Submitted:
21 May 2025
Posted:
22 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Unsupervised machine learning is important in this kind of study because it allows the model to create its classification based on data correlations, instead of assigning the classifications beforehand.
- At the time of the study, NSDUH 2019 was a more recent database with 56,137 entries and half of the data size was used. DSM 5 is more current and is widely used by mental health professionals in classifying SUDs.
- The result of our study is an automated classification in terms of SUD based on DSM 5 and is validated by a mental health professional.
2. Methodology
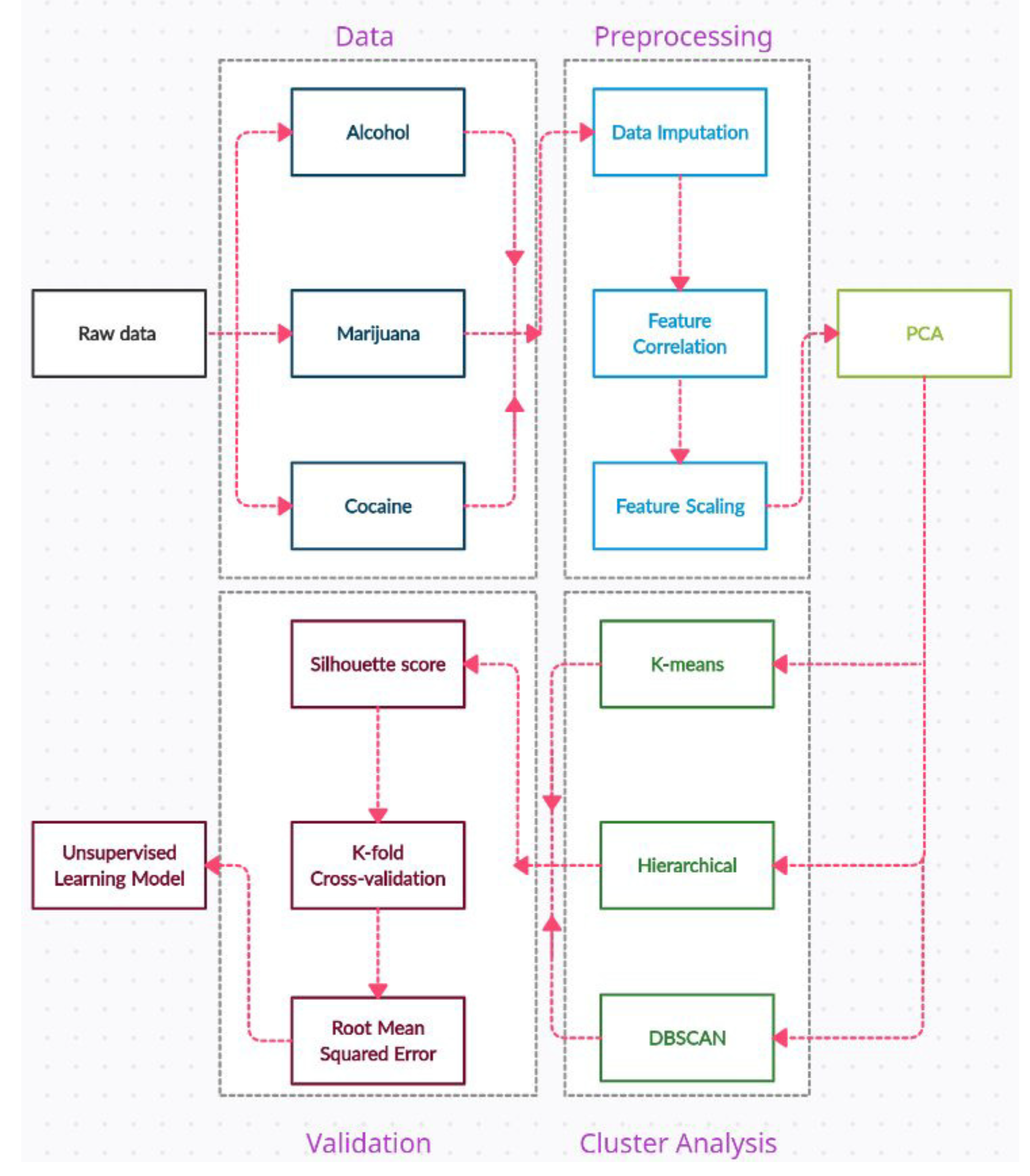
3. Materials and Methods
