Submitted:
24 May 2025
Posted:
26 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Genetic Diversity Within Populations
2.2. Analysis of Molecular Variance, Genetic Differentiation and Phylogenetic Relationship Among the Populations
2.2.1. Analysis of Molecular Variance
2.2.2. Genetic Differentiations
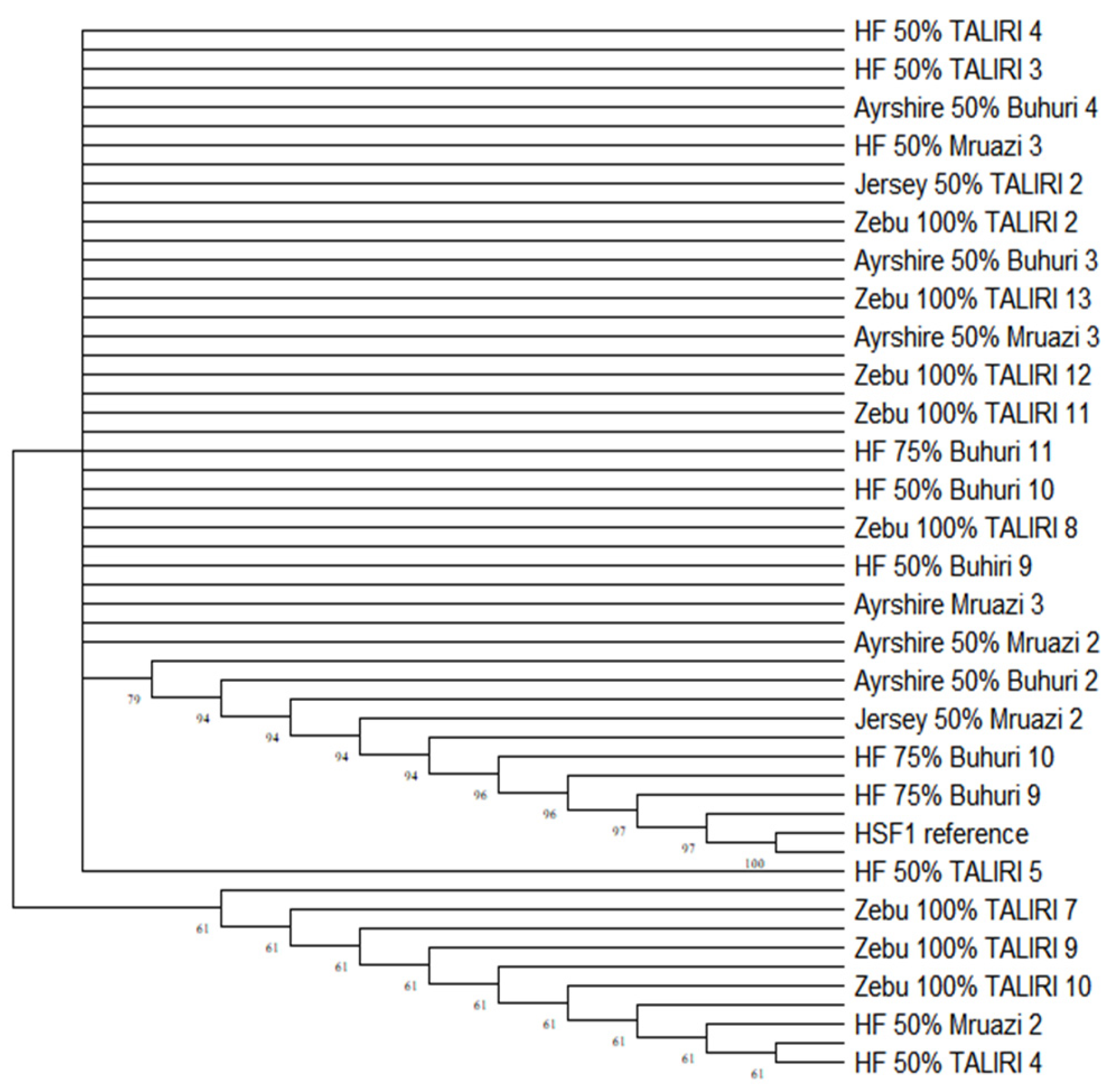
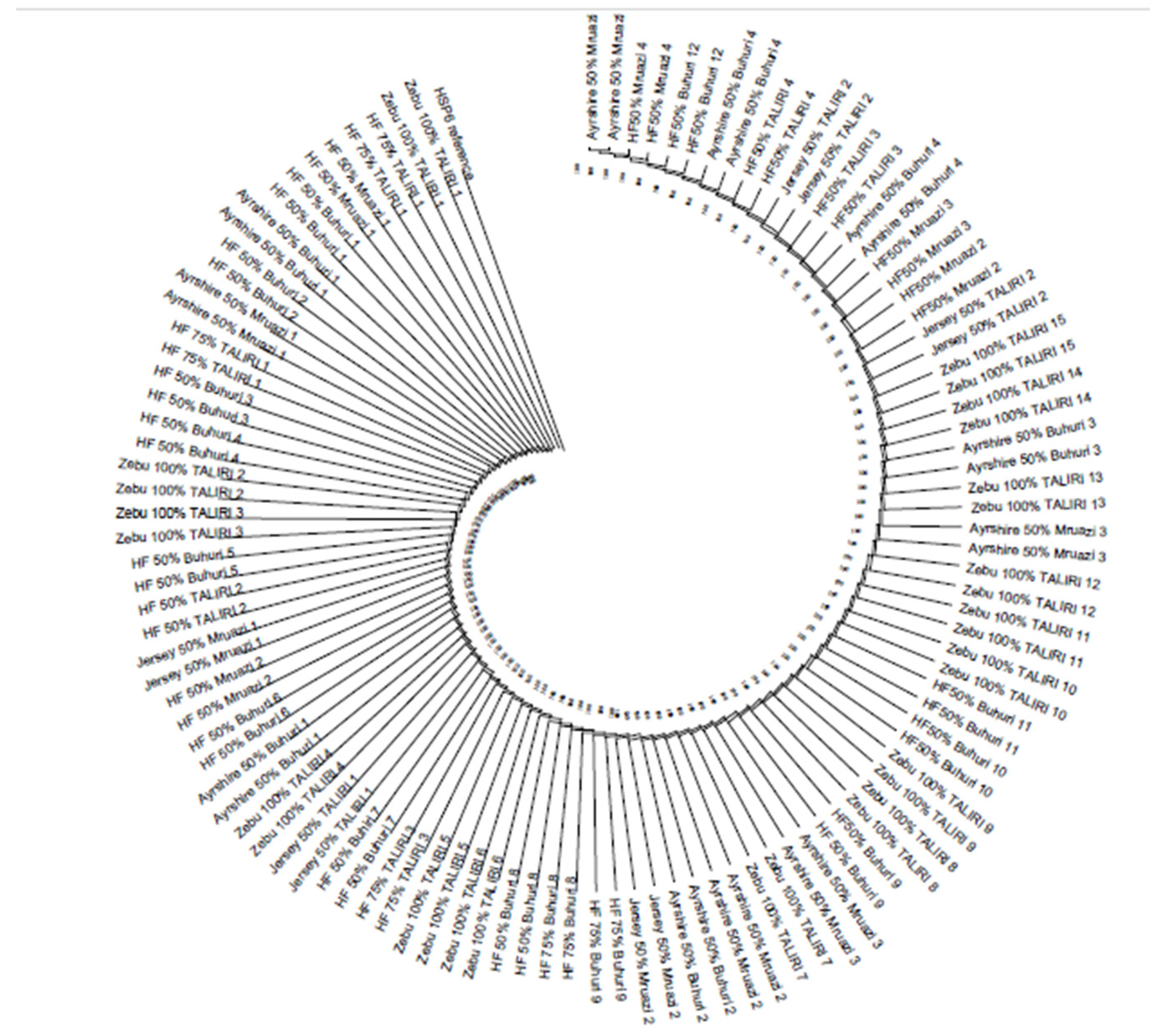
2.2.3. Phylogenetic Relationships Among the Populations
2.2.4. Demographic History
3. Discussion
3.1. Genetic Diversity Within Populations
3.2. Genetic Differentiation and Phylogenetic Relations Among the Populations
3.3. Demographic History Within the Populations
4. Materials and Methods
4.1. Ethics Statement
4.2. Study Site
4.3. Animal Breeds and Blood Sampling
4.4. DNA Extraction
4.5. DNA Quantification
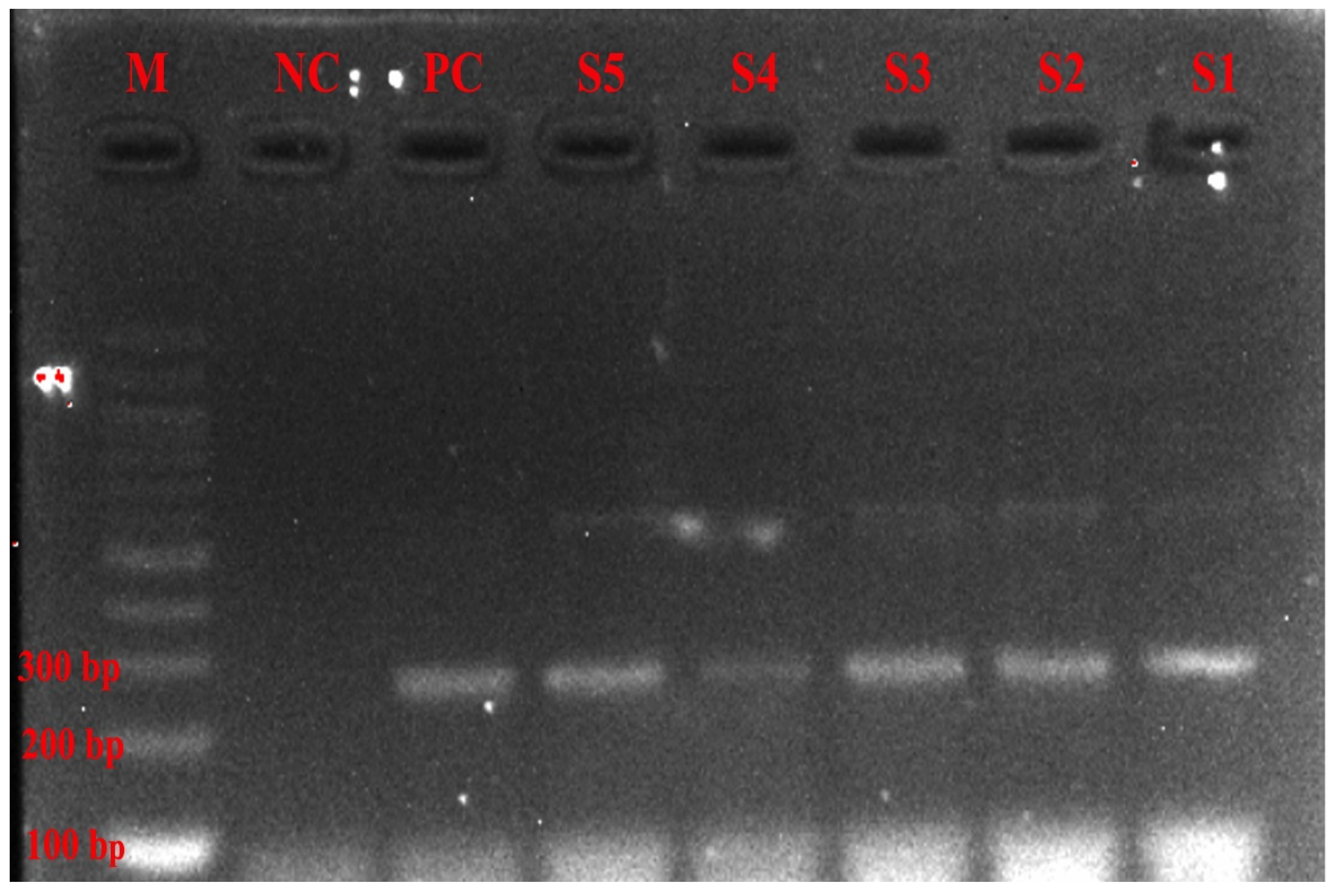
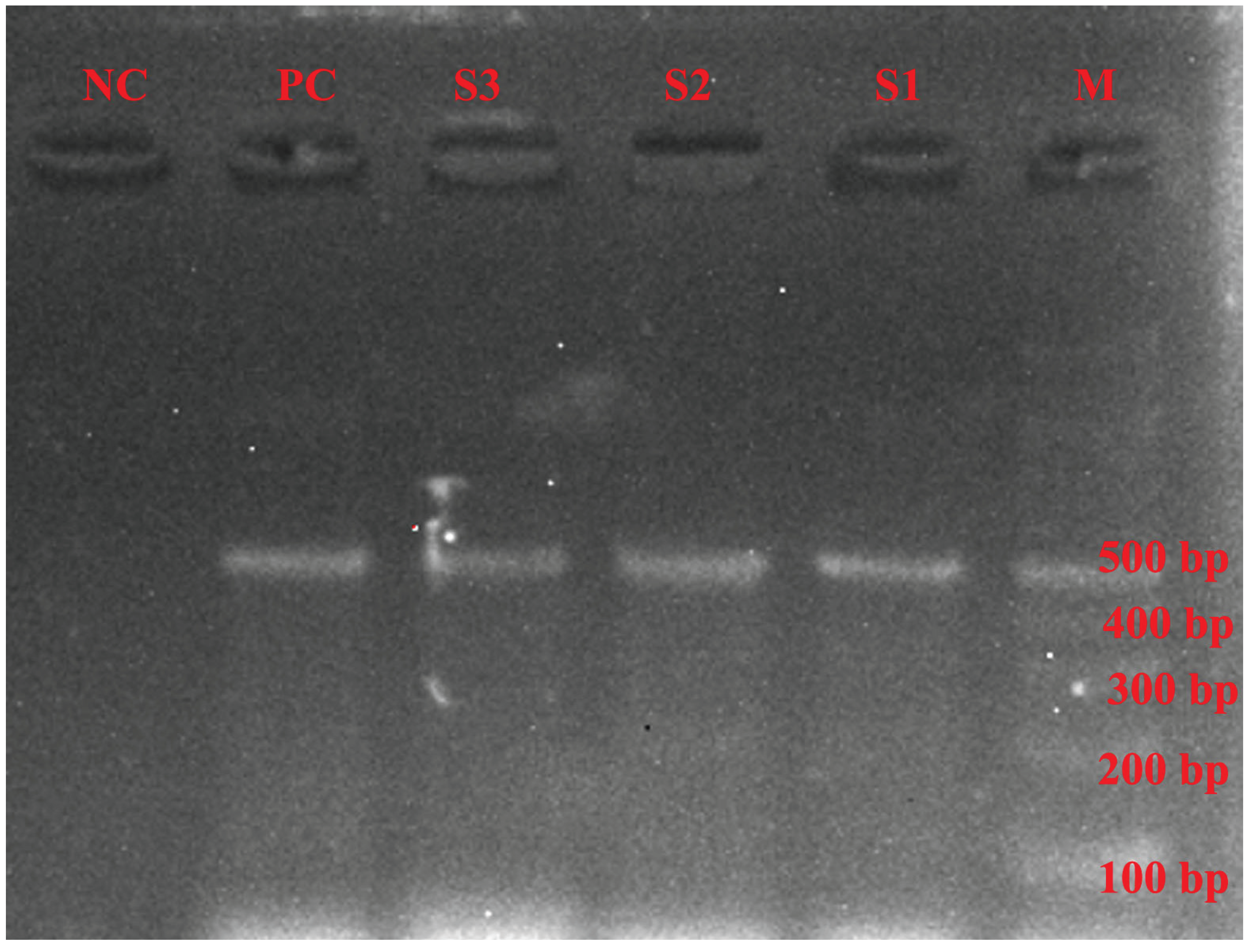
4.6. DNA Amplification and Gel Electrophoresis of PCR Products
4.7. PCR Purification and Sequencing
4.8. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interests
References
- Msalya, G.; Kim, E.-S.; Laisser, E.L.K.; Kipanyula, M.J.; Karimuribo, E.D.; Kusiluka, L.J.M.; Chenyambuga, S.W.; Rothschild, M.F. Determination of Genetic Structure and Signatures of Selection in Three Strains of Tanzania Shorthorn Zebu, Boran and Friesian Cattle by Genome-Wide SNP Analyses. PLOS ONE 2017, 12, e0171088. [Google Scholar] [CrossRef] [PubMed]
- Cheruiyot, E.K.; Bett, R.C.; Amimo, J.O.; Zhang, Y.; Mrode, R.; Mujibi, F.D.N. Signatures of Selection in Admixed Dairy Cattle in Tanzania. Front. Genet. 2018, 9, 607. [Google Scholar] [CrossRef] [PubMed]
- Kibona, C.A.; Yuejie, Z.; Tian, L. Towards developing a beef meat export oriented policy in Tanzania: -Exploring the factors that influence beef meat exports-. PLOS ONE 2022, 17, e0270146. [Google Scholar] [CrossRef] [PubMed]
