supervised learning. Through attribute reconstruction and multi-view contrast, these two approaches work synergistically to enhance model performance in imbalanced classification tasks.
2. Materials and Methods
2.1. Datasets
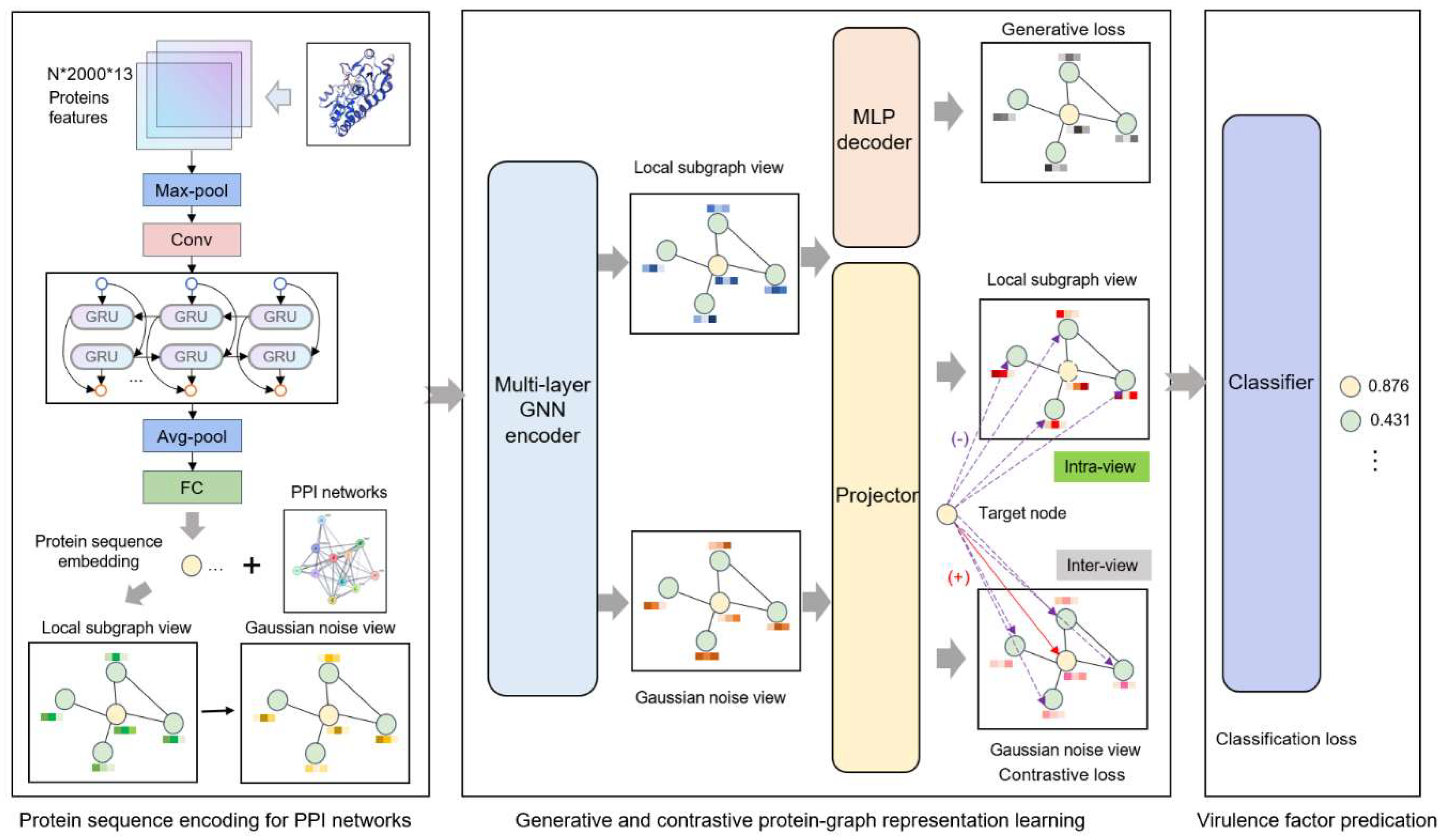
2.2. Protein Sequence Encoding
2.3. Generative and Contrastive Protein Representation Learning
2.3.1. Graph View Establishment
2.3.2. Generative Attribute Reconstruction
2.3.3. Multi-View Local-Local Contrasting
2.4. VF Prediction
3. Results
3.1. Experimental Settings
| Algorithm 1. The key algorithm for the proposed GC-VF framework |
| Input: PPI graph , Initial protein embeddings Xi, Maximum number of training epochs T, Batch size B |
| Output: VF prediction probability |
| 1: for each training epoch
do: 2: Randomly divide the protein nodes V into batches of size B. |
| 3: for each batch
do: 4: for each node vi in b do: |
| 5: Randomly sample a second-order subgraph of vi as , and generate by adding Gaussian noise to the features. 6: Compute the protein embeddings and from the GNN encoder using the embeddings of both views, and , via Eq. (3). 7: Perform attribute reconstruction on via Eq. (4) to obtain the reconstructed protein attribute . 8: Calculate the reconstruction loss using Eq. (5) between and . 9: Project the GNN-encoded target node embeddings from the and by the projection head to obtain latent embeddings via Eq. (6) and Eq. (7), respectively. 10: Perform contrastive learning using and to compute contrastive loss via Eq. (8). 11: Calculate the classification loss for VF prediction via Eq. (12). 12: Update the model parameters by backpropagating the total loss via Eq. (13). 13: end for 14: end for 15: end for 16: Predict the virulence factor probability for node via Eq. (11). |
