Submitted:
20 May 2025
Posted:
20 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
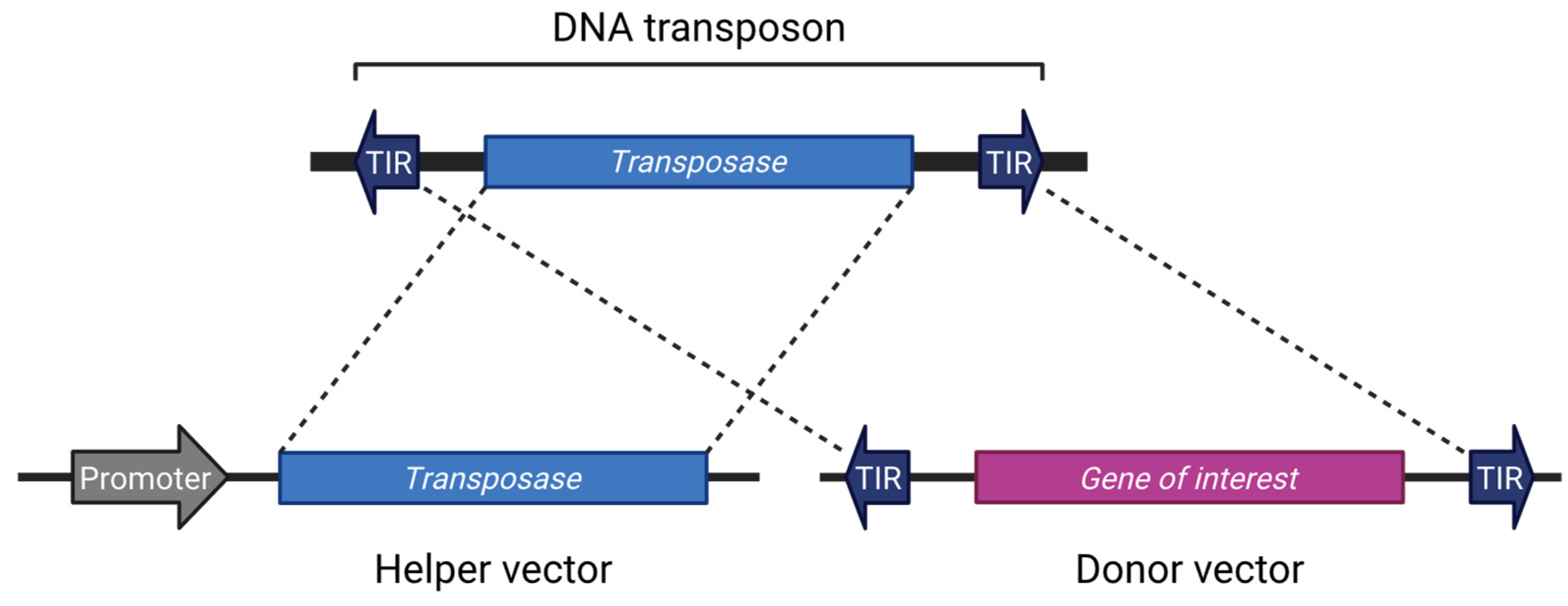
2. Cut-and-Paste DNA Transposons: Structural Overview and Applications in Research
2.1. TIR
2.2. Enzymatic Activity of Transposases Used in Research
2.3. Sequence-Specific Targeting Bias of Transposases
2.4. Discovery and Improvement of Transposase
3. Randomness and Its Value
3.1. Serendipitous Discoveries Brought About by Chance
3.2. From Chance to Strategy: The Research Value of Transposon Randomness
3.3. Historical Development and Impact of Transposon Randomness
3.4. Transition to Mammalian Systems
3.5. Transposon Applications in Vertebrate Species: Advances in Mammalian Mutagenesis
4. Gene Screening Utilizing Randomness: Proof-of-Concept Studies Based on Our Transposon-Mediated Approaches
4.1. Fusion of Genes with Reporter Proteins: Applications in Protein–Protein Interaction Analysis
4.2. Fusion of Promoters/Enhancers with Reporter Genes
4.3. High-Sensitivity Trap Vector-Based Random Screening
4.3.1. Isolation of c-Myc-Responsive Cells
4.3.2. Isolation of Cells Responsive to ER Stresses
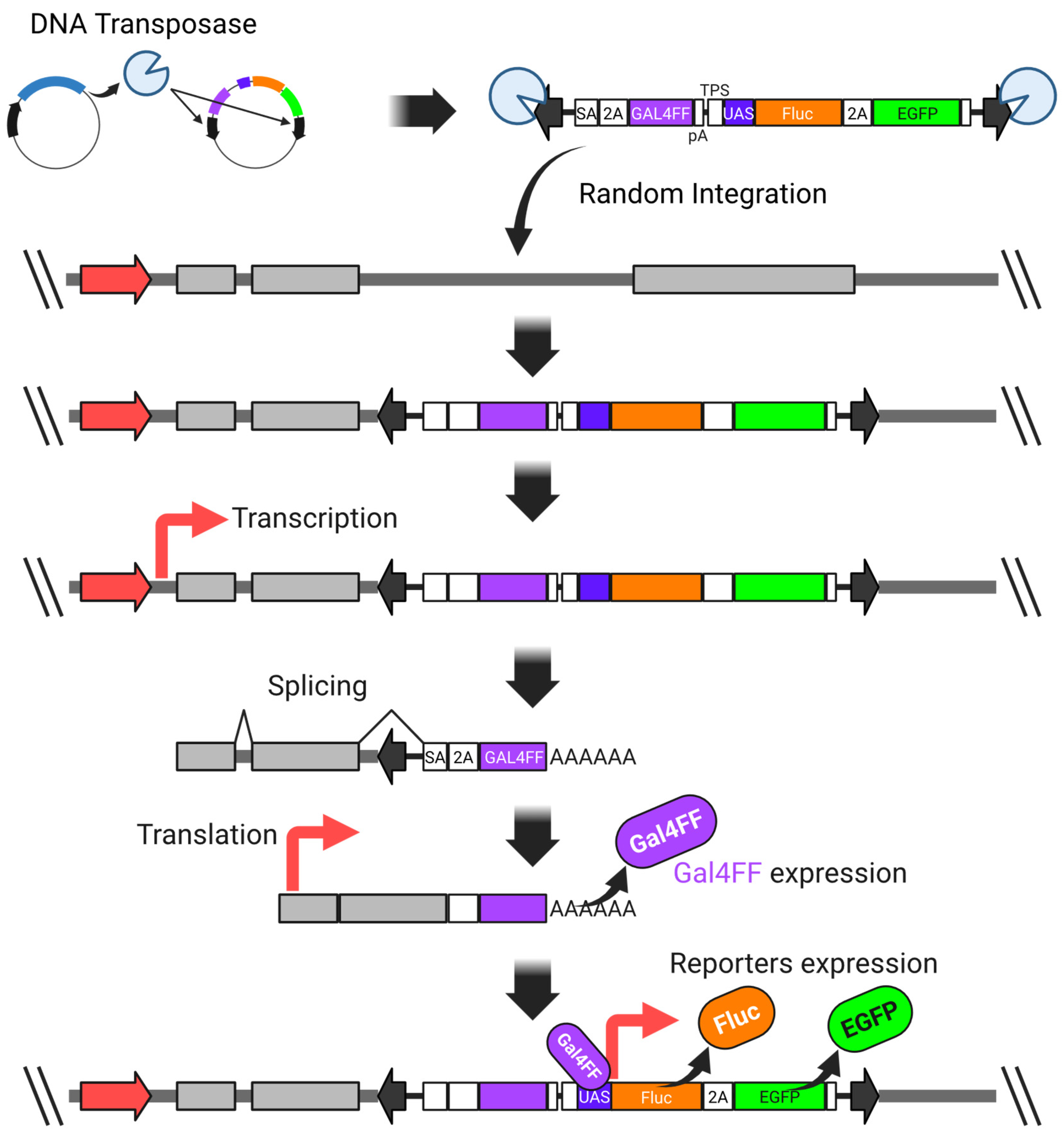
4.3.3. Cross-Pathway Compound Profiling Using Randomly Generated Reporter Cell Lines for Vitamins and Forskolin
4.3.4. Reporter Cells Revealing Hidden Cell Cycle Dynamics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Cas | CRISPR associated protein |
| LEDLGF | Lens epithelium-derived growth factor |
| UTR | untranslated region |
| LTR | long terminal repeat |
| TIR | Terminal inverted repeats |
| TSS | Target site sequence |
| CHO | Chinese hamster ovary |
| Blm | Bloom syndrome homologue |
| ES | embryonic stem |
| BiFC | Bimolecular fluorescence complementation |
| GFP | Green Fluorescent Protein |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| UAS | upstream activating sequence |
| c-Myc | Cellular myelocytomatosis oncogene |
| BiP | Binding immunoglobulin protein |
| OSBPL9 | Oxysterol binding protein-like 9 |
| LINC-PINT | Long intragenic non-coding RNA p53-induced transcript |
| TISPL | transcript induced by stressors from LINC-PINT locus |
| CYP24A1 | Cytochrome P450 family 24 subfamily A member 1 |
| BDKRB2 | Bradykinin receptor B2 |
| TSKU | Tsukushi, Small Leucine Rich Proteoglycan |
| cAMP | Cyclic adenosine monophosphate |
References
- McCLINTOCK, B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A 1950, 36, 344-355. 15430309. [CrossRef]
- Jones, R.N. McClintock’s controlling elements: the full story. Cytogenet Genome Res 2005, 109, 90-103. 15753564. [CrossRef]