- Sajjanar, B.; Aalam, M.T.; Khan, O.; Tanuj, G.N.; Sahoo, A.P.; Manjunathareddy, G.B.; Gandham, R.K.; Dhara, S.K.; Gupta, P.K.; Mishra, B.P.; et al. Genome-wide expression analysis reveals different heat shock responses in indigenous (Bos indicus) and crossbred (Bos indicus X Bos taurus) cattle. Genes Environ. 2023, 45, 1–15. [Google Scholar] [CrossRef]
- Shija, D. S. , Okeyo, M. A., Migwi, P. K., Kelya, N. J., and Bebe, B. O. (2022). Assessing Animal Disease Prevalence and Mortality in Smallholder Dairy Farms under Contrasting Management Practices and Stressful Environments in Tanzania. Open Journal of Veterinary Medicine 12, 117–134. [CrossRef]
- Habimana, V., Nguluma, A. S., Nziku, Z. C., Ekine-Dzivenu, C. C., Morota, G., Mrode, R., and Chenyambuga, S. W. (2024). Heat Stress Effects on Physiological and Milk Yield Traits of Lactating Holstein Friesian Crossbreds Reared in Tanga Region, Tanzania. Animals, 14(13). https://doi.org/10.3390/ ani14131914. [CrossRef]
- Abbas, Z.; Hu, L.; Fang, H.; Sammad, A.; Kang, L.; Brito, L.F.; Xu, Q.; Wang, Y. Association Analysis of Polymorphisms in the 5′ Flanking Region of the HSP70 Gene with Blood Biochemical Parameters of Lactating Holstein Cows under Heat and Cold Stress. Animals 2020, 10, 2016. [Google Scholar] [CrossRef]
- Gill, J.K.; Arora, J.S.; Kumar, B.V.S.; Mukhopadhyay, C.S.; Kaur, S.; Kashyap, N. Cellular thermotolerance is independent of HSF 1 expression in zebu and crossbred non-lactating cattle. Int. J. Biometeorol. 2017, 61, 1687–1693. [Google Scholar] [CrossRef]