3.2. Methods Comparison
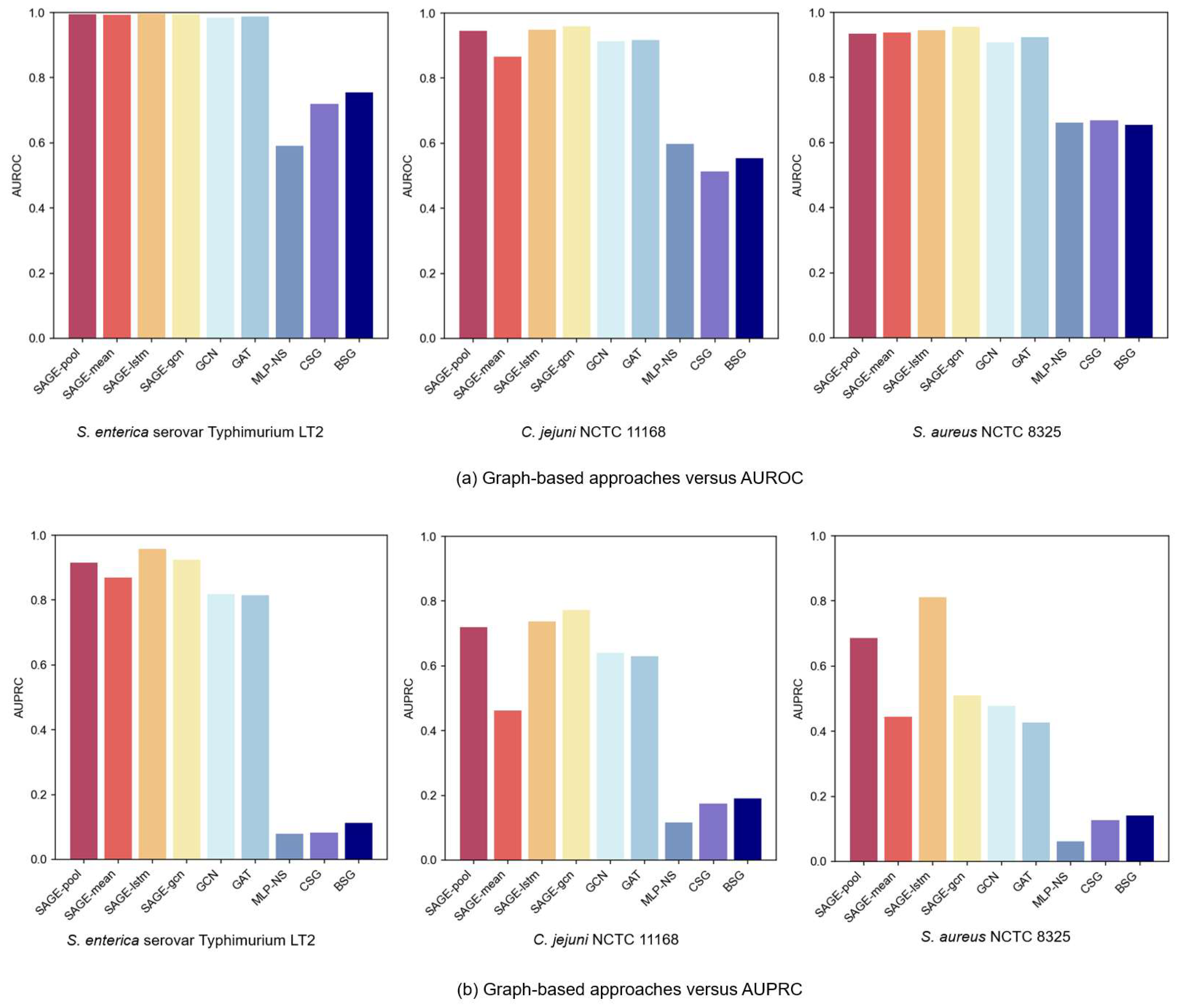
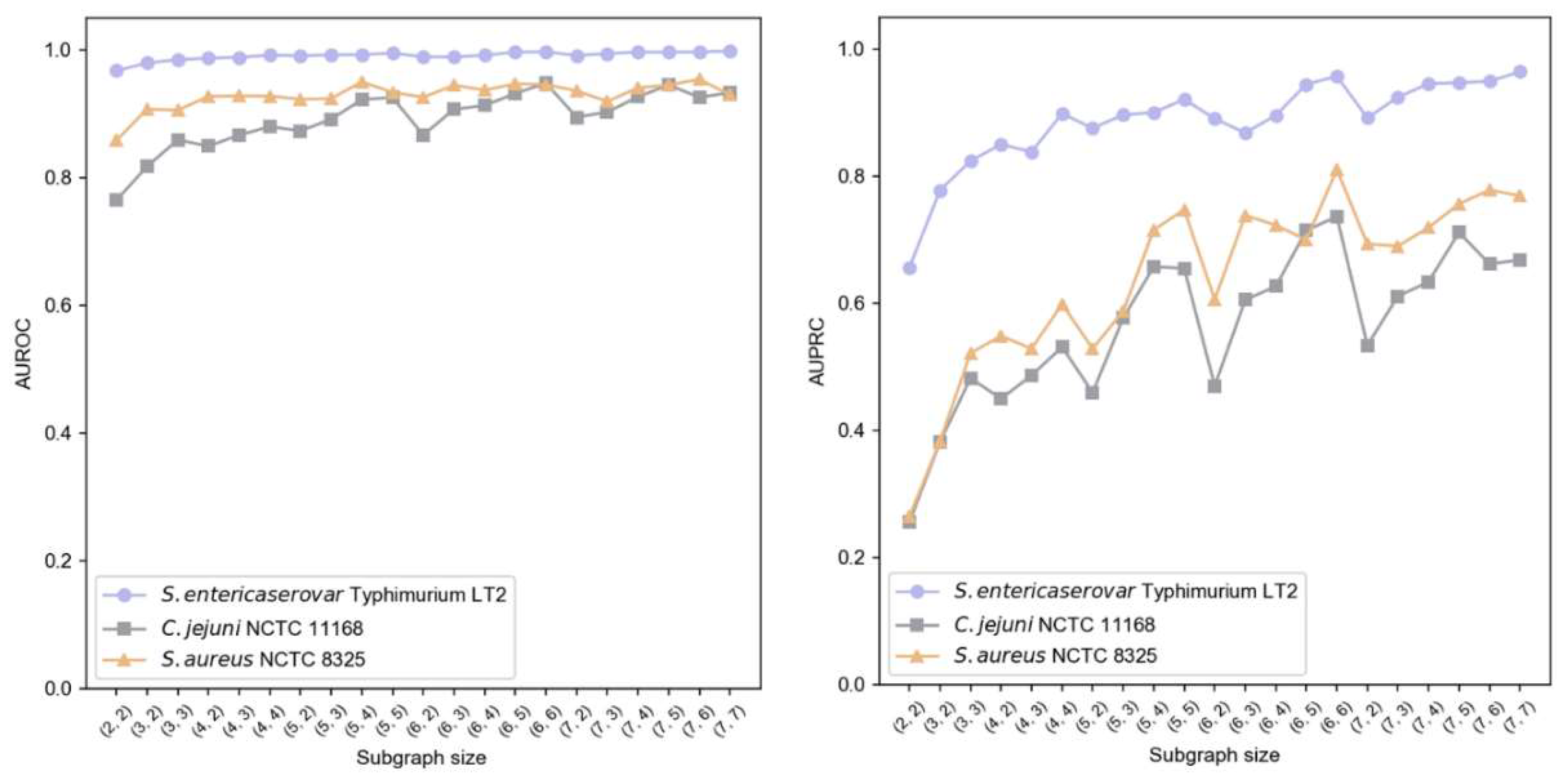
3.3. Analysis of Graph-Based Approaches
3.4. Analysis of Graph-Based Approaches
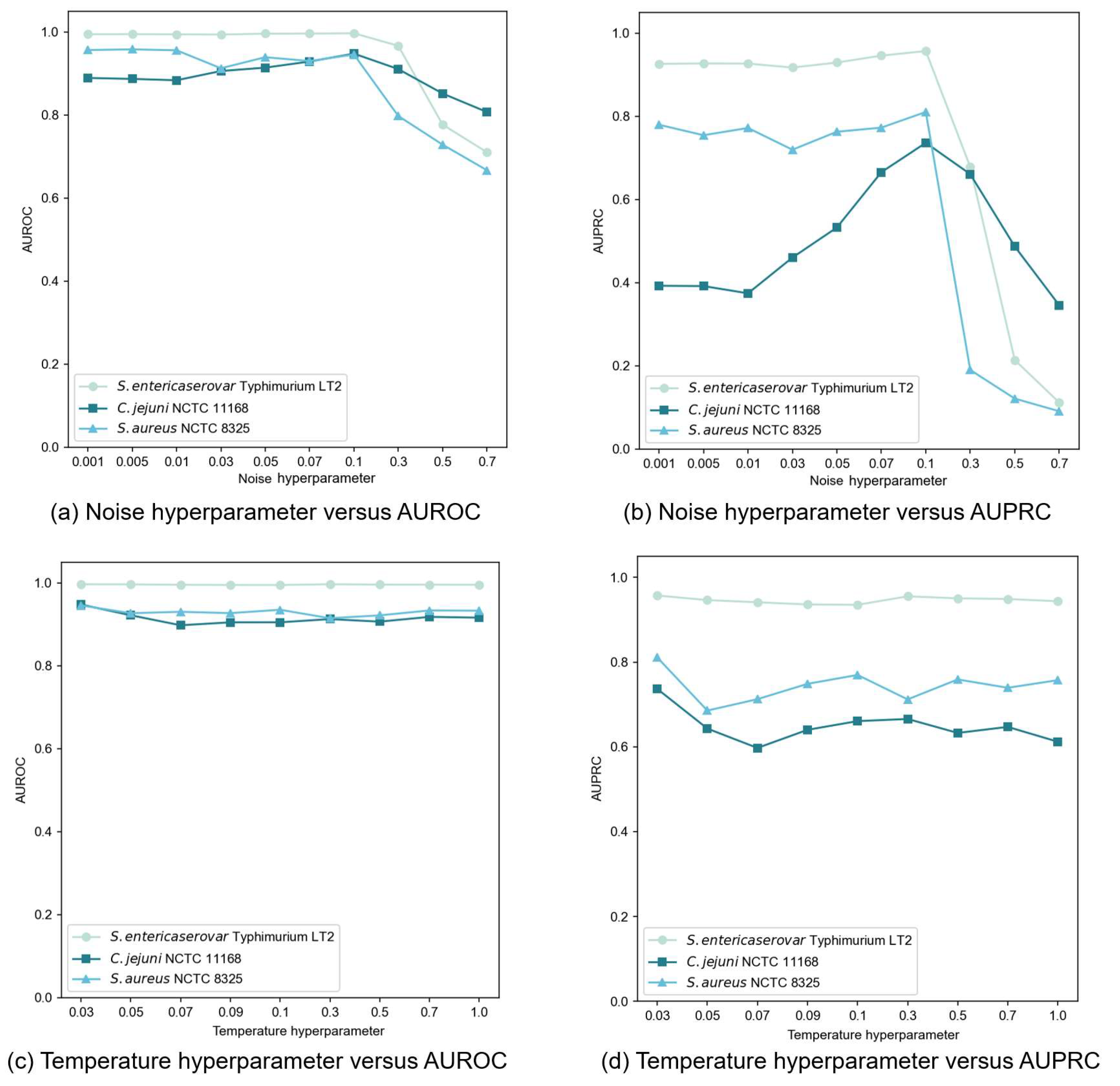
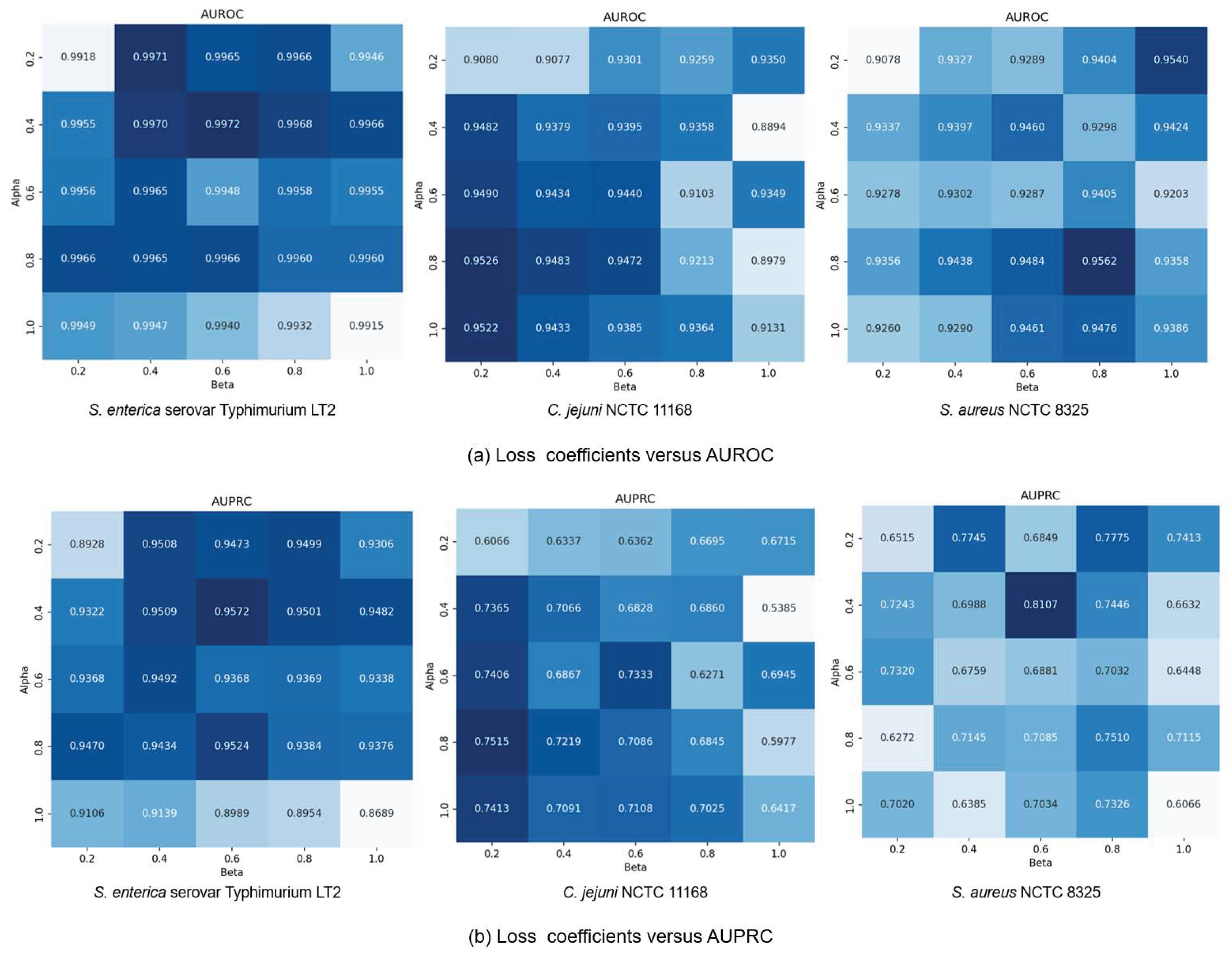
3.5. Ablation Study
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muteeb, G.; Rehman, M. T.; Shahwan, M.; Aatif, M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals 2023, 16(11), 1615. [Google Scholar] [CrossRef]
- Dance, A. Five ways science is tackling the antibiotic resistance crisis. Nature 2024, 632(8025), 494–496. [Google Scholar] [CrossRef]
- Dehbanipour, R.; Ghalavand, Z. Anti-virulence therapeutic strategies against bacterial infections: recent advances. Germs 2022, 12(2), 262. [Google Scholar] [CrossRef]
- Gupta, A.; Kapil, R.; Dhakan, D. B.; Sharma, V. K. MP3: a software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PloS One 2014, 9(4), e93907. [Google Scholar] [CrossRef]
- Altschul, S. F.; Gish, W.; Miller, W.; Myers, E. W.; Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215(3), 403–410. [Google Scholar] [CrossRef]
- Garg, A.; Gupta, D. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinformatics 2008, 9, 1–12. [Google Scholar] [CrossRef]
