Submitted:
22 May 2025
Posted:
25 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Maternal Nutritional Data
2.3. Collection of Human Milk Samples
2.4. DNA Extraction
2.5. 16S rRNA Amplicon Sequencing
2.6. Data Analysis and Availability
2.7. Statistical Analysis
3. Results
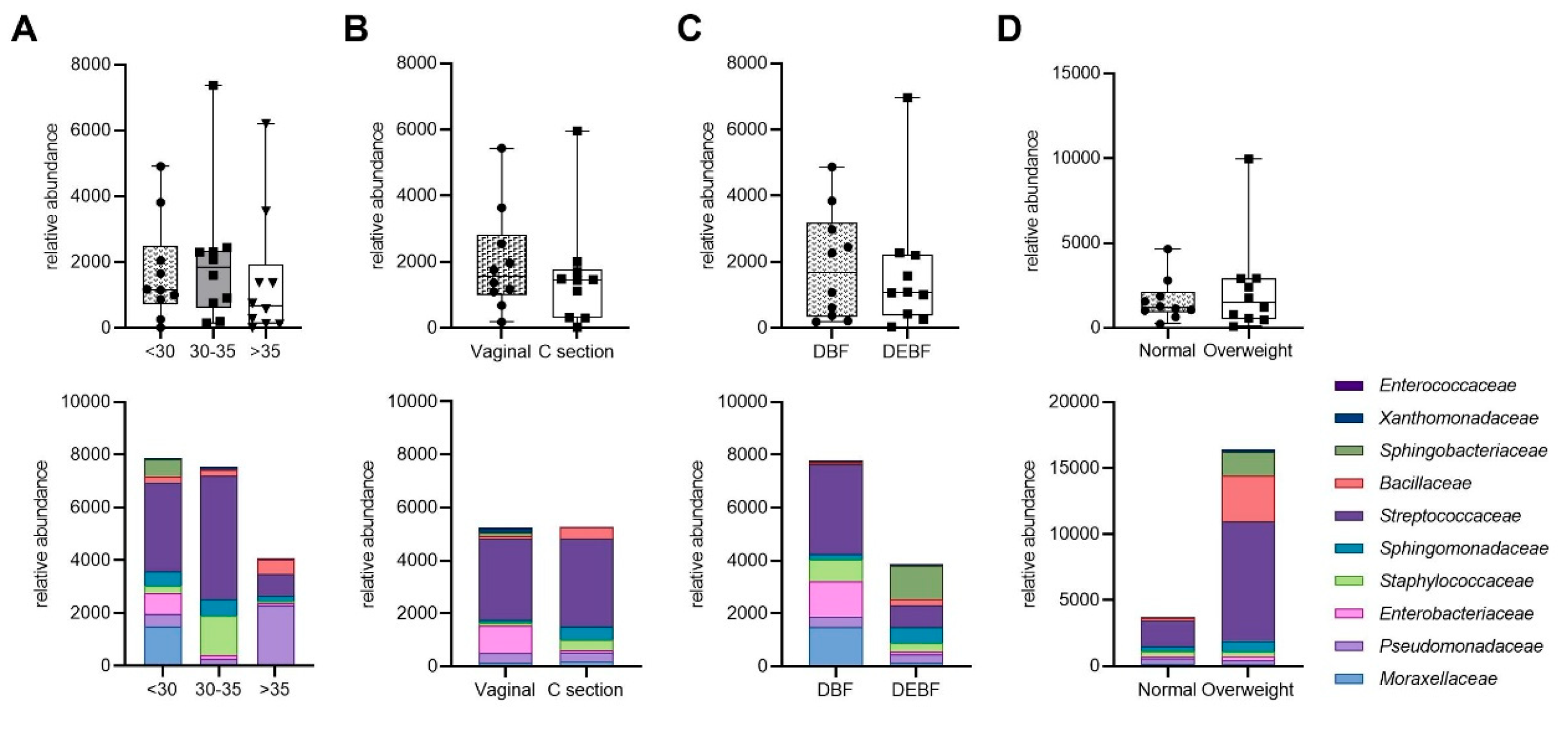
3.1. The Characteristics of the Study Group
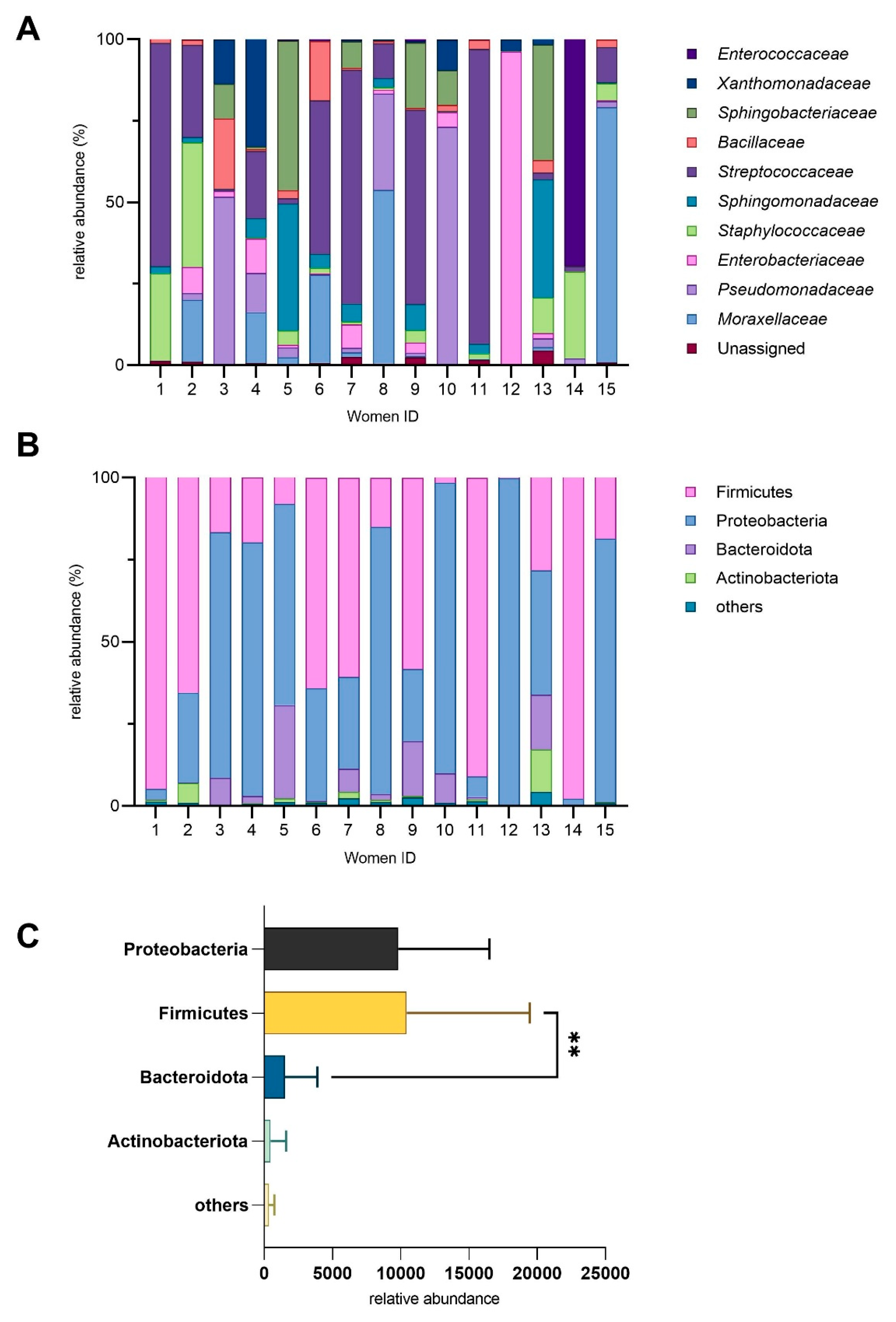
3.2. Microbial Diversity in Human Milk Samples
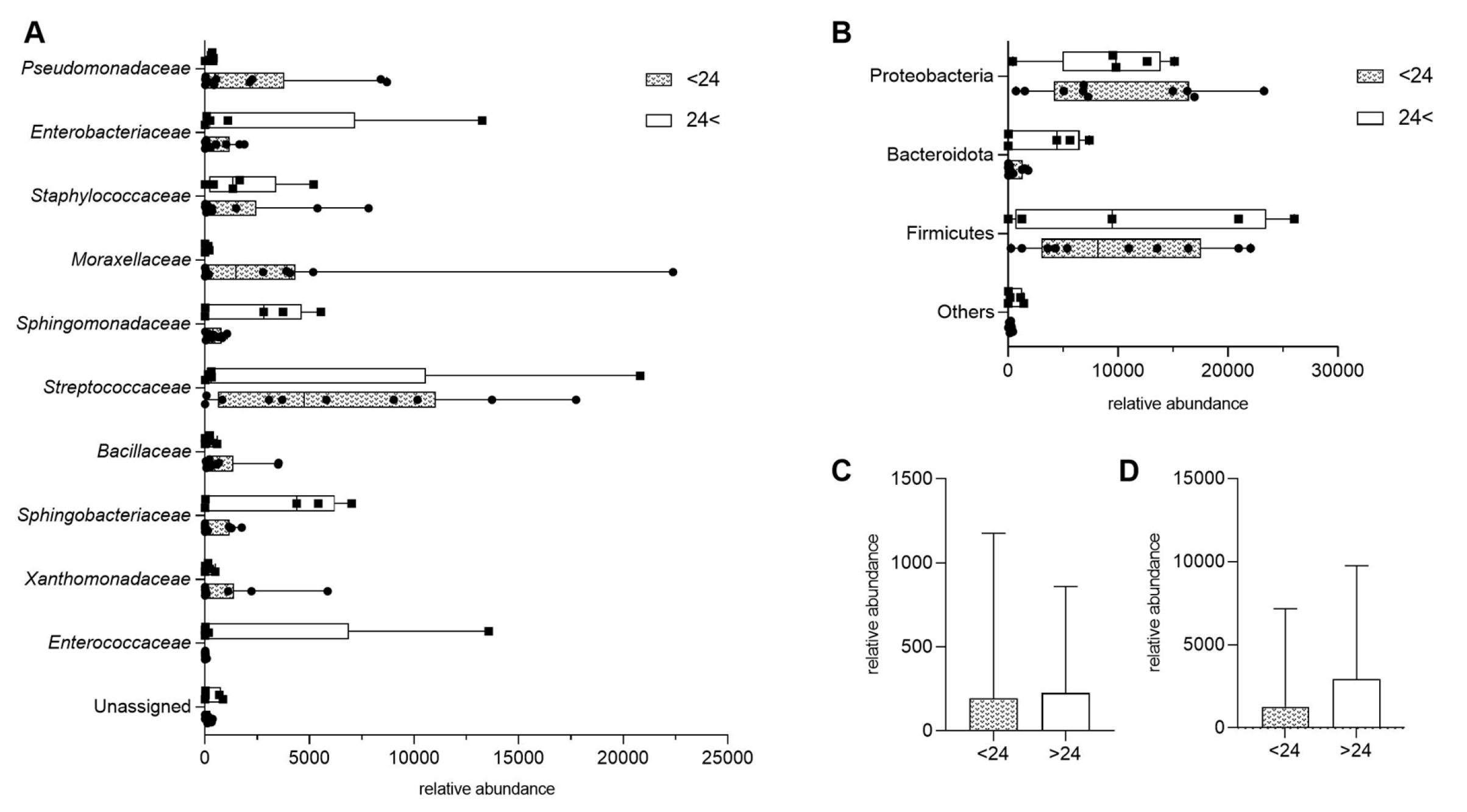
3.3. The Impact of Dietary Fiber Intake on Human Milk Microbiota
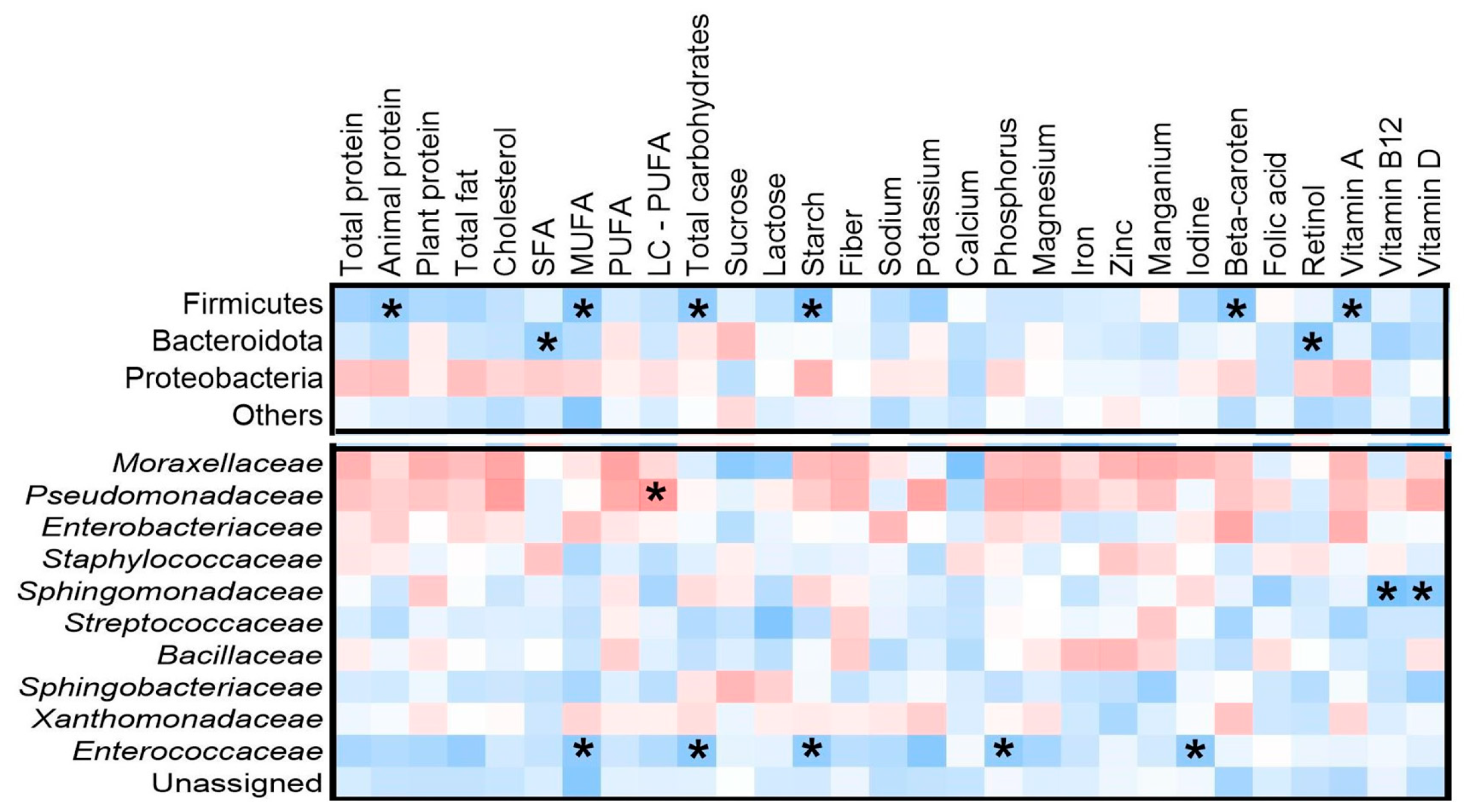
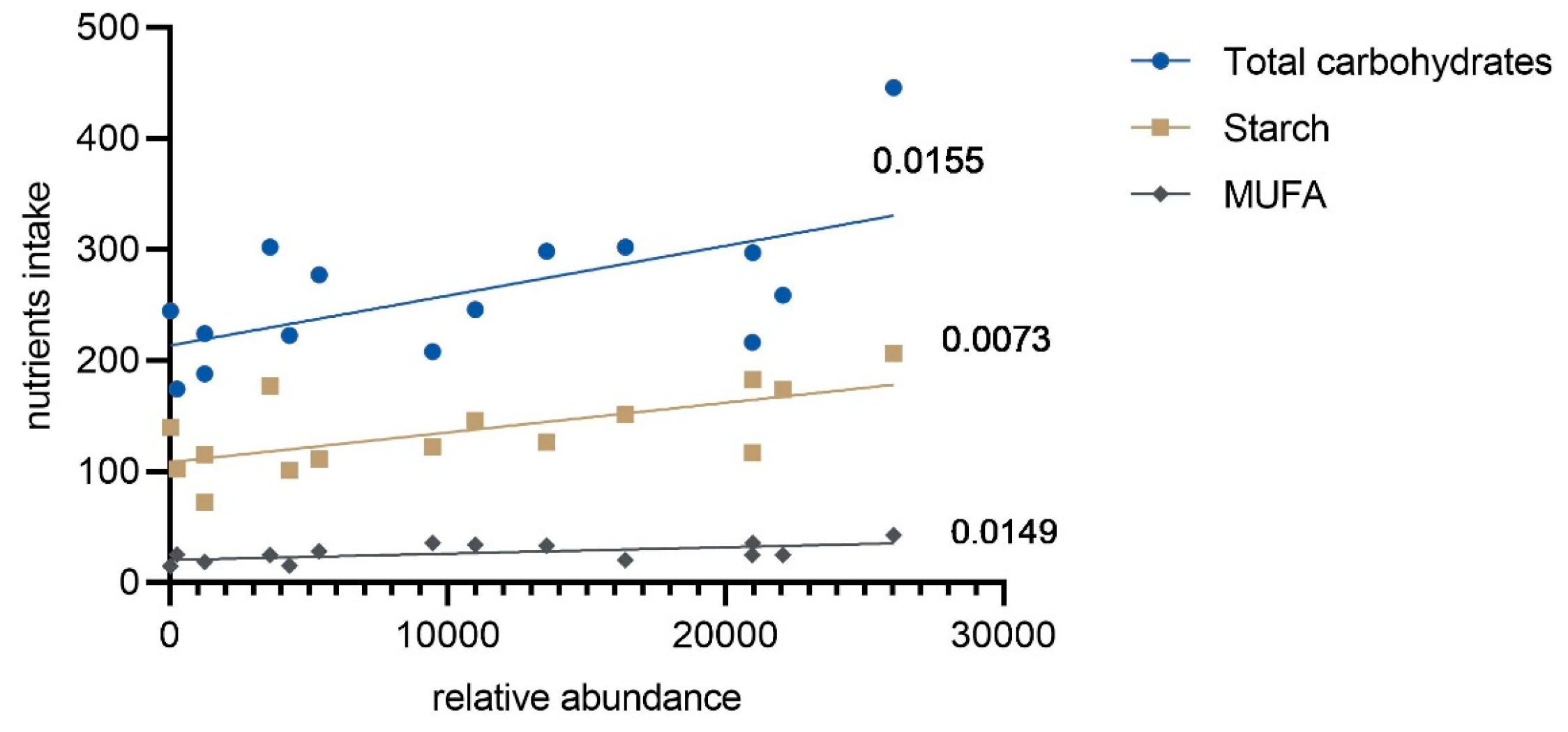
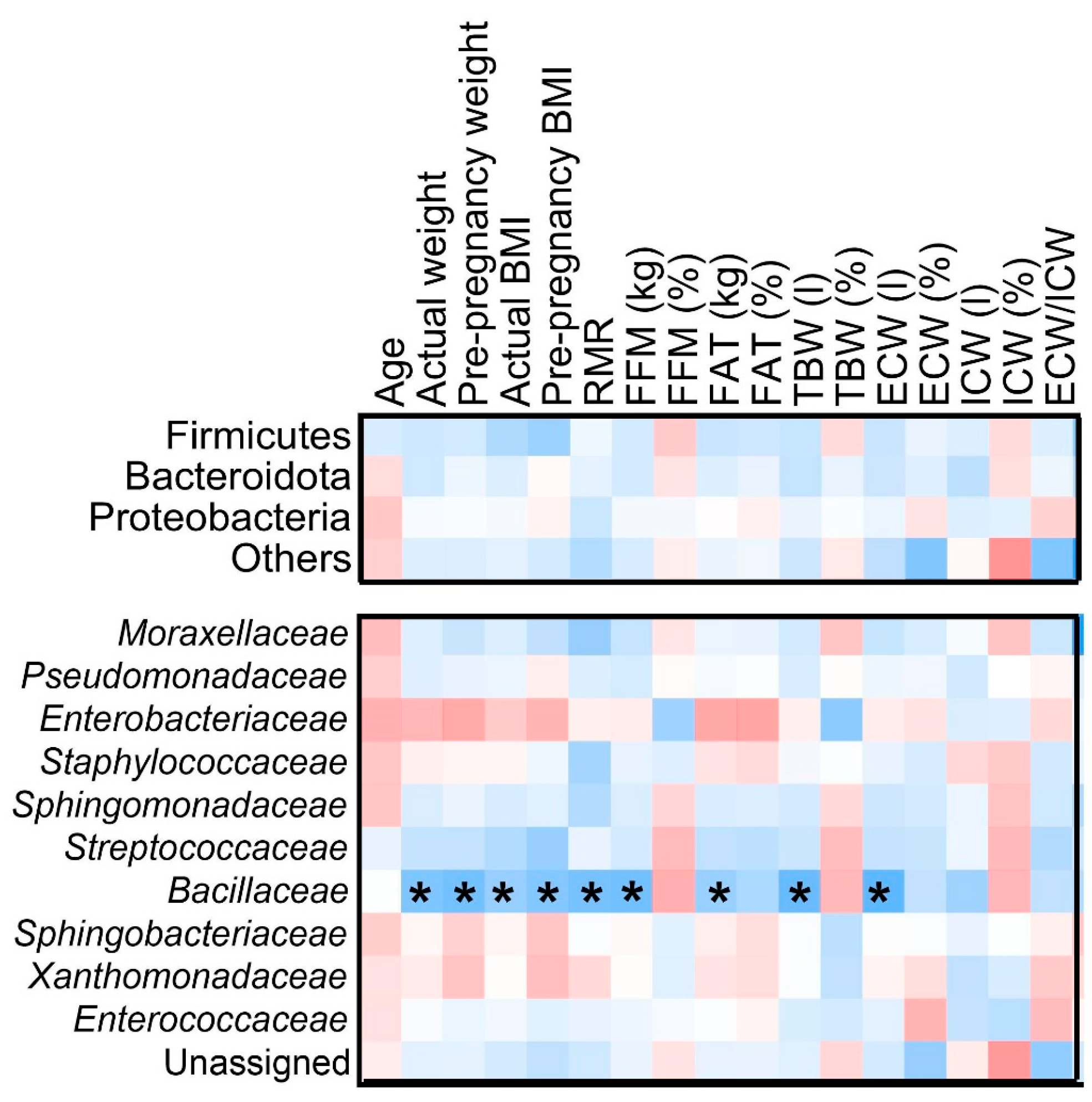
3.4. Associations Between Selected Maternal Nutrient Intake and Microbial Composition
3.5. Associations Between Food Frequency Intake and Bacterial Taxa