- Rocha, R.d.F.B.; Baena, M.M.; Estopa, A.d.C.; Gervásio, I.C.; Ibelli, A.M.G.; Gionbelli, T.R.S.; Gionbelli, M.P.; de Freitas, R.T.F.; Meirelles, S.L.C. Differential expression of HSF1 and HSPA6 genes and physiological responses in Angus and Simmental cattle breeds. J. Therm. Biol. 2019, 84, 92–98. [Google Scholar] [CrossRef]
- Rojas-Downing, M.M.; Nejadhashemi, A.P.; Harrigan, T.; Woznicki, S.A. Climate change and livestock: Impacts, adaptation, and mitigation. Clim. Risk Manag. 2017, 16, 145–163. [Google Scholar] [CrossRef]
- Corredor, F.-A.; Figueroa, D.; Estrada, R.; Salazar, W.; Quilcate, C.; Vásquez, H.V.; Gonzales, J.; Maicelo, J.L.; Medina, P.; Arbizu, C.I. Genetic diversity and population structure of a Peruvian cattle herd using SNP data. Front. Genet. 2023, 14, 1073843. [Google Scholar] [CrossRef]
- Martínez-Rocha, R.; Hidalgo, J.; Cesarani, A.; Ramírez-Valverde, R.; Núñez-Domínguez, R.; García-Muñiz, J.G.; Domínguez-Viveros, J. Assessment of Genetic Diversity, Runs of Homozygosity, and Signatures of Selection in Tropical Milking Criollo Cattle Using Pedigree and Genomic Data. Genes 2022, 13, 1896. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Cao, Y.; Wu, Z.; Huang, M.; Zhang, G.; Lei, C.; Zhao, Y. A Missense Mutation of the HSPB7 Gene Associated with Heat Tolerance in Chinese Indicine Cattle. Animals 2019, 9, 554. [Google Scholar] [CrossRef] [PubMed]
- Elayadeth-Meethal, M.; Tiambo, C.K.; Naseef, P.P.; Kuruniyan, M.S.; Maloney, S.K. The profile of HSPA1A gene expression and its association with heat tolerance in crossbred cattle and the tropically adapted dwarf Vechur and Kasaragod. J. Therm. Biol. 2022, 111, 103426. [Google Scholar] [CrossRef]
- Han, X.; Jin, S.; Shou, C.; Han, Z. Hsp70 Gene Family in Sebastiscus marmoratus: The Genome-Wide Identification and Transcriptome Analysis under Thermal Stress. Genes 2023, 14, 1779. [Google Scholar] [CrossRef]