- Rentzsch, R.; Deneke, C.; Nitsche, A.; Renard, B. Y. Predicting bacterial virulence factors–evaluation of machine learning and negative data strategies. Brief. Bioinform. 2020, 21(5), 1596–1608. [Google Scholar] [CrossRef]
- Xie, R.; Li, J.; Wang, J.; Dai, W.; Leier, A.; Marquez-Lago, T. T.; Akutsu, T.; Lithgow, T.; Song, J.; et al. DeepVF: a deep learning-based hybrid framework for identifying virulence factors using the stacking strategy. Brief. Bioinform. 2021, 22(3), bbaa125. [Google Scholar] [CrossRef]
- Singh, S.; Le, N. Q. K.; Wang, C. VF-Pred: Predicting virulence factor using sequence alignment percentage and ensemble learning models. Comput. Biol. Med. 2024, 168, 107662. [Google Scholar] [CrossRef]
- Ofer, D.; Brandes, N.; Linial, M. The language of proteins: NLP, machine learning & protein sequences. Comput. Struct. Biotechnol. J. 2021, 19, 1750–1758. [Google Scholar]
- Ferruz, N.; Höcker, B. Controllable protein design with language models. Nat. Mach. Intell. 2022, 4(6), 521–532. [Google Scholar] [CrossRef]
- Sun, J.; Yin, H.; Ju, C.; Wang, Y.; Yang, Z. DTVF: A User-Friendly Tool for Virulence Factor Prediction Based on ProtT5 and Deep Transfer Learning Models. Genes 2024, 15(9), 1170. [Google Scholar] [CrossRef]
- Elnaggar, A.; Heinzinger, M.; Dallago, C.; Rehawi, G.; Wang, Y.; Jones, L.; Gibbs, T.; Feher, T.; Angerer, C.; Steinegger, M.; et al. ProtTrans: Toward Understanding the Language of Life Through Self-Supervised Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44(10), 7112–7127. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Bai, P.; Chen, J.; Liang, C. Identifying virulence factors using graph transformer autoencoder with ESMFold-predicted structures. Comput. Biol. Med. 2024, 170, 108062. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379(6637), 1123–1130. [Google Scholar] [CrossRef]