- Modzelewski, A.J., Gan, Chong J., Wang, T., He, L. Mammalian genome innovation through transposon domestication. Nat Cell Biol 2022, 24, 1332-1340. 36008480. [CrossRef]
- Sandoval-Villegas, N., Nurieva, W., Amberger, M., Ivics, Z. Contemporary Transposon Tools: A Review and Guide through Mechanisms and Applications of Sleeping Beauty, piggyBac and Tol2 for Genome Engineering. Int J Mol Sci 22: 2021, , . 34064900. [CrossRef]
- Balciunas, D., Wangensteen, K.J., Wilber, A., Bell, J., Geurts, A., Sivasubbu, S., Wang, X., Hackett, P.B., Largaespada, D.A., McIvor, R.S., Ekker, S.C. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet 2006, 2, e169. 17096595. [CrossRef]
- Suster, M.L., Sumiyama, K., Kawakami, K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics 2009, 10, 477. 19832998. [CrossRef]
- Li, M.A., Turner, D.J., Ning, Z., Yusa, K., Liang, Q., Eckert, S., Rad, L., Fitzgerald, T.W., Craig, N.L., Bradley, A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res 2011, 39, e148. 21948799. [CrossRef]
- Rostovskaya, M., Fu, J., Obst, M., Baer, I., Weidlich, S., Wang, H., Smith, A.J., Anastassiadis, K., Stewart, A.F. Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res 2012, 40, e150. 22753106. [CrossRef]
- Turchiano, G., Latella, M.C., Gogol-Döring, A., Cattoglio, C., Mavilio, F., Izsvák, Z., Ivics, Z., Recchia, A. Genomic analysis of Sleeping Beauty transposon integration in human somatic cells. PLoS One 2014, 9, e112712. 25390293. [CrossRef]
- Tagaya, H., Ishikawa, K., Hosokawa, Y., Kobayashi, S., Ueoka, Y., Shimada, M., Ohashi, Y., Mikami, H., Yamamoto, M., Ihara, T., Kumazawa, K., Sugihara, K., Goshima, N., Watanabe, S., Semba, K. A method of producing genetically manipulated mouse mammary gland. Breast Cancer Res 2019, 21, 1. 30611295. [CrossRef]
- Darquet, A.M., Cameron, B., Wils, P., Scherman, D., Crouzet, J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther 1997, 4, 1341-1349. 9472558. [CrossRef]
- Garcia-Garcia, L., Recalde, S., Hernandez, M., Bezunartea, J., Rodriguez-Madoz, J.R., Johnen, S., Diarra, S., Marie, C., Izsvák, Z., Ivics, Z., Scherman, D., Kropp, M., Thumann, G., Prosper, F., Fernandez-Robredo, P., Garcia-Layana, A. Long-Term PEDF Release in Rat Iris and Retinal Epithelial Cells after Sleeping Beauty Transposon-Mediated Gene Delivery. Mol Ther Nucleic Acids 2017, 9, 1-11. 29246287. [CrossRef]