- Rehman, S.U.; Nadeem, A.; Javed, M.; Hassan, F.-U.; Luo, X.; Khalid, R.B.; Liu, Q. Genomic Identification, Evolution and Sequence Analysis of the Heat-Shock Protein Gene Family in Buffalo. Genes 2020, 11, 1388. [Google Scholar] [CrossRef]
- Trivedi, R.; Jurivich, D.A. A molecular perspective on age-dependent changes to the heat shock axis. Exp. Gerontol. 2020, 137, 110969. [Google Scholar] [CrossRef]
- Elrobh, M.S.; Alanazi, M.S.; Khan, W.; Abduljaleel, Z.; Al-Amri, A.; Bazzi, M.D. Molecular Cloning and Characterization of cDNA Encoding a Putative Stress-Induced Heat-Shock Protein from Camelus dromedarius. Int. J. Mol. Sci. 2011, 12, 4214–4236. [Google Scholar] [CrossRef]
- Banerjee, D.; Upadhyay, R.C.; Chaudhary, U.B.; Kumar, R.; Singh, S.; Ashutosh; G., J.M.; Polley, S.; Mukherjee, A.; Das, T.K.; et al. Seasonal variation in expression pattern of genes under HSP70. Cell Stress Chaperon- 2014, 19, 401–408. [CrossRef]
- Kim, W.-S.; Nejad, J.G.; Roh, S.-G.; Lee, H.-G. Heat-Shock Proteins Gene Expression in Peripheral Blood Mononuclear Cells as an Indicator of Heat Stress in Beef Calves. Animals 2020, 10, 895. [Google Scholar] [CrossRef]
- Rong, Y.; Zeng, M.; Guan, X.; Qu, K.; Liu, J.; Zhang, J.; Chen, H.; Huang, B.; Lei, C. Association of HSF1 Genetic Variation with Heat Tolerance in Chinese Cattle. Animals 2019, 9, 1027. [Google Scholar] [CrossRef]
- Miyayo, S.F.; Owili, P.O.; Muga, M.A.; Lin, T.-H. Analysis of Pneumonia Occurrence in Relation to Climate Change in Tanga, Tanzania. Int. J. Environ. Res. Public Heal. 2021, 18, 4731. [Google Scholar] [CrossRef]