- Wan, X. F.; Xu, D. Computational methods for remote homolog identification. Curr. Prot. Pept. Sci. 2005, 6(6), 527–546. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Chen, L.; Huang, T.; Gao, Q.; Jiang, M.; Zhang, N.; Zheng, L.; Feng, K.; Cai, Y.; Wang, H. Computationally identifying virulence factors based on KEGG pathways. Mol. Biosyst 2013, 9(6), 1447–1452. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S. S.; Riley, P. F.; Vinyals, O.; Dahl, G. E. Neural message passing for quantum chemistry. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; 1263-1272. [Google Scholar]
- Kipf, T. N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the 5th International Conference on Learning Representations, Toulon, France, 24–26 April 2017; 2713-2726. [Google Scholar]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph Attention Networks. In Proceedings of the 6th International Conference on Learning Representations, Vancouver, Canada, 30 April–3 May 2018; 2920-2931. [Google Scholar]
- Hamilton, W. L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Proceedings of the 31st Annual Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; 1025-1035. [Google Scholar]
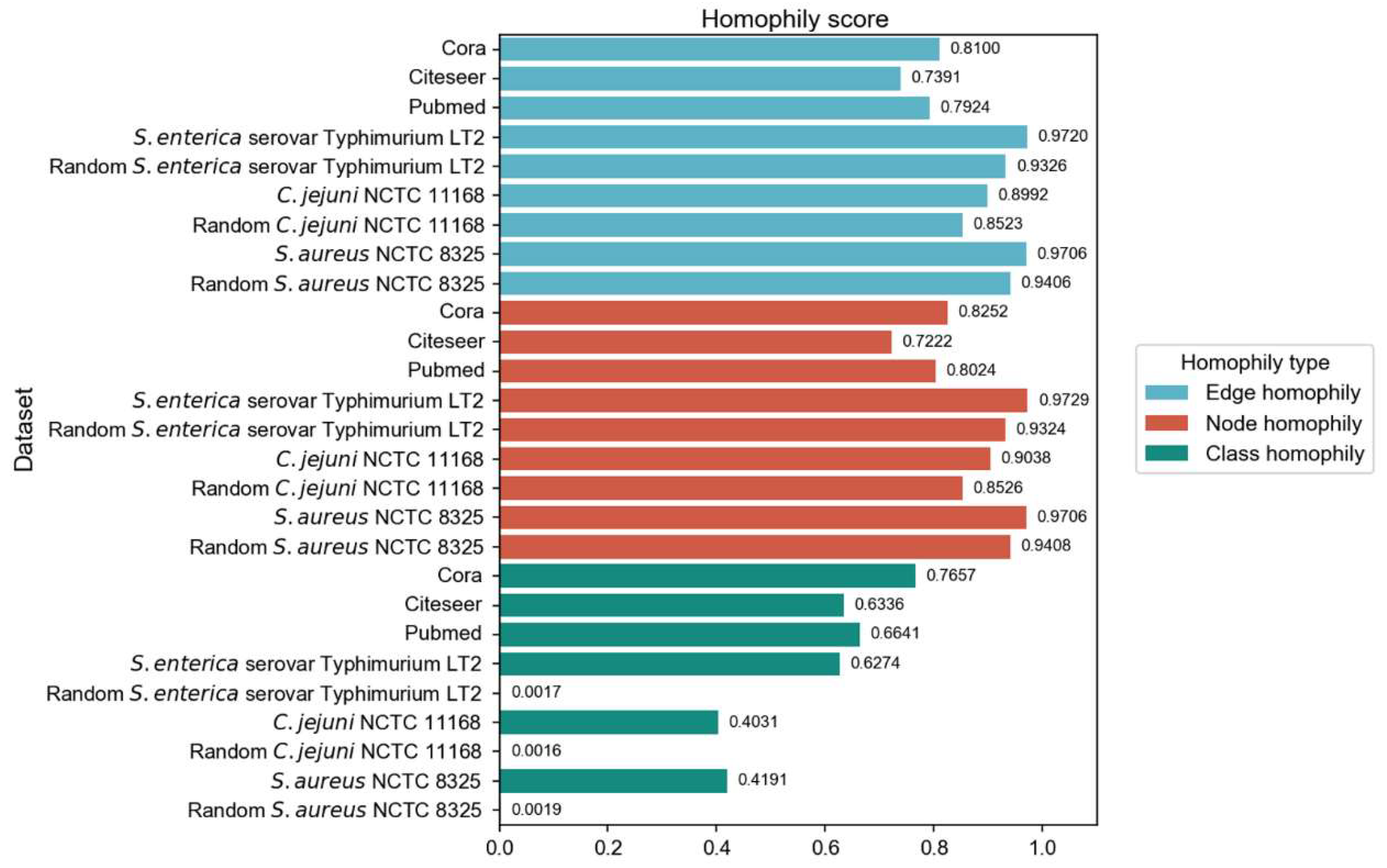
- McPherson, M.; Smith-Lovin, L.; Cook, J. M. Birds of a Feather: Homophily in Social Networks. Annu. Rev. Sociol. 2001, 27(1), 415–444. [Google Scholar] [CrossRef]