- Hernandez, M., Recalde, S., Garcia-Garcia, L., Bezunartea, J., Miskey, C., Johnen, S., Diarra, S., Sebe, A., Rodriguez-Madoz, J.R., Pouillot, S., Marie, C., Izsvák, Z., Scherman, D., Kropp, M., Prosper, F., Thumann, G., Ivics, Z., Garcia-Layana, A., Fernandez-Robredo, P. Preclinical Evaluation of a Cell-Based Gene Therapy Using the Sleeping Beauty Transposon System in Choroidal Neovascularization. Mol Ther Methods Clin Dev 2019, 15, 403-417. 31890733. [CrossRef]
- Walters, A.A., Kinnear, E., Shattock, R.J., McDonald, J.U., Caproni, L.J., Porter, N., Tregoning, J.S. Comparative analysis of enzymatically produced novel linear DNA constructs with plasmids for use as DNA vaccines. Gene Ther 2014, 21, 645-652. 24830436. [CrossRef]
- Bishop, D.C., Caproni, L., Gowrishankar, K., Legiewicz, M., Karbowniczek, K., Tite, J., Gottlieb, D.J., Micklethwaite, K.P. CAR T Cell Generation by piggyBac Transposition from Linear Doggybone DNA Vectors Requires Transposon DNA-Flanking Regions. Mol Ther Methods Clin Dev 2020, 17, 359-368. 32071928. [CrossRef]
- Lamers, C.H., Willemsen, R., van, Elzakker P., van, Steenbergen-Langeveld S., Broertjes, M., Oosterwijk-Wakka, J., Oosterwijk, E., Sleijfer, S., Debets, R., Gratama, J.W. Immune responses to transgene and retroviral vector in patients treated with ex vivo-engineered T cells. Blood 2011, 117, 72-82. 20889925. [CrossRef]
- Kawakami, K., Shima, A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 1999, 240, 239-244. 10564832. [CrossRef]
- Wilber, A., Frandsen, J.L., Geurts, J.L., Largaespada, D.A., Hackett, P.B., McIvor, R.S. RNA as a source of transposase for Sleeping Beauty-mediated gene insertion and expression in somatic cells and tissues. Mol Ther 2006, 13, 625-630. 16368272. [CrossRef]
- Wilber, A., Wangensteen, K.J., Chen, Y., Zhuo, L., Frandsen, J.L., Bell, J.B., Chen, Z.J., Ekker, S.C., McIvor, R.S., Wang, X. Messenger RNA as a source of transposase for sleeping beauty transposon-mediated correction of hereditary tyrosinemia type I. Mol Ther 2007, 15, 1280-1287. 17440442. [CrossRef]
- Karikó, K., Buckstein, M., Ni, H., Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165-175. 16111635. [CrossRef]
- Kormann, M.S., Hasenpusch, G., Aneja, M.K., Nica, G., Flemmer, A.W., Herber-Jonat, S., Huppmann, M., Mays, L.E., Illenyi, M., Schams, A., Griese, M., Bittmann, I., Handgretinger, R., Hartl, D., Rosenecker, J., Rudolph, C. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 2011, 29, 154-157. 21217696. [CrossRef]
- Galla, M., Schambach, A., Falk, C.S., Maetzig, T., Kuehle, J., Lange, K., Zychlinski, D., Heinz, N., Brugman, M.H., G旦hring, G., Izsv叩k, Z., Ivics, Z., Baum, C. Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res 2011, 39, 7147-7160. 21609958. [CrossRef]