3.6. Relationships Between Maternal Factors and Microbial Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front Microbiol 2016, 20(7), 492. [Google Scholar] [CrossRef] [PubMed]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum Dev. 2015, 91(11), 629–35. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A. Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations. Nutrients. 2021, 13(4), 1123. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird’s-eye view. Front. Microbiol. 2017, 8, 1388. [Google Scholar] [CrossRef]
- Stinson, L.F.; Boyce, M.C.; Payne, M.S.; Keelan, J.A. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Aldrovandi, G.M. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatrics 2017, 171(7), 647–654. [Google Scholar] [CrossRef]
- Lopez Leyva, L.; Gonzalez, E.; Li, C.; Ajeeb, T.; Solomons, N.W.; Agellon, L.B.; Scott, M.E.; Koski, K.G. Human Milk Microbiota in an Indigenous Population Is Associated with Maternal Factors, Stage of Lactation, and Breastfeeding Practices. Curr Dev Nutr. 2021, 5(4), nzab013. [Google Scholar] [CrossRef]
- Demmelmair, H.; Jiménez, E.; Collado, M.C.; Salminen, S.; McGuire, M.K. Maternal and Perinatal Factors Associated with the Human Milk Microbiome. Curr Dev Nutr. 2020, 4(4), nzaa027. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Moraes, T.J.; Lefebvre, D.L.; Sears, M.R.; Khafipour, E.; Azad, M.B. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 13(25(2)), 324–335.e4. [Google Scholar] [CrossRef]
- Fitzstevens, J.L.; Smith, K.C.; Hagadorn, J.I.; Caimano, M.J.; Matson, A.P.; Brownell, E.A. Systematic review of the human milk microbiota. Nutrients 2017, 9(12), 1261. [Google Scholar] [CrossRef]
- Rodriguez, J.M. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr. 2014, 5(6), 779–84. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, D.T.; Kent, J.C.; Owens, R.A.; Hartmann, P.E. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics. 2004, 113(2), 361–367. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Toit Edu Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; Salminen, S. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Grossi, E.; Drago, L. Role of the Human Breast Milk-Associated Microbiota on the Newborns’ Immune System: A Mini Review. Front Microbiol. 2017, 8, 2100. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012, 22(9), 1147–1162. [Google Scholar] [CrossRef]
- Marcobal, A.; Sonnenburg, J.L. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012, 18 Suppl 4(0 4), 12–5. [Google Scholar] [CrossRef]
- Cortes-Macías, E.; Selma-Royo, M.; García-Mantrana, I.; Calatayud, M.; González, S.; Martínez-Costa, C.; Collado, M.C. Maternal Diet Shapes the Breast Milk Microbiota Composition and Diversity: Impact of Mode of Delivery and Antibiotic Exposure. J Nutr. 2021, 151(2), 330–340. [Google Scholar] [CrossRef]
- Padilha, M.; Danneskiold-samsøe, N.B.; Brejnrod, A.; Hoffmann, C.; Cabral, V.P.; Iaucci, J.; de, M.; Sales, C.H.; Fisberg, R.M.; Cortez, R.V.; Brix Set, a.l. The human milk microbiota is modulated by maternal diet. Microorganisms. 2019, 7. [Google Scholar] [CrossRef]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.J.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western diet leads to distinct mammary gland microbiome populations. Cell Rep. 2018, 25(1), 47–56. [Google Scholar] [CrossRef]
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J Dairy Sci 2018, 101(2), 889–899. [Google Scholar] [CrossRef]
- Heyward, V.H.; Stolarczyk, L.M. Applied Body Composition Assessment; Human Kinetics Publisher: Champaign, IL, USA, 1996; pp. 1–215. [Google Scholar]
- World Health Organization (WHO). Fourth WHO Coordinated Survey of Human milk for persistent organic pollutants in cooperation with UNEP. Guidelines for developing a national protocol. 2007 [28 March 2025]. Available online: http://www.who.int/foodsafety/chem/POPprotocol.pdf.