- Abu-El-Haija, S.; Perozzi, B.; Kapoor, A.; Alipourfard, N.; Lerman, K.; Harutyunyan, H.; Steeg, G. V.; Galstyan, A. Mixhop: Higher-order Graph Convolutional Architectures via Sparsified Neighborhood Mixing. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; 21-29. [Google Scholar]
- Zhu, J.; Yan, Y.; Zhao, L.; Heimann, M.; Akoglu, L.; Koutra, D. Beyond Homophily in Graph Neural Networks: Current Limitations and Effective Designs. In Proceedings of the 34th Conference on Neural Information Processing Systems, Vancouver, Canada, 6–12 December 2020; 7793-7804. [Google Scholar]
- Pei, H.; Wei, B.; Chang, K. C. C.; Yang, B.; Lei, Y. GEOM-GCN: GEOMETRIC GRAPH CONVOLUTIONAL NETWORKS. In Proceedings of the 8th International Conference on Learning Representations, Addis Ababa, Ethiopia, 26–30 April 2020; 10247-10258. [Google Scholar]
- Hamilton, W. L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Proceedings of the 31st Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; 1025-1035. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A. L.; Fang, T.; Doncheva, N. T.; Pyysalo, S.; et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51 (D1), D638–D646. [Google Scholar] [CrossRef]
- Ericsson, L.; Gouk, H.; Loy, C. C.; Hospedales, T. M. Self-supervised representation learning: Introduction, advances, and challenges. IEEE Signal Process. Mag. 2022, 39(3), 42–62. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, Z. Rethinking the Value of Labels for Improving Class-Imbalanced Learning. In Proceedings of the 34th Conference on Neural Information Processing Systems, Vancouver, Canada, 6–12 December 2020; 19290-19301. [Google Scholar]