- Holstein, M., Mesa-Nuñez, C., Miskey, C., Almarza, E., Poletti, V., Schmeer, M., Grueso, E., Ordóñez, Flores J.C., Kobelt, D., Walther, W., Aneja, M.K., Geiger, J., Bonig, H.B., Izsvák, Z., Schleef, M., Rudolph, C., Mavilio, F., Bueren, J.A., Guenechea, G., Ivics, Z. Efficient Non-viral Gene Delivery into Human Hematopoietic Stem Cells by Minicircle Sleeping Beauty Transposon Vectors. Mol Ther 2018, 26, 1137-1153. 29503198. [CrossRef]
- Kebriaei, P., Singh, H., Huls, M.H., Figliola, M.J., Bassett, R., Olivares, S., Jena, B., Dawson, M.J., Kumaresan, P.R., Su, S., Maiti, S., Dai, J., Moriarity, B., Forget, M.A., Senyukov, V., Orozco, A., Liu, T., McCarty, J., Jackson, R.N., Moyes, J.S., Rondon, G., Qazilbash, M., Ciurea, S., Alousi, A., Nieto, Y., Rezvani, K., Marin, D., Popat, U., Hosing, C., Shpall, E.J., Kantarjian, H., Keating, M., Wierda, W., Do, K.A., Largaespada, D.A., Lee, D.A., Hackett, P.B., Champlin, R.E., Cooper, L.J. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J Clin Invest 2016, 126, 3363-3376. 27482888. [CrossRef]
- Hudecek, M., Izsvak, Z., Johnen, S., Renner, M., Thumann, G., Ivics, Z. Going non-viral: the Sleeping Beauty transposon system breaks on through to the clinical side. Crit Rev Biochem Mol Biol 1-26 2017, , . 28402189. [CrossRef]
- Hodge, R., Narayanavari, S.A., Izsvák, Z., Ivics, Z. Wide Awake and Ready to Move: 20 Years of Non-Viral Therapeutic Genome Engineering with the Sleeping Beauty Transposon System. Hum Gene Ther 2017, 28, 842-855. 28870121. [CrossRef]
- Tipanee, J., Chai, Y.C., VandenDriessche, T., Chuah, M.K. Preclinical and clinical advances in transposon-based gene therapy. Biosci Rep 37: 2017, , . 29089466. [CrossRef]
- Monjezi, R., Miskey, C., Gogishvili, T., Schleef, M., Schmeer, M., Einsele, H., Ivics, Z., Hudecek, M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 2017, 31, 186-194. 27491640. [CrossRef]
- Morero, N.R., Zuliani, C., Kumar, B., Bebel, A., Okamoto, S., Guynet, C., Hickman, A.B., Chandler, M., Dyda, F., Barabas, O. Targeting IS608 transposon integration to highly specific sequences by structure-based transposon engineering. Nucleic Acids Res 2018, 46, 4152-4163. 29635476. [CrossRef]
- Klompe, S.E., Vo, P.L.H., Halpin-Healy, T.S., Sternberg, S.H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 2019, 571, 219-225. 31189177. [CrossRef]