- Klindworth, A.; Pruesse, E.; Schweer, T.; et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013, 41(1), e1. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. . Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; et al. . Milk Microbiota: What Are We Exactly Talking About? Front Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Breast milk microbiota: A review of the factors that influence composition. J Infect. 2020, 81(1), 17–47. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011, 6(6), e21313. [Google Scholar] [CrossRef]
- Mantrana, I.; Calatayud, M.; González, S.; Martínez-Costa, C.; Collado, M.C. Maternal Diet Shapes the Breast Milk Microbiota Composition and Diversity: Impact of Mode of Delivery and Antibiotic Exposure. J Nutr. 2021, 151(2), 330–340. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; et al. . Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 2017, 17(1), 120. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016, 4, 1. [Google Scholar] [CrossRef]
- Chavoya-Guardado, M.A.; Vasquez-Garibay, E.M.; Ruiz-Quezada, S.L.; Ramírez-Cordero, M.I.; Larrosa-Haro, A.; Castro-Albarran, J. Firmicutes, Bacteroidetes and Actinobacteria in Human Milk and Maternal Adiposity. Nutrients. 2022, 14(14), 2887. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006, 444(7122), 1022–1023. [Google Scholar] [CrossRef]
- McGuire, M.K.; McGuire, M.A. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr Opin Biotechnol. 2017, 44, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Dombrowska-Pali, A.; Wiktorczyk-Kapischke, N.; Chrustek, A.; Olszewska-Słonina, D.; Gospodarek-Komkowska, E.; Socha, M.W. Human Milk Microbiome-A Review of Scientific Reports. Nutrients. 2024, 16(10), 1420. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Al Mohannadi, N.; Murugesan, S.; et al. . Unveiling the dynamics of the breast milk microbiome: impact of lactation stage and gestational age. J Transl Med 2023, 21(1), 784. [Google Scholar] [CrossRef]
- Bode, L.; McGuire, M.; Rodriguez, J.M.; et al. . It’s alive: microbes and cells in human milk and their potential benefits to mother and infant. Adv Nutr. 2014, 5(5), 571–573. [Google Scholar] [CrossRef] [PubMed]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martinez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014, 34(8), 599–605. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012, 96(3), 544–51. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Mira-Pascual, L.; Mira, A.; Collado, M.C. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016, 7(1), 54–60. [Google Scholar] [CrossRef]
- Sakwinska, O.; Moine, D.; Delley, M.; Combremont, S.; Rezzonico, E.; Descombes, P.; Vinyes-Pares, G.; Zhang, Y.; Wang, P.; Thakkar, S.K. Microbiota in breast milk of Chinese lactating mothers. PLoS One. 2016, 11(8), e0160856. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Peroni, D.G.; Grossi, E.; Facchin, V.; Comberiati, P.; Drago, L. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 2017, 17(1), 205. [Google Scholar] [CrossRef]
- Santacruz, A.; Collado, M.C.; Garcia-Valdes, L.; Segura, M.T.; Martin-Lagos, J.A.; Anjos, T.; Marti-Romero, M.; Lopez, R.M.; Florido, J.; Campoy Cet, a.l. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010, 104(1), 83–92. [Google Scholar] [CrossRef]
- Lundgren, S.N.; Madan, J.C.; Karagas, M.R.; Morrison, H.G.; Hoen, A.G.; Christensen, B.C. Microbial communities in human milk relate to measures of maternal weight. Front Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Jiang, J.; Lu, M.; et al. Human milk microbiota development during lactation and its relation to maternal geographic location and gestational hypertensive status. Gut Microbes. 2020, 11(5), 1438–1449. [CrossRef]