- Liu, H.; HaoChen, J. Z.; Gaidon, A.; Ma, T. Self-supervised Learning is More Robust to Dataset Imbalance. In Proceedings of the 10th International Conference on Learning Representations, Virtual Conference, 25–29 April 2022. [Google Scholar]
- Wu, L.; Lin, H.; Tan, C.; Gao, Z.; Li, S. Z. Self-Supervised Learning on Graphs: Contrastive, Generative, or Predictive. IEEE Trans. Knowl. Data Eng. 2023, 35(4), 4216–4226. [Google Scholar] [CrossRef]
- You, Y.; Chen, T.; Sui, Y.; Chen, T.; Wang, Z.; Shen, Y. Graph Contrastive Learning with Augmentations. In Proceedings of the 34th Conference on Neural Information Processing Systems, Vancouver, Canada, 6–12 December 2020; 5812-5823. [Google Scholar]
- Veličković, P.; Fedus, W.; Hamilton, W. L.; Liò, P.; Bengio, Y.; Hjelm, R. D. Deep Graph Infomax. In Proceedings of the 7th International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Qiu, J.; Chen, Q.; Dong, Y.; Zhang, J.; Yang, H.; Ding, M.; Wang, K.; Tang, J. GCC: Graph Contrastive Coding for Graph Neural Network Pre-Training. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Conference, 23–27 August 2020; 1150-1160. [Google Scholar]
- You, Y.; Chen, T.; Wang, Z.; Shen, Y. When Does Self-supervision Help Graph Convolutional Networks? In Proceedings of the 37th International Conference on Machine Learning, Vienna, Austria, 13–18 July 2020; 10871-10880.