- Onasanya, G.O.; Msalya, G.M.; Thiruvenkadan, A.K.; Sreekumar, C.; Tirumurugaan, G.K.; O Fafiolu, A.; A Adeleke, M.; Yakubu, A.; Ikeobi, C.O.N.; Okpeku, M. Heterozygous Single-Nucleotide Polymorphism Genotypes at Heat Shock Protein 70 Gene Potentially Influence Thermo-Tolerance Among Four Zebu Breeds of Nigeria. Front. Genet. 2021, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Masila, E.M.; Ogada, S.O.; Ogali, I.N.; Kennedy, G.M.; Too, E.K.; Ommeh, C.S. Mitochondrial DNA D-Loop Polymorphisms among the Galla Goats Reveals Multiple Maternal Origins with Implication on the Functional Diversity of the HSP70 Gene. Genet. Res. 2024, 2024, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Makina, S.O.; Muchadeyi, F.C.; van Marle-Kã¶Ster, E.; MacNeil, M.D.; Maiwashe, A. Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Front. Genet. 2014, 5, 333–333. [Google Scholar] [CrossRef]
- Baena, M.M.; Tizioto, P.C.; Meirelles, S.L.C.; Regitano, L.C.d.A. HSF1 and HSPA6 as functional candidate genes associated with heat tolerance in Angus cattle. Rev. Bras. de Zootec. 2018, 47. [Google Scholar] [CrossRef]
- Aboul-Naga, A.M.; Alsamman, A.M.; El Allali, A.; Elshafie, M.H.; Abdelal, E.S.; Abdelkhalek, T.M.; Abdelsabour, T.H.; Mohamed, L.G.; Hamwieh, A. Genome-wide analysis identified candidate variants and genes associated with heat stress adaptation in Egyptian sheep breeds. Front. Genet. 2022, 13, 898522. [Google Scholar] [CrossRef]
- Bora, S.K.; Tessema, T.S.; Girmay, G. Genetic Diversity and Population Structure of Selected Ethiopian Indigenous Cattle Breeds Using Microsatellite Markers. Genet. Res. 2023, 2023, 1–12. [Google Scholar] [CrossRef]
- Mao, Y.; Chang, H.; Yang, Z.; Zhang, L.; Xu, M.; Chang, G.; Sun, W.; Song, G.; Ji, D. The analysis of genetic diversity and differentiation of six Chinese cattle populations using microsatellite markers. J. Genet. Genom. 2008, 35, 25–32. [Google Scholar] [CrossRef]