- Li C, Gonzalez E, Solomons NW, Scott ME, Koski KG. Human breast milk microbiota is Influenced by maternal age and BMI, stage of lactation and infant feeding practices. FASEB J. 2017, 31(S1), 965.25–965.25. [CrossRef]
- Ojo-Okunola A, Claassen-Weitz S, Mwaikono KS, Gardner-Lubbe S, Stein DJ, Zar HJ, Nicol MP, du Toit E. Influence of Socio-Economic and Psychosocial Profiles on the Human Breast Milk Bacteriome of South African Women. Nutrients. 2019, 11(6), 1390. [CrossRef]
- Cortés-Macías, E.; Selma-Royo, M.; Martínez-Costa, C.; Collado, M.C. Breastfeeding Practices Influence the Breast Milk Microbiota Depending on Pre-Gestational Maternal BMI and Weight Gain over Pregnancy. Nutrients. 2021, 13(5), 1518. [Google Scholar] [CrossRef] [PubMed]
- Reyes, S.M.; Allen, D.L.; Williams, J.E.; et al. Pumping supplies alter the microbiome of pumped human milk: An in-home, randomized, crossover trial. Am J Clin Nutr. 2021, 114(6), 1960-1970. [CrossRef]
- Sun, Q.; Zhou, Q.; Ge, S.; Liu, L.; Li, P.; Gu, Q. Effects of Maternal Diet on Infant Health: A Review Based on Entero-Mammary Pathway of Intestinal Microbiota. Mol. Nutr. Food Res. 2024, 68, e2400077. [Google Scholar] [CrossRef]
- LeMay-Nedjelski, L.; Asbury, M.R.; Butcher, J.; Ley, S.H.; Hanley, A.J.; Kiss, A.; Unger, S.; Copeland, J.K.; Wang, P.W.; Stintzi, A.; O’Connor, D.L. Maternal Diet and Infant Feeding Practices Are Associated with Variation in the Human Milk Microbiota at 3 Months Postpartum in a Cohort of Women with High Rates of Gestational Glucose Intolerance. J Nutr. 2021, 151(2), 320–329. [Google Scholar] [CrossRef]
- Sindi, A.S.; Stinson, L.F.; Gridneva, Z.; Leghi, G.E.; Netting, M.J.; Wlodek, M.E.; Muhlhausler, B.S.; Rea, A.; Trevenen, M.L.; Geddes, D.T.; Payne, M.S. Maternal dietary intervention during lactation impacts the maternal faecal and human milk microbiota. J Appl Microbiol. 2024, 135(5), lxae024. [Google Scholar] [CrossRef]
- Dzidic, M.; Mira, A.; Artacho, A.; Abrahamsson, T.R.; Jenmalm, M.C.; Collado, M.C. Allergy development is associated with consumption of breastmilk with a reduced microbial richness in the first month of life. Pediatr Allergy Immunol. 2020, 31, 250–257. [Google Scholar] [CrossRef]
- Marsh, A.J.; Azcarate-Peril, M.A.; Aljumaah, M.R.; Neville, J.; Perrin, M.T.; Pawlak, R. Fatty acid profle driven by maternal diet is associated with the composition of humannmilk microbiota. Front. Microbiom. 2022, 1, 1041752. [Google Scholar] [CrossRef]
- Abrante-Pascual, S.; Nieva-Echevarría, B.; Goicoechea-Oses, E. Vegetable Oils and Their Use for Frying: A Review of Their Compositional Differences and Degradation. Foods. 2024, 13(24), 4186. [Google Scholar] [CrossRef] [PubMed]
- Chrustek, A.; Dombrowska-Pali, A.; Olszewska-Słonina, D.; et al. Human Milk Microbiome from Polish Women Giving Birth via Vaginal Delivery-Pilot Study. Biology (Basel). 2025, 14(4), 332. [CrossRef]
- Stinson, L.F.; Sindi, A.S.M.; Cheema, A.S.; et al. The human milk microbiome: who, what, when, where, why, and how? Nutr Rev. 2021, 79(5), 529–543. [Google Scholar] [CrossRef]
| Parameters | Mean ± SD | Median (IQR) |
|---|---|---|
| Maternal characteristic | ||
| Age (years) | 32.1 ± 5.0 | 31 (29 - 33) |
| Pre-pregnancy BMI (kg/m2) | 21.2 ± 2.7 | 20.8 (19.1 - 22.6) |
| Weight gain during pregnancy (kg) | 14.0 ± 2.6 | 14 (12 - 16) |
| Actual BMI (kg/ m2) | 22.4 ± 2.8 | 22.3 (19.8 - 23.8) |
| Fat free mass (kg) | 44.0 ± 3.2 | 44.6 (42.4 - 45.6) |
| Fat free mass (%) | 71.8 ± 6 | 70.7 (67.8 - 75.7) |
| Fat mass (kg) | 17.9 ± 6.1 | 18 (13.4 - 21.7) |
| Fat mass (%) | 28.2 ± 6.0 | 29.3 (24.3 - 32.2) |
| Total body water (L) | 30.7 ± 2.7 | 30.5 (28.9 - 32) |
| Total body water (%) | 49.9 ± 3.9 | 49.5 (47.1 - 51.5) |
| Extracellular body water (L) | 14.2 ± 1.5 | 14.5 (12.9 - 15.3) |