- Hu, Z.; Dong, Y.; Wang, K.; Chang, K. W.; Sun, Y. GPT-GNN: Generative Pre-Training of Graph Neural Networks. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Conference, 23–27 August 2020; 1857-1867. [Google Scholar]
- Sun, K.; Lin, Z.; Zhu, Z. Multi-Stage Self-Supervised Learning for Graph Convolutional Networks on Graphs with Few Labeled Nodes. In Proceedings of the 34th AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; 5892-5899. [Google Scholar]
- Rong, Y.; Bian, Y.; Xu, T.; Xie, W.; Wei, Y.; Huang, W.; Huang, J. Self-Supervised Graph Transformer on Large-Scale Molecular Data. In Proceedings of the 34th Conference on Neural Information Processing Systems, Vancouver, Canada, 6–12 December 2020; 12559-12571. [Google Scholar]
- Juan, X.; Zhou, F.; Wang, W.; Jin, W.; Tang, J.; Wang, X. INS-GNN: Improving Graph Imbalance Learning with Self-supervision. Inf. Sci. 2023, 637, 118935. [Google Scholar] [CrossRef]
- Wu, L.; Xia, J.; Gao, Z.; Lin, H.; Tan, C.; Li, S. Z. GraphMixup: Improving Class-Imbalanced Node Classification by Reinforcement Mixup and Self-supervised Context Prediction. In Proceedings of the 2022 European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases, Grenoble, France, 19–23 September 2022; 519-535. [Google Scholar]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50 (D1), D912–D917. [Google Scholar] [CrossRef]
- Zhang, Y.; Kang, B.; Hooi, B.; Yan, S.; Feng, J. Deep Long-Tailed Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45(9), 10795–10816. [Google Scholar] [CrossRef]
- Wang, Y.; You, Z. H.; Yang, S.; Li, X.; Jiang, T. H.; Zhou, X. A High Efficient Biological Language Model for Predicting Protein-Protein Interactions. Cells 2019, 8(2), 122. [Google Scholar] [CrossRef]
- Slam, S. A.; Heil, B. J.; Kearney, C. M.; Baker, E. J. Protein Classification Using Modified N-Grams and Skip-Grams. Bioinformatics 2018, 34(9), 1481–1487. [Google Scholar]
- Chen, M.; Ju, C. J. T.; Zhou, G.; Chen, X.; Zhang, T.; Chang, K. W.; Zaniolo, C.; Wang, W. Multifaceted Protein-Protein Interaction Prediction Based on Siamese Residual RCNN. Bioinformatics 2019, 35(14), i305–i314. [Google Scholar] [CrossRef] [PubMed]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed Representations of Words and Phrases and Their Compositionality. In Proceedings of the 27th Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013; 3111-3119. [Google Scholar]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting Protein–Protein Interactions Based Only on Sequences Information. Proceedings of the National Academy of Sciences 2007, 104(11), 4337–4341. [Google Scholar] [CrossRef]
- Lv, G.; Hu, Z.; Bi, Y.; Zhang, S. Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction. In Proceedings of the 30th International Joint Conference on Artificial Intelligence (IJCAI), Montreal, Canada, 19–26 August 2021; 3677-3683. [Google Scholar]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How Powerful are Graph Neural Networks? In Proceedings of the 7th International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019; 9104-9120.
- Glorot, X.; Bordes, A.; Bengio, Y. Deep Sparse Rectifier Neural Networks. In Proceedings of the 14th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; 315-323. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the 37th International Conference on Machine Learning, Virtual Conference, 13–18 July 2020; 1597-1607. [Google Scholar]
- Zhu, Y.; Xu, Y.; Yu, F.; Liu, Q.; Wu, S.; Wang, L. Deep Graph Contrastive Representation Learning. In Proceedings of the 37th International Conference on Machine Learning Workshop on Graph Representation Learning and Beyond, Virtual Conference, 13–18 July 2020. [Google Scholar]
- Lin, T. Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; 2980-2988. [Google Scholar]
- Sharma, A.; Garg, A.; Ramana, J.; Gupta, D. VirulentPred 2.0: An Improved Method for Prediction of Virulent Proteins in Bacterial Pathogens. Protein Sci. 2023, 32 (12), e4808.