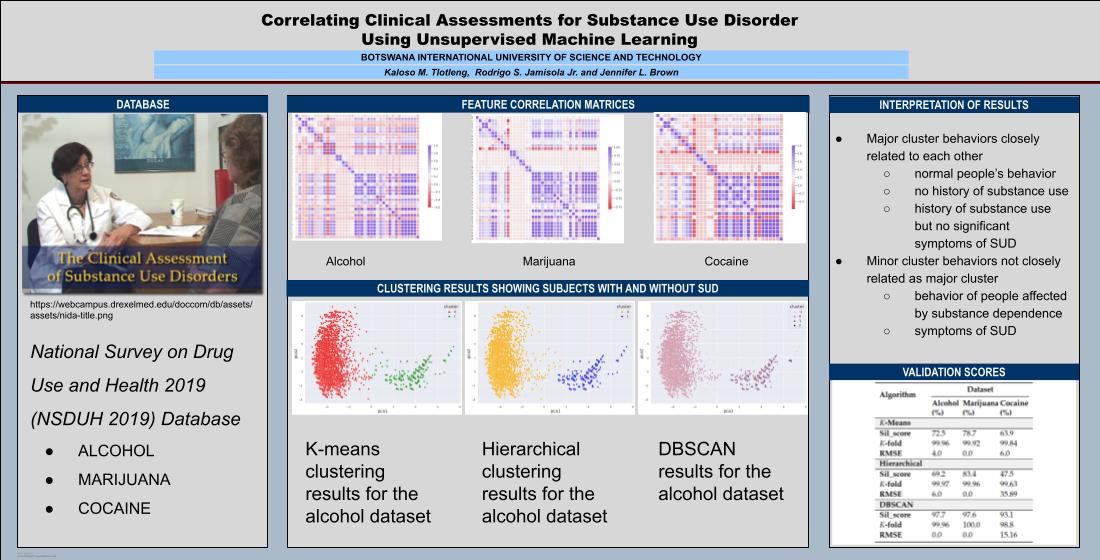
3.1. Dataset
3.2. Data Preprocessing
3.2.1. Data Cleaning
3.2.2. Feature Correlation
3.2.3. Feature Scaling
3.2.4. Dimensionality Reduction
3.3. Cluster Analysis
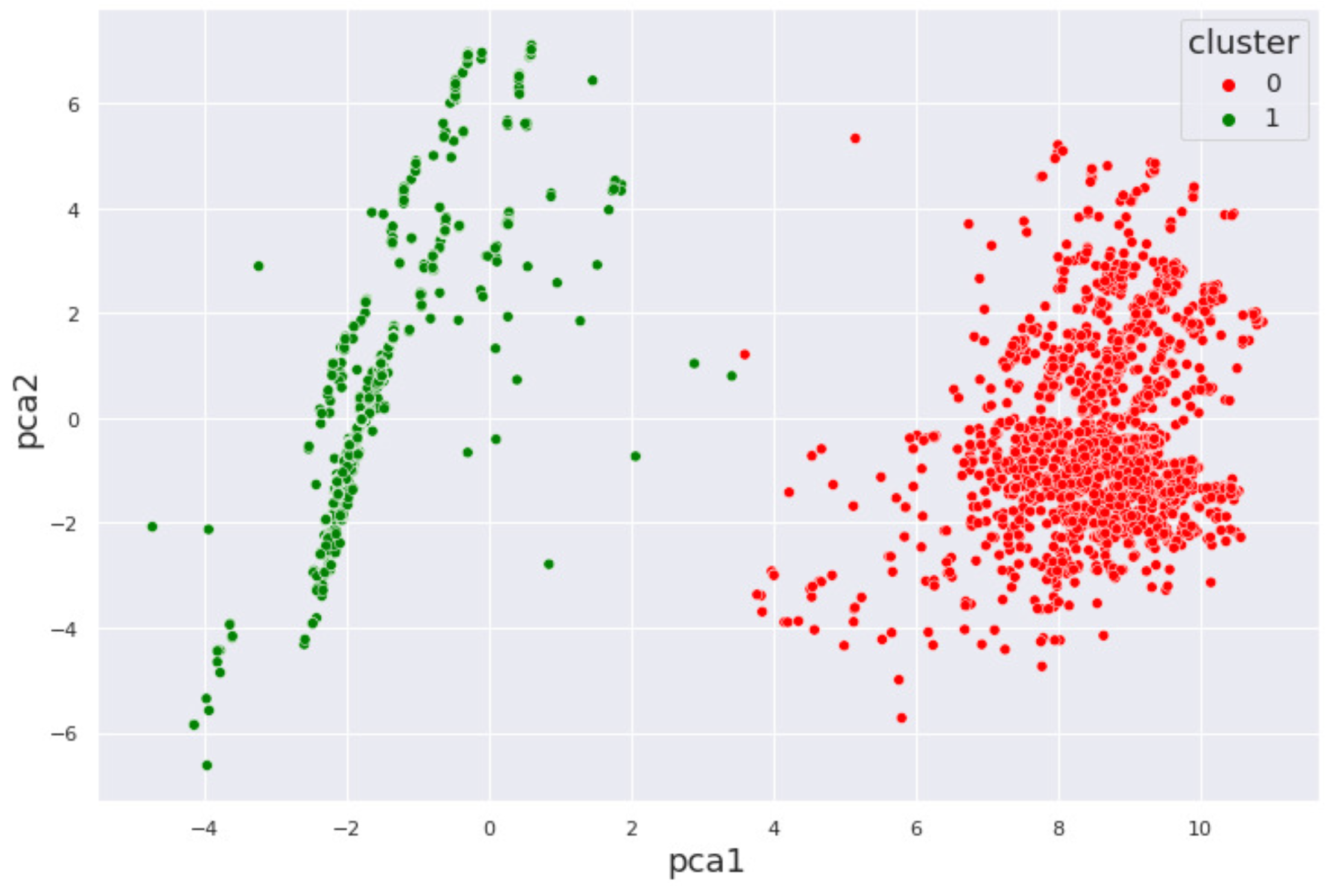
3.3.1. K-means Clustering
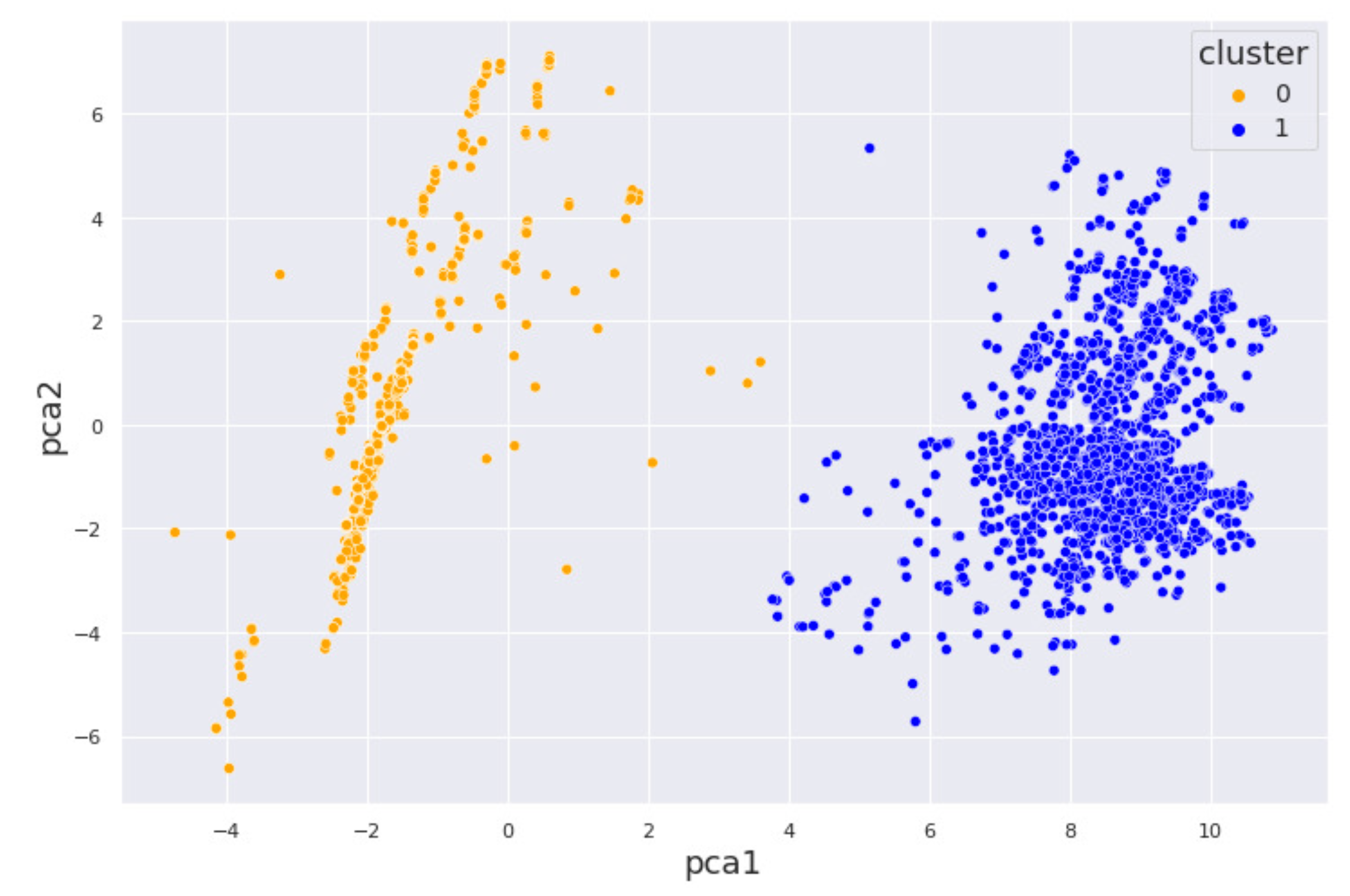
3.3.2. Hierarchical Clustering
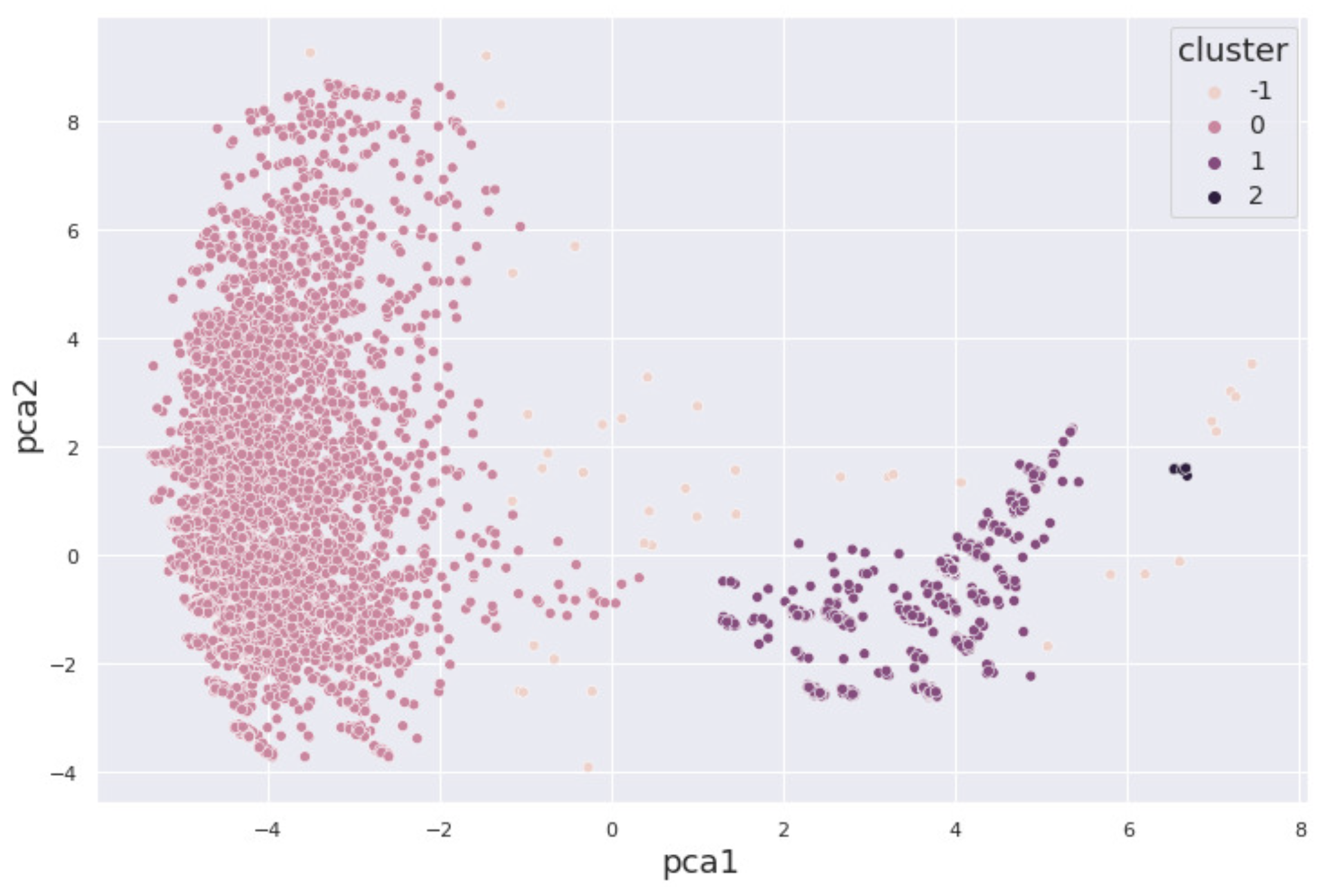
3.3.3. DBSCAN
3.4. Validation
3.4.1. Internal validation
3.4.2. External Validation
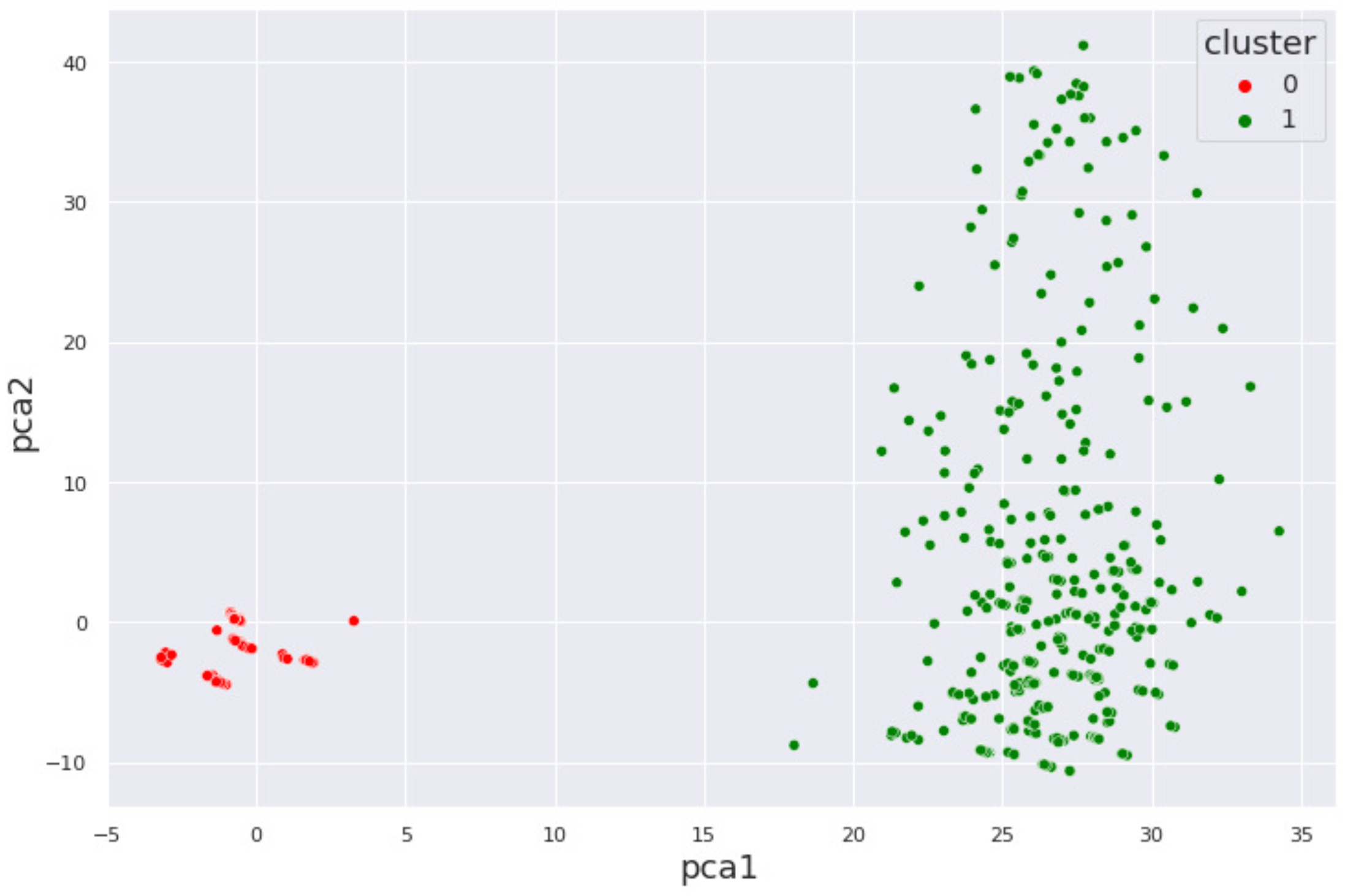
4. Results and Discussion
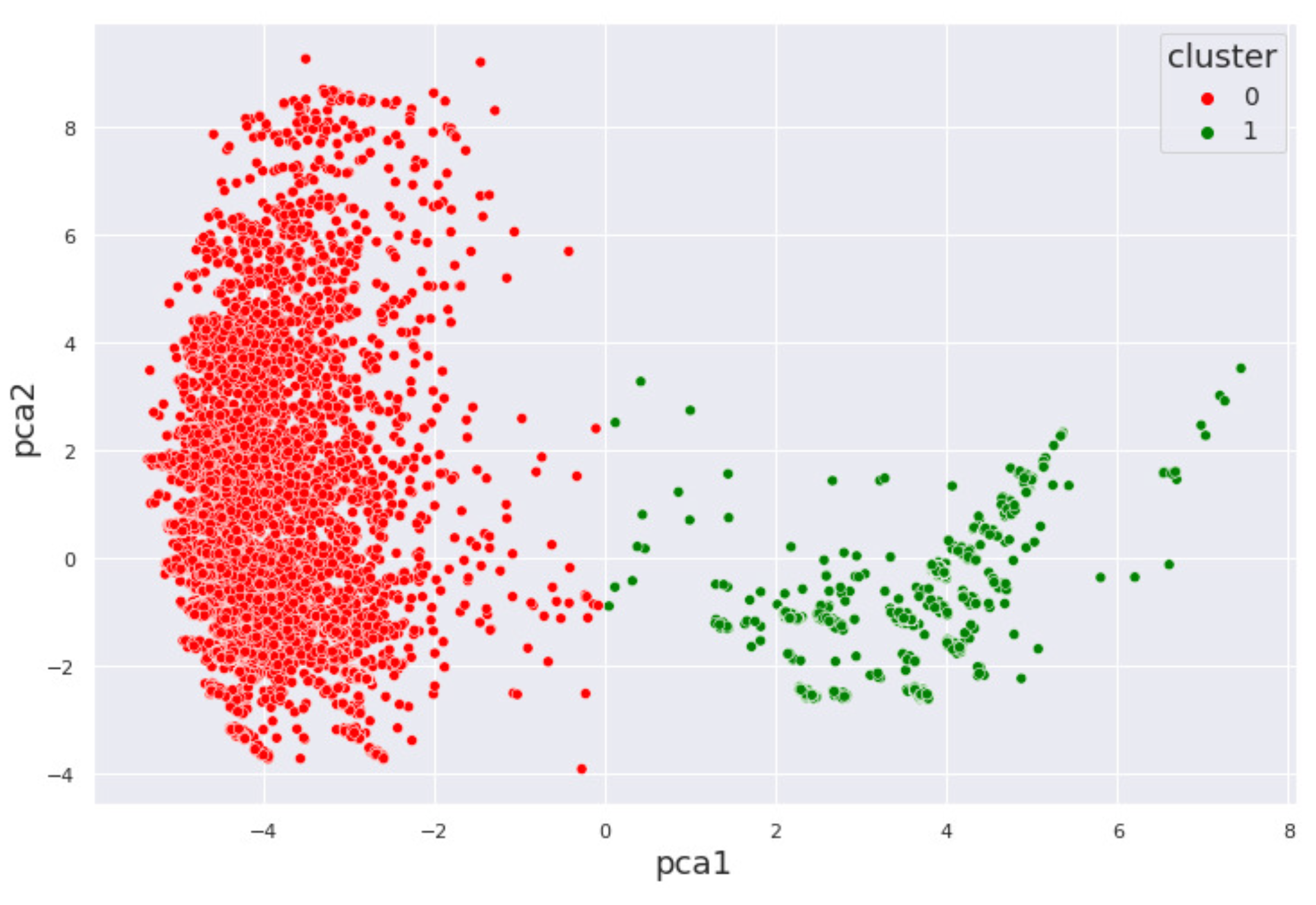
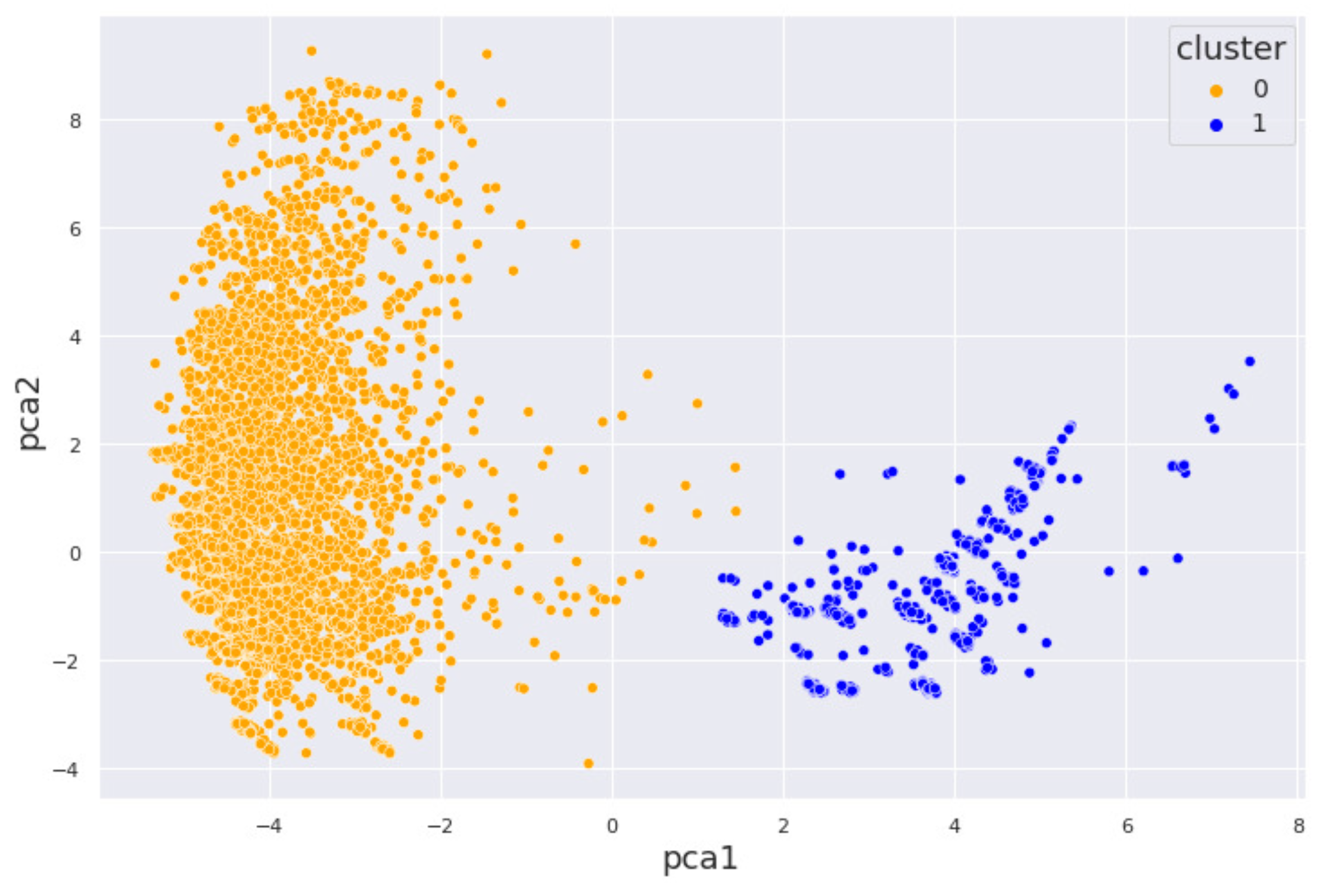
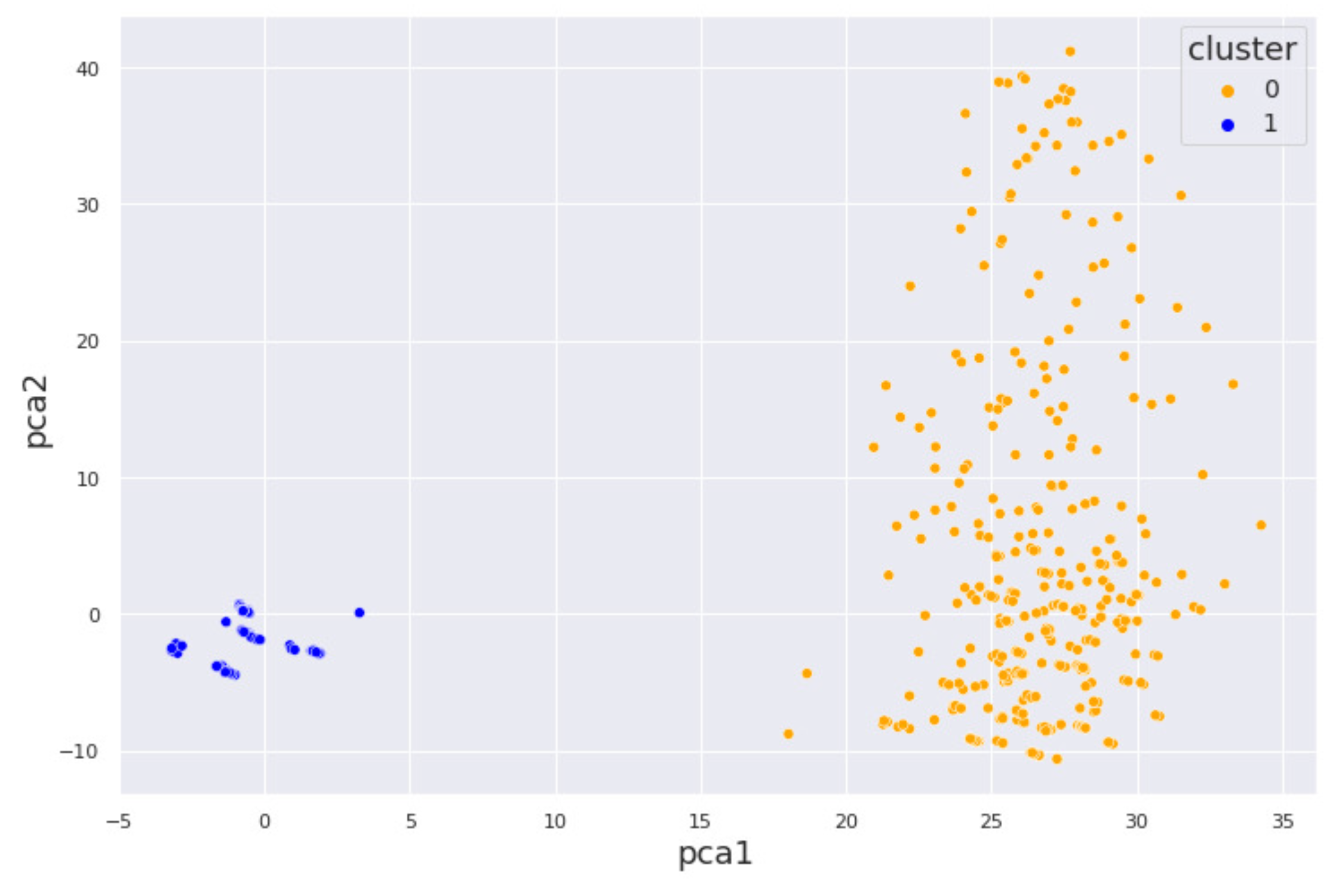
4.1. Alcohol Dataset
4.2. Cocaine Dataset
4.3. Validation Results