- Strecker, J., Ladha, A., Gardner, Z., Schmid-Burgk, J.L., Makarova, K.S., Koonin, E.V., Zhang, F. RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019, 365, 48-53. 31171706. [CrossRef]
- Saito, M., Xu, P., Faure, G., Maguire, S., Kannan, S., Altae-Tran, H., Vo, S., Desimone, A., Macrae, R.K., Zhang, F. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 2023, 620, 660-668. 37380027. [CrossRef]
- Liu, P., Panda, K., Edwards, S.A., Swanson, R., Yi, H., Pandesha, P., Hung, Y.H., Klaas, G., Ye, X., Collins, M.V., Renken, K.N., Gilbertson, L.A., Veena, V., Hancock, C.N., Slotkin, R.K. Transposase-assisted target-site integration for efficient plant genome engineering. Nature 2024, 631, 593-600. 38926583. [CrossRef]
- Mikkelsen, J.G., Yant, S.R., Meuse, L., Huang, Z., Xu, H., Kay, M.A. Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther 2003, 8, 654-665. 14529839. [CrossRef]
- Mátés, L., Izsvák, Z., Ivics, Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol 8 Suppl 2007, 1, S1. 18047686. [CrossRef]
- Ivics, Z., Li, M.A., Mates, L., Boeke, J.D., Nagy, A., Bradley, A., Izsvak, Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods 2009, 6, 415-422. 19478801. [CrossRef]
- Muñoz-López, M., García-Pérez, J.L. DNA transposons: nature and applications in genomics. Curr Genomics 2010, 11, 115-128. 20885819. [CrossRef]
- Yusa, K. piggyBac Transposon. Microbiol Spectr 2015, 3, MDNA3-0028-2014. 26104701. [CrossRef]
- Sato, M., Inada, E., Saitoh, I., Watanabe, S., Nakamura, S. piggyBac-Based Non-Viral In Vivo Gene Delivery Useful for Production of Genetically Modified Animals and Organs. Pharmaceutics 2020, 12, 277. 32204422. [CrossRef]
- Zhang, T., Tan, S., Tang, N., Li, Y., Zhang, C., Sun, J., Guo, Y., Gao, H., Cai, Y., Sun, W., Wang, C., Fu, L., Ma, H., Wu, Y., Hu, X., Zhang, X., Gee, P., Yan, W., Zhao, Y., Chen, Q., Guo, B., Wang, H., Zhang, Y.E. Heterologous survey of 130 DNA transposons in human cells highlights their functional divergence and expands the genome engineering toolbox. Cell 2024, 187, 3741-3760.e30. 38843831. [CrossRef]
- Yang, G., Zhang, F., Hancock, C.N., Wessler, S.R. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2007, 104, 10962-10967. 17578919. [CrossRef]
- Hancock, C.N., Zhang, F., Wessler, S.R. Transposition of the Tourist-MITE mPing in yeast: an assay that retains key features of catalysis by the class 2 PIF/Harbinger superfamily. Mob DNA 2010, 1, 5. 20226077. [CrossRef]
- Huang, X., Haley, K., Wong, M., Guo, H., Lu, C., Wilber, A., Zhou, X. Unexpectedly high copy number of random integration but low frequency of persistent expression of the Sleeping Beauty transposase after trans delivery in primary human T cells. Hum Gene Ther 2010, 21, 1577-1590. 20528476. [CrossRef]