- Ganin, Y.; Ustinova, E.; Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; March, M.; Lempitsky, V. Domain-Adversarial Training of Neural Networks. J. Mach. Learn. Res. 2016, 17(59), 1–35. [Google Scholar]
- Long, M.; Cao, Z.; Wang, J.; Jordan, M. I. Conditional Adversarial Domain Adaptation. In Proceedings of the 32nd Conference on Neural Information Processing Systems, Montreal, Canada, 2–8 December 2018; 1647-1657. [Google Scholar]

| Dataset | Nodes | Edges | VFs | Imbalance Ratio |
|---|---|---|---|---|
| S. enterica serovar Typhimurium LT2 | 4451 | 86605 | 156 | 27.53 |
| C. jejuni NCTC 11168 | 1623 | 81710 | 130 | 11.48 |
| S. aureus NCTC 8325 | 2847 | 79578 | 87 | 31.72 |
| Dataset | Method | Accuracy | Sensitivity | Specificity | F1-score | MCC | AUPRC | AUROC |
|---|---|---|---|---|---|---|---|---|
| S. enterica serovar Typhimurium LT2 | BLAST | 0.9560 | 0.4839 | 0.9731 | 0.4348 | 0.4145 | 0.3440 | |
| VirulentPred 2.0 | 0.6979 | 0.9677 | 0.6881 | 0.1829 | 0.2552 | 0.0989 | ||
| DeepVF | 0.9008 | 0.3548 | 0.9208 | 0.2018 | 0.1788 | 0.2593 | 0.7942 | |
| DT-VF | 0.9651 | 0.3548 | 0.9871 | 0.4151 | 0.4038 | 0.2761 | 0.8678 | |
| GC-VF | 0.9941 | 0.9058 | 0.9973 | 0.9140 | 0.9119 | 0.9572 | 0.9972 | |
| C. jejuni NCTC 11168 | BLAST | 0.8738 | 0.3462 | 0.9197 | 0.3051 | 0.2388 | 0.3356 | 0.8738 |
| VirulentPred 2.0 | 0.4831 | 0.9231 | 0.4448 | 0.2222 | 0.2025 | 0.1228 | ||
| DeepVF | 0.6440 | 0.7308 | 0.6364 | 0.2484 | 0.2045 | 0.4510 | 0.6695 | |
| DT-VF | 0.6800 | 0.8077 | 0.6689 | 0.2877 | 0.2679 | 0.1817 | 0.7622 | |
| GC-VF | 0.9588 | 0.6877 | 0.9824 | 0.7276 | 0.7107 | 0.7365 | 0.9482 | |
| S. aureus NCTC 8325 | BLAST | 0.9579 | 0.3529 | 0.9765 | 0.3333 | 0.3122 | 0.3440 | |
| VirulentPred 2.0 | 0.4614 | 1.0000 | 0.4448 | 0.0997 | 0.1528 | 0.0525 | ||
| DeepVF | 0.5554 | 0.8750 | 0.5456 | 0.1041 | 0.1427 | 0.4670 | 0.7021 | |
| DT-VF | 0.9035 | 0.4706 | 0.9168 | 0.2254 | 0.2250 | 0.1678 | 0.8171 | |
| GC-VF | 0.9889 | 0.7524 | 0.9961 | 0.8005 | 0.7990 | 0.8107 | 0.9460 |
| Dataset | Model | Accuracy | Sensitivity | Specificity | F1-score | MCC | AUPRC | AUROC |
|---|---|---|---|---|---|---|---|---|
| S. enterica serovar Typhimurium LT2 | GC-VF w/o Con | 0.9906 | 0.8458 | 0.9959 | 0.8625 | 0.8596 | 0.9106 | 0.9929 |
| GC-VF w/o Gen | 0.9922 | 0.8632 | 0.9969 | 0.8855 | 0.8831 | 0.9151 | 0.9889 | |
| GC-VF w/o SL | 0.9894 | 0.8055 | 0.9961 | 0.8414 | 0.8380 | 0.8754 | 0.9813 | |
| GC-VF w/o EW | 0.9873 | 0.8084 | 0.9938 | 0.8168 | 0.8132 | 0.8689 | 0.9915 | |
| GC-VF w/ BCE | 0.9775 | 0.7100 | 0.9872 | 0.6886 | 0.6823 | 0.7333 | 0.9824 | |
| C. jejuni NCTC 11168 | GC-VF | 0.9941 | 0.9058 | 0.9973 | 0.9140 | 0.9119 | 0.9572 | 0.9972 |
| GC-VF w/o Con | 0.8803 | 0.6046 | 0.9042 | 0.4482 | 0.4087 | 0.3591 | 0.8688 | |
| GC-VF w/o Gen | 0.9447 | 0.5323 | 0.9806 | 0.6070 | 0.5928 | 0.5921 | 0.8748 | |
| GC-VF w/o SL | 0.9287 | 0.4519 | 0.9701 | 0.5065 | 0.4937 | 0.4833 | 0.8323 | |
| GC-VF w/o EW | 0.9325 | 0.7012 | 0.9526 | 0.6249 | 0.6000 | 0.6013 | 0.9365 | |
| S. aureus NCTC 8325 | GC-VF w/ BCE | 0.9268 | 0.4646 | 0.9670 | 0.5061 | 0.4873 | 0.8428 | 0.4646 |
| GC-VF | 0.9588 | 0.6877 | 0.9824 | 0.7276 | 0.7107 | 0.7365 | 0.9482 | |
| GC-VF w/o Con | 0.9856 | 0.6612 | 0.9955 | 0.7327 | 0.7401 | 0.7457 | 0.9535 | |
| GC-VF w/o Gen | 0.9806 | 0.7382 | 0.9880 | 0.6942 | 0.6877 | 0.6613 | 0.9296 | |
| GC-VF w/o SL | 0.9827 | 0.6835 | 0.9919 | 0.7035 | 0.7038 | 0.7301 | 0.9379 | |
| GC-VF w/o EW | 0.9841 | 0.6759 | 0.9936 | 0.7172 | 0.7141 | 0.6978 | 0.9342 | |
| GC-VF w/ BCE | 0.9867 | 0.7124 | 0.9951 | 0.7610 | 0.7585 | 0.7538 | 0.9013 | |
| GC-VF | 0.9889 | 0.7524 | 0.9961 | 0.8005 | 0.7990 | 0.8107 | 0.9460 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