5. Interpretation of Results
5.1. By Machine Learning
5.2. By a Mental Health Professional
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahn, W.-Y.; Vassileva, J. Machine-learning identifies substance-specific behavioral markers for opiate and stimulant dependence. Drug and Alcohol Dependence 2016, 161, 247–257. [Google Scholar] [CrossRef]
- Al Sukar, M.; Sleit, A.; Abu-Dalhoum, A.; Al-Kasasbeh, B. Identifying a drug addict person using artificial neural networks. International Journal of Computer and Information Engineering 2016, 10(3), 611–616. [Google Scholar]
- Mak, K.K.; Lee, K.; Park, C. Applications of machine learning in addiction studies: A systematic review. Psychiatry Research 2019, 275, 53–60. [Google Scholar] [CrossRef]
- Robbins, T.W.; Clark, L. Behavioral addictions. Current Opinion in Neurobiology 2015, 30, 66–72. [Google Scholar] [CrossRef]
- Srividya, M.; Mohanavalli, S.; Bhalaji, N. Behavioral modeling for mental health using machine learning algorithms. Journal of Medical Systems 2018, 42(5), 1–12. [Google Scholar] [CrossRef]
- Kharabsheh, M.; Meqdadi, O.; Alabed, M.; Veeranki, S.; Abbadi, A.; Alzyoud, S. A machine learning approach for predicting nicotine dependence. International Journal of Advanced Computer Science and Applications 2019, 10(3), 179–184. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Wiil, U.K.; Andersen, K.; Mansourvar, M.; Nielsen, A.S. A predictive machine learning model to