- Ayalew, W.; Wu, X.-Y.; Tarekegn, G.M.; Chu, M.; Liang, C.-N.; Tessema, T.S.; Yan, P. Signatures of positive selection for local adaptation of African native cattle populations: A review. J. Integr. Agric. 2023, 22, 1967–1984. [Google Scholar] [CrossRef]
- Nwachukwu, E.N.; Kalla, D.J.U.; Ukwu, H.O.; Ogbu, C.C.; Ezea, J.; Udoh, U.H.; Ekumankama, O.O. Genetic diversity and population structure of four Nigerian indigenous cattle breeds. Trop. Anim. Heal. Prod. 2022, 54, 1–11. [Google Scholar] [CrossRef]
- Ning, Q.; Qu, K.; Hanif, Q.; Jia, Y.; Cheng, H.; Zhang, J.; Chen, N.; Chen, H.; Huang, B.; Lei, C. MTOR Variation Related to Heat Resistance of Chinese Cattle. Animals 2019, 9, 915. [Google Scholar] [CrossRef]
- Tarekegn, G.M.; Ji, X.-Y.; Bai, X.; Liu, B.; Zhang, W.; Birungi, J.; Djikeng, A.; Tesfaye, K. Variations in mitochondrial cytochrome b region among Ethiopian indigenous cattle populations assert Bos taurus maternal origin and historical dynamics. Asian-Australasian J. Anim. Sci. 2018, 31, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E. M., Gebrehiwot, N. Z., Swaminathan, M., Joshi, S., Al Kalaldeh, M., and Gibson, J. P. (2021). Genetic diversity and effective population sizes of thirteen Indian cattle breeds. Genetics Selection Evolution, 53(1), 1–17.
- Li, M.H.; Zerabruk, M.; Vangen, O.; Olsaker, I.; Kantanen, J. Reduced genetic structure of north Ethiopian cattle revealed by Y-chromosome analysis. Heredity 2007, 98, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Samuel, B.; Mengistie, D.; Assefa, E.; Kang, M.; Park, C.; Dadi, H.; Dinka, H. Genetic diversity of DGAT1 gene linked to milk production in cattle populations of Ethiopia. BMC Genet. 2022, 23, 1–10. [Google Scholar] [CrossRef]