| Extracellular body water (%) | 46.3 ± 1.8 | 47 (45.4 - 47.6) |
| Intracellular body water (L) | 16.5 ± 1.5 | 16.3 (15.9 - 16.9) |
| Intracellular body water (%) | 53.7 ± 1.8 | 52.9 (52.3 - 54.6) |
| Protein (kg) | 9.5 ± 0.8 | 9.7 (8.7 - 10.1) |
| Minerals (kg) | 3.9 ± 0.3 | 4 (3.5 - 4.1) |
| Muscle mass (kg) | 18.9 ± 2.0 | 19.6 (18.4 - 19.9) |
| Resting metabolic rate (kcal) | 1509.5 ± 49.7 | 1513 (1481.5 - 1532) |
| Infant characteristic | ||
| Birth weight (g) | 3553.7 ± 401.3 | 3550 (3185 - 3827.5) |
| Birth height (cm) | 55.3 ± 1 | 55 (54.5 - 56) |
| Number of day feedings | 8.8 ± 3.5 | 8 (7 - 9.5) |
| Number of night feedings | 1.9 ± 0.6 | 2 (2 - 2) |
| Mean ± SD | Median (IQR) | |
|---|---|---|
| Macronutrients | ||
| Total protein (g) | 81.91 ± 24.2 | 74.39 (62.91 - 102.5) |
| Animal protein (g) | 50.6 ± 17.26 | 44.89 (37.44 - 64.21) |
| Plant protein (g) | 30.75 ± 10.36 | 27.22 (23.14 - 32.6) |
| Total fat (g) | 68.7 ± 17.91 | 63.92 (57.91 - 82.95) |
| Cholesterol (mg) | 289 ± 123.5 | 289.2 (208 - 343.8) |
| Saturated fatty acids (SFA) (g) | 25.91 ± 8.34 | 26.05 (19.06 - 31.5) |
| Monounsaturated fatty acids (MUFA) (g) | 26.28 ± 8.467 | 24.89 (18.69 - 34.05) |
| Polyunsaturated fatty acids (PUFA) (g) | 11.61 ± 5.284 | 10.7 (8.94 - 13.15) |
| Long chain polyunsaturated fatty acids (g) | 0.3385 ± 0.6199 | 0.073 (0.03 - 0.2773) |
| Total carbohydrates (g) | 260.5 ± 66.3 | 245.9 (216.4 - 298.6) |
| Sucrose (g) | 41.57 ± 20.11 | 39.56 (26.71 - 55.49) |
| Lactose (g) | 10.99 ± 9.232 | 10.48 (2.54 - 17.98) |
| Starch (g) | 136.4 ± 36.55 | 126.6 (111.5 - 173.8) |
| Fiber (g) | 22.28 ± 6.821 | 21.21 (16.77 - 25.13) |
| Energy from protein (%) | 17.06 ± 2.726 | 17.3 (15.35 - 19.23) |
| Energy from fat (%) | 31.54 ± 4.606 | 31.11 (27.81 - 32.57) |
| Energy from carbohydrates (%) | 51.09 ± 5.751 | 50.73 (47.68 - 56.84) |
| Micronutrients | ||
| Sodium (mg) | 2862 ± 1043 | 2892 (1962 - 3548) |
| Potassium (mg) | 3202 ± 893.5 | 3092 (2485 - 3573) |
| Calcium (mg) | 818 ± 354.3 | 723.1 (590.4 - 1030) |
| Phosphorus (mg) | 1402 ± 386.8 | 1230 (1182 - 1620) |
| Magnesium (mg) | 347.3 ± 129.1 | 301.1 (262.4 - 391.6) |
| Iron (mg) | 16.06 ± 14.79 | 11.55 (10.04 - 14.55) |
| Zinc (mg) | 11.95 ± 6.289 | 10.95 (7.276 - 12.79) |
| Manganese (mg) | 5.287 ± 2.204 | 4.61 (3.54 - 6.53) |
| Iodine (μg) | 118 ± 47.3 | 107.4 (75.31 - 154.2) |
| Beta-carotene (μg) | 4542 ± 2402 | 3786 (3345 - 5568) |
| Folic acid (μg) | 380.7 ± 206.8 | 308.7 (262.6 - 417.8) |
| Retinol (μg) | 361.3 ± 130.9 | 359 (285.2 - 407.8) |
| Vitamin A (μg) | 1110 ± 484.9 | 1007 (780.4 - 1284) |
| Vitamin B6 (mg) | 2.167 ± 0.9334 | 1.729 (1.459 - 2.44) |
| Vitamin B12 (μg) | 3.758 ± 2.187 | 2.915 (2.44 - 4.8) |
| Vitamin C (mg) | 137.3 ± 100.9 | 123.2 (87.63 - 151) |
| Vitamin D (μg) | 4.266 ± 3.71 | 2.549 (1.63 - 5.618) |
| Vitamin E (mg) | 11.46 ± 4.27 | 10.87 (7.54 - 14.75) |
| Thiamine (mg) | 1.401 ± 0.6372 | 1.147 (0.91 - 1.56) |
| Riboflavin (mg) | 1.886 ± 0.6132 | 1.64 (1.53 - 2.038) |
| Niacin (mg) | 17.98 ± 7.996 | 15.09 (10.52 - 23.5) |
| Food group | Bacteria family | P value |
|---|---|---|
| Animal milk | Streptococcaceae | 0.072 |
| Sphingomonadaceae | 0.005 | |
| Plant oils | Streptococcaceae | 0.088 |
| Staphylococcaceae | 0.002 | |
| Oily fishes | Bacillaceae | 0.069 |
| Lean fishes | Bacillaceae | 0.052 |
| Sphingobacteriaceae | 0.131 | |
| Butter | Bacillaceae | 0.103 |
| Staphylococcaceae | 0.073 | |
| Wholegrain | Sphingobacteriaceae | 0.111 |
| Xanthomonadaceae | 0.059 | |
| Enterobacteriaceae | 0.018 | |
| Buckweats | Sphingobacteriaceae | 0.035 |
| peantuts | Xanthomonadaceae | 0.027 |
| Sphingobacteriaceae | 0.029 | |
| Poultry | Enterococcaceae | 0.038 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