- Fraser, M.J., Ciszczon, T., Elick, T., Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol 1996, 5, 141-151. 8673264. [CrossRef]
- Vigdal, T.J., Kaufman, C.D., Izsvak, Z., Voytas, D.F., Ivics, Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol 2002, 323, 441-452. 12381300. [CrossRef]
- Kondrychyn, I., Garcia-Lecea, M., Emelyanov, A., Parinov, S., Korzh, V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics 2009, 10, 418. 19737393. [CrossRef]
- Wilson, M.H., Coates, C.J., George, A.L. Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther 2007, 15, 139-145. 17164785. [CrossRef]
- Galvan, D.L., Nakazawa, Y., Kaja, A., Kettlun, C., Cooper, L.J., Rooney, C.M., Wilson, M.H. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother 2009, 32, 837-844. 19752750. [CrossRef]
- Ikeda, R., Kokubu, C., Yusa, K., Keng, V.W., Horie, K., Takeda, J. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol Cell Biol 2007, 27, 1665-1676. 17178833. [CrossRef]
- Liang, Q., Kong, J., Stalker, J., Bradley, A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 2009, 47, 404-408. 19391106. [CrossRef]
- Huang, X., Guo, H., Tammana, S., Jung, Y.C., Mellgren, E., Bassi, P., Cao, Q., Tu, Z.J., Kim, Y.C., Ekker, S.C., Wu, X., Wang, S.M., Zhou, X. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther 2010, 18, 1803-1813. 20606646. [CrossRef]
- Grabundzija, I., Irgang, M., Mates, L., Belay, E., Matrai, J., Gogol-Doring, A., Kawakami, K., Chen, W., Ruiz, P., Chuah, M.K., VandenDriessche, T., Izsvák, Z., Ivics, Z. Comparative analysis of transposable element vector systems in human cells. Mol Ther 2010, 18, 1200-1209. 20372108. [CrossRef]
- Meir, Y.J., Weirauch, M.T., Yang, H.S., Chung, P.C., Yu, R.K., Wu, S.C. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol 2011, 11, 28. 21447194. [CrossRef]
- Gogol-Doring, A., Ammar, I., Gupta, S., Bunse, M., Miskey, C., Chen, W., Uckert, W., Schulz, T.F., Izsvak, Z., Ivics, Z. Genome-wide Profiling Reveals Remarkable Parallels Between Insertion Site Selection Properties of the MLV Retrovirus and the piggyBac Transposon in Primary Human CD4(+) T Cells. Mol Ther 2016, 24, 592-606. 26755332. [CrossRef]
- Yoshida, J., Akagi, K., Misawa, R., Kokubu, C., Takeda, J., Horie, K. Chromatin states shape insertion profiles of the piggyBac, Tol2 and Sleeping Beauty transposons and murine leukemia virus. Sci Rep 2017, 7, 43613. 28252665. [CrossRef]