- Meseret, S.; Mekonnen, Y.A.; Brenig, B.; Schütz, E.; Hanotte, O.; Gültas, M.; Schmitt, A.O. Genetic diversity and population structure of six ethiopian cattle breeds from different geographical regions using high density single nucleotide polymorphisms. Livest. Sci. 2020, 234, 103979. [Google Scholar] [CrossRef]
- Alam Bhuiyan, M.S.; Lee, S.-H.; Hossain, S.M.J.; Deb, G.K.; Afroz, M.F.; Lee, S.-H.; Bhuiyan, A.K.F.H. Unraveling the Genetic Diversity and Population Structure of Bangladeshi Indigenous Cattle Populations Using 50K SNP Markers. Animals 2021, 11, 2381. [Google Scholar] [CrossRef]
- Tapsoba, A.S.R.; (LaBioSA), I.d.l.e.d.R.A. (.L.d.B.e.S.a.; Sawadogo, S.E.; Yougbaré, B.; Traoré, F.G.; Béré, F.; Sanou, M.; Tamboura, H.H.; Bayala, B.; Pichler, R.; et al. Genetic Diversity and Population Structure of Taurine Cattle Using STR Markers in Burkina Faso, West Africa. Trop. Anim. Sci. J. 2024, 47, 131–140. [Google Scholar] [CrossRef]
- Ariyaraphong, N.; Laopichienpong, N.; Singchat, W.; Panthum, T.; Ahmad, S.F.; Jattawa, D.; Duengkae, P.; Muangmai, N.; Suwanasopee, T.; Koonawootrittriron, S.; et al. High-Level Gene Flow Restricts Genetic Differentiation in Dairy Cattle Populations in Thailand: Insights from Large-Scale Mt D-Loop Sequencing. Animals 2021, 11, 1680. [Google Scholar] [CrossRef]
- Mujibi, F.D.N.; Rao, J.; Agaba, M.; Nyambo, D.; Cheruiyot, E.K.; Kihara, A.; Zhang, Y.; Mrode, R. Performance Evaluation of Highly Admixed Tanzanian Smallholder Dairy Cattle Using SNP Derived Kinship Matrix. Front. Genet. 2019, 10, 375. [Google Scholar] [CrossRef]
- Mandefro, A.; Sisay, T.; Edea, Z.; Uzzaman, R.; Kim, K.-S.; Dadi, H. Genetic assessment of BoLA-DRB3 polymorphisms by comparing Bangladesh, Ethiopian, and Korean cattle. J. Anim. Sci. Technol. 2021, 63, 248–261. [Google Scholar] [CrossRef]