determine alcohol use disorder. In 2020 IEEE Symposium on Computers and Communications (ISCC) (pp. 1-7); IEEE, 2020. [Google Scholar]
- Stinchfield, R.; McCready, J.; Turner, N.E.; Jimenez-Murcia, S.; Petry, N.M.; Grant, J. ... & Winters, K. C. Reliability, validity, and classification accuracy of the DSM-5 diagnostic criteria for gambling disorder. Journal of Gambling Studies 2016, 32(3), 905–922. [Google Scholar] [CrossRef]
- Tarekegn, A.N.; Michalak, K.; Giacobini, M. Cross-validation approach to evaluate clustering algorithms: An experimental study using multi-label datasets. SN Computer Science 2020, 1(5), 1–9. [Google Scholar] [CrossRef]
- Stevens, E.; Dixon, D.R.; Novack, M.N.; Granpeesheh, D.; Smith, T.; Linstead, E. Identification and analysis of behavioral phenotypes in autism spectrum disorder via unsupervised machine learning. International Journal of Medical Informatics 2019, 129, 29–36. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5®); American Psychiatric Pub, 2013. [Google Scholar]
- Hasin, D.S.; O’Brien, C.P.; Auriacombe, M.; Borges, G.; Bucholz, K.; Budney, A. .. & Petry, N.M. DSM-5 criteria for substance use disorders: Recommendations and rationale. American Journal of Psychiatry 2013, 170(8), 834–851. [Google Scholar]
- Substance Abuse and Mental Health Services Administration. (2019). National survey on drug use and health. Available online: https://www.samhsa.gov/data/ (accessed on 1 January 2023).
- Chowdhry, A.K.; Gondi, V.; Pugh, S.L. Missing data in clinical studies. International Journal of Radiation Oncology* Biology* Physics 2021. [Google Scholar] [CrossRef] [PubMed]
- Kherif, F.; Latypova, A. Principal component analysis. In Machine Learning (pp. 209-225); Elsevier, 2020. [Google Scholar]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature selection in machine learning: A new perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Ali, P.J.M.; Faraj, R.H.; Koya, E.; Ali, P.J.M.; Faraj, R.H. Data normalization and standardization: A technical report. Mach Learn Tech Rep 2014, 1(1), 1–6. [Google Scholar]
- Chakraborty, S.; Paul, D.; Das, S.; Xu, J. Entropy regularized power k-means clustering. arXiv preprint 2020, arXiv:2001.03452. [Google Scholar]
- Saha, T.D.; Chou, S.P.; Grant, B.F. The performance of DSM-5 alcohol use disorder and quantity-frequency of alcohol consumption criteria: An item response theory analysis. Drug and Alcohol Dependence 2020, 216, 108299. [Google Scholar] [CrossRef] [PubMed]
- Alcohol, Research; Staff, C.R.E. Drinking patterns and their definitions. Alcohol Research: Current Reviews 2018, 39(1), 17. [Google Scholar]
- Zhang, M. Use density-based spatial clustering of applications with noise (DBSCAN) algorithm to identify galaxy cluster members. In IOP Conference Series: Earth and Environmental Science (Vol. 252, No. 4, p. 042033); IOP Publishing, 2019. [Google Scholar]
- Iniesta, R.; Stahl, D.; McGuffin, P. Machine learning, statistical learning and the future of biological research in psychiatry. Psychological Medicine 2016, 46(12), 2455–2465. [Google Scholar] [CrossRef]
- Hauke, J.; Kossowski, T. Comparison of values of Pearson’s and Spearman’s correlation coefficient on the same sets of data. Quaestiones Geographicae 2011, 30(2), 87–93. [Google Scholar] [CrossRef]
- Pedersen, A.B.; Mikkelsen, E.M.; Cronin-Fenton, D.; Kristensen, N.R.; Pham, T.M.; Pedersen, L.; Petersen, I. Missing data and multiple imputation in clinical epidemiological research. Clinical Epidemiology 2017, 9, 157–165. [Google Scholar] [CrossRef]
- Wong, T.-T.; Yang, N.-Y. Dependency analysis of accuracy estimates in k-fold cross validation. IEEE Transactions on Knowledge and Data Engineering 2017, 29(11), 2417–2427. [Google Scholar] [CrossRef]
- Salkind, N.J. Root mean square error. Encyclopedia of Research Design. 2010.
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. SN Computer Science 2021, 2(3), 1–21. [Google Scholar] [CrossRef]
- Wetherill, L.; Agrawal, A.; Kapoor, M.; Bertelsen, S.; Bierut, L.J.; Brooks, A. .. & others. Association of substance dependence phenotypes in the COGA sample. Addiction Biology 2015, 20(3), 617–627. [Google Scholar]
- Choi, J.; Jung, H.T.; Choi, J. Marijuana addiction prediction models by gender in young adults using random forest. Online Journal of Nursing Informatics (OJNI) 2021, 25(2). [Google Scholar]
- Choi, J.; Chung, J.; Choi, J. Exploring impact of marijuana (cannabis) abuse on adults using machine learning. International Journal of Environmental Research and Public Health 2021, 18(19), 10357. [Google Scholar] [CrossRef]
- Grant, B.F.; Saha, T.D.; Ruan, W.J.; Goldstein, R.B.; Chou, S.P.; Jung, J. ; ... & others. Epidemiology of DSM-5 drug use disorder: Results from the National Epidemiologic Survey on Alcohol and Related Conditions-III. JAMA Psychiatry 2016, 73(1), 39–47. [Google Scholar]
- Hayley, A.C.; Stough, C.; Downey, L.A. DSM-5 cannabis use disorder, substance use and DSM-5 specific substance-use disorders: Evaluating comorbidity in a population-based sample. European Neuropsychopharmacology 2017, 27(8), 732–743. [Google Scholar] [CrossRef]
- Mumtaz, W.; Vuong, P.L.; Xia, L.; Malik, A.S.; Rashid, R.B.A. An EEG-based machine learning method to screen alcohol use disorder. Cognitive Neurodynamics 2017, 11(2), 161–171. [Google Scholar] [CrossRef] [PubMed]
- Substance Abuse and Mental Health Services Administration. (2020). Key substance use and mental health indicators in the United States: Results from the 2019 National Survey on Drug Use and Health. Available online: https://www.samhsa.gov/data/ (accessed on 1 January 2023).