- Shija, D.S.; Mwai, O.A.; Ojango, J.M.K.; Komwihangilo, D.M.; Bebe, B.O. Assessing Lactation Curve Characteristics of Dairy Cows Managed under Contrasting Husbandry Practices and Stressful Environments in Tanzania. World 2022, 3, 1032–1052. [Google Scholar] [CrossRef]
| Population | N | Haplotypes | Polymorphic Sites | Hd ± SD | π ± SD |
| Holstein-Friesian crosses | 8 | 3 | 2 | 0.34 ± 0.14 | 0.22 ± 0.23 |
| Ayrshire crosses | 7 | 1 | 0 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Jersey crosses | 3 | 2 | 1 | 0.33 ± 0.21 | 0.16 ± 0.21 |
| Zebu | 9 | 2 | 1 | 0.47 ± 0.08 | 0.23 ± 0.24 |
| Population | N | MNA | He ± SD | K |
| Holstein Friesian crosses | 8 | 2.00 ± 0.00 | 0.22 ± 0.14 | 0.797 |
| Jersey crosses | 3 | 1.50 ± 0.70 | 0.16 ± 0.23 | 0.592 |
| Ayrshire crosses | 7 | 1.00 ± 0.00 | 0.00 ± 0.00 | 0.000 |
| Zebu | 9 | 1.50 ± 0.70 | 0.23 ± 0.33 | 0.331 |
| Source of Variation | Degrees of Freedom | Sum of Squares | Variation | Percentage of Variation | P-Value |
| Among populations | 3 | 0.921 | 0.01101 | 6.28 | 0.37830 |
| Within populations | 50 | 8.208 | 0.16417 | 93.72 | 0.37830 |
| Total | 53 | 9.130 | 0.17517 |
| Breed | Ayrshire | Jersey | Holstein Friesian | Zebu |
| Ayrshire | - | 0.000** | 0.025** | 0.098** |
| Jersey | 0.153** | - | -0.037** | -0.013** |
| Holstein Friesian | 0.086** | -0.116** | - | -0.001** |
| Zebu | 0.261* | -0.050** | -0.004** | - |
| Population | Sample Size | Fu’s Fs | Tajima’s D |
| Holstein Friesian crosses | 8 | -0.571 * | -0.648* |
| Jersey crosses | 3 | -0.002 * | -0.933* |
| Ayrshire crosses | 7 | - | - |
| Zebu | 9 | 1.214 * | 1.166 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