- Jing, Y.; Hu, Z.; Fan, P.; Xue, Y.; Wang, L.; Tarter, R.E. .. & Xie, X.-Q. Analysis of substance use and its outcomes by machine learning I. Childhood evaluation of liability to substance use disorder. Drug and Alcohol Dependence 2020, 206, 107605. [Google Scholar] [PubMed]
- Mannes, Z.L.; Shmulewitz, D.; Livne, O.; Stohl, M.; Hasin, D.S. Correlates of mild, moderate, and severe Alcohol Use Disorder among adults with problem substance use: Validity implications for DSM-5. Alcoholism: Clinical and Experimental Research 2021, 45(10), 2118–2129. [Google Scholar] [CrossRef]
- Mintz, C.M.; Hartz, S.M.; Fisher, S.L.; Ramsey, A.T.; Geng, E.H.; Grucza, R.A.; Bierut, L.J. A cascade of care for alcohol use disorder: Using 2015-2019 National Survey on Drug Use and Health data to identify gaps in past 12-month care. Alcoholism: Clinical and Experimental Research 2021, 45(6), 1276–1286. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Therneau, T.M.; Atkinson, E.J.; Tafti, A.P.; Zhang, N. .. & Liu, H. Unsupervised machine learning for the discovery of latent disease clusters and patient subgroups using electronic health records. Journal of Biomedical Informatics 2020, 102, 103364. [Google Scholar]
- Lopez, C.; Tucker, S.; Salameh, T.; Tucker, C. An unsupervised machine learning method for discovering patient clusters based on genetic signatures. Journal of Biomedical Informatics 2018, 85, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ma, J.; Jiang, J.; Guo, X. Robust feature matching using spatial clustering with heavy outliers. IEEE Transactions on Image Processing 2019, 29, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Birant, D.; Kut, A. ST-DBSCAN: An algorithm for clustering spatial-temporal data. Data & Knowledge Engineering 2007, 60(1), 208–221. [Google Scholar]
- Franzwa, F.; Harper, L.A.; Anderson, K.G. Examination of social smoking classifications using a machine learning approach. Addictive Behaviors 2022, 126, 107175. [Google Scholar] [CrossRef]
- Dell, N.A.; Srivastava, S.P.; Vaughn, M.G.; Salas-Wright, C.; Hai, A.H.; Qian, Z. Binge drinking in early adulthood: A machine learning approach. Addictive Behaviors 2022, 124, 107122. [Google Scholar] [CrossRef]
- Waddell, J.T. Age-varying time trends in cannabis- and alcohol-related risk perceptions 2002-2019. Addictive Behaviors 2022, 124, 107091. [Google Scholar] [CrossRef]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New data preprocessing trends based on ensemble of multiple preprocessing techniques. TrAC Trends in Analytical Chemistry 2020, 132, 116045. [Google Scholar] [CrossRef]
- Khatun, J.; Azad, T.; Seaum, S.Y.; Jabiullah, M.I.; Habib, M.T. (2020, November). Betel nut addiction detection using machine learning. In 2020 4th International Conference on Electronics, Communication and Aerospace Technology (ICECA); IEEE, 2020; pp. 1435–1440. [Google Scholar]
- Bleeker, S.E.; Moll, H.A.; Steyerberg, E.W.; Donders, A.R.T.; Derksen-Lubsen, G.; Grobbee, D.E.; Moons, K.G.M. External validation is necessary in prediction research: A clinical example. Journal of Clinical Epidemiology 2003, 56(9), 826–832. [Google Scholar] [CrossRef]
- Saha, T.D.; Chou, S.P.; Grant, B.F. The performance of DSM-5 alcohol use disorder and quantity-frequency of alcohol consumption criteria: An item response theory analysis. Drug and Alcohol Dependence 2020, 216, 108299. [Google Scholar] [CrossRef]


| Author & Year | Title (First Three Words) | Mental Health Issue | Machine Learning Model Used | Dataset used |
|---|---|---|---|---|
| Srividya et al., 2018 | Behavioral Modeling Mental | State of mental health | Unsupervised learning, Supervised learning, SVM, Naive Bayes, KNN, Logistic Regression | Survey (20 questions), Population 1: 300 subjects, Population 2: 356 subjects |
| Tarekeng et al., 2018 | Cross-Validation Approach | No specific mental health issue analyzed | Unsupervised learning, K-means, K-fold cross-validation, RMSE, MAPE | Chronic diseases, Emotions, Yeast dataset |
| Stevens et al., 2020 | Identification Analysis Behavioral | Autism Spectrum Disorder | Unsupervised learning, Hierarchical clustering | 2400 children from community centers across the U.S. |
| Jing et al., 2020 | Analysis of Substance Use | Substance Use Disorder (SUD) | Supervised learning, Random Forest | Recruited via advertisement, public service announcements, random digital calls, posters; Age: 10-22 years |
| Mannes et al., 2021 | Correlates of Mild, Moderate | Alcohol Use Disorder (AUD) | Supervised learning, Statistical analysis, Multinomial Logistic Regression | 150 participants (suburban inpatient addiction treatment), 438 participants (urban medical center), Age: 18+ years |
| Mintz et al., 2021 | A Cascade of Care for Alcohol | Alcohol Use Disorder (AUD) | Supervised learning, Statistical analysis, Multinomial Logistic Regression | 150 participants (suburban inpatient addiction treatment), 438 participants (urban medical center), Age: 18+ years |
| Hayley et al., 2017 | DSM-5 Cannabis Use Disorder | Cannabis Use Disorder (CaUD) | Supervised learning, Logistic Regression | NESARC-III (n=36,309), Age: 18+ years |
| Algorithm | Dataset | ||
|---|---|---|---|
| Alcohol (%) | Marijuana (%) | Cocaine (%) | |
| K-Means | |||
| Sil_score | 72.5 | 78.7 | 63.9 |
| K-fold | 99.96 | 99.92 | 99.84 |
| RMSE | 4.0 | 0.0 | 6.0 |
| Hierarchical | |||
| Sil_score | 69.2 | 83.4 | 47.5 |
| K-fold | 99.97 | 99.96 | 99.63 |
| RMSE | 6.0 | 0.0 | 35.89 |
| DBSCAN | |||
| Sil_score | 97.7 | 97.6 | 93.1 |
| K-fold | 99.96 | 100.0 | 98.8 |
| RMSE | 0.0 | 0.0 | 15.16 |
| Algorithm | Dataset | ||
|---|---|---|---|
| Alcohol (%) | Marijuana (%) | Cocaine (%) | |
| K-Means++ | |||
| Sil_score | 78.7 | 83.4 | 95.56 |
| K-fold | 99.99 | 99.99 | 100 |
| RMSE | 0.0 | 0.0 | 0.0 |
| BIRCH | |||
| Sil_score | 68.7 | 61.2 | 96.6 |
| K-fold | 99.6 | 99.87 | 98.8 |
| RMSE | 8.9 | 8.0 | 15.12 |
| HDBSCAN | |||
| Sil_score | 38.2 | 23.7 | 37.1 |
| K-fold | 93.7 | 92.3 | 86.4 |
| RMSE | 7.5 | 19.5 | 57.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