- Elling, U., Wimmer, R.A., Leibbrandt, A., Burkard, T., Michlits, G., Leopoldi, A., Micheler, T., Abdeen, D., Zhuk, S., Aspalter, I.M., Handl, C., Liebergesell, J., Hubmann, M., Husa, A.M., Kinzer, M., Schuller, N., Wetzel, E., van, de Loo N., Martinez, J.A.Z., Estoppey, D., Riedl, R., Yang, F., Fu, B., Dechat, T., Ivics, Z., Agu, C.A., Bell, O., Blaas, D., Gerhardt, H., Hoepfner, D., Stark, A., Penninger, J.M. A reversible haploid mouse embryonic stem cell biobank resource for functional genomics. Nature 2017, 550, 114-118. 28953874. [CrossRef]
- Weidhaas, J.B., Angelichio, E.L., Fenner, S., Coffin, J.M. Relationship between retroviral DNA integration and gene expression. J Virol 2000, 74, 8382-8389. 10954538. [CrossRef]
- Schröder, A.R., Shinn, P., Chen, H., Berry, C., Ecker, J.R., Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521-529. 12202041. [CrossRef]
- Wu, X., Li, Y., Crise, B., Burgess, S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003, 300, 1749-1751. 12805549. [CrossRef]
- Narezkina, A., Taganov, K.D., Litwin, S., Stoyanova, R., Hayashi, J., Seeger, C., Skalka, A.M., Katz, R.A. Genome-wide analyses of avian sarcoma virus integration sites. J Virol 2004, 78, 11656-11663. 15479807. [CrossRef]
- Mitchell, R.S., Beitzel, B.F., Schroder, A.R., Shinn, P., Chen, H., Berry, C.C., Ecker, J.R., Bushman, F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2004, 2, E234. 15314653. [CrossRef]
- Maertens, G., Cherepanov, P., Pluymers, W., Busschots, K., De, Clercq E., Debyser, Z., Engelborghs, Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 2003, 278, 33528-33539. 12796494. [CrossRef]
- Yant, S.R., Wu, X., Huang, Y., Garrison, B., Burgess, S.M., Kay, M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol 2005, 25, 2085-2094. 15743807. [CrossRef]
- Derse, D., Crise, B., Li, Y., Princler, G., Lum, N., Stewart, C., McGrath, C.F., Hughes, S.H., Munroe, D.J., Wu, X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J Virol 2007, 81, 6731-6741. 17409138. [CrossRef]
- Walisko, O., Schorn, A., Rolfs, F., Devaraj, A., Miskey, C., Izsvák, Z., Ivics, Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther 2008, 16, 359-369. 18071335. [CrossRef]
- Wei, M., Mi, C.L., Jing, C.Q., Wang, T.Y. Progress of Transposon Vector System for Production of Recombinant Therapeutic Proteins in Mammalian Cells. Front Bioeng Biotechnol 2022, 10, 879222. 35600890. [CrossRef]
- Matasci, M., Bachmann, V., Baldi, L., Hacker, D.L., De, Jesus M., Wurm, F.M. CHO cell lines generated by PiggyBac transposition. BMC Proc 5 Suppl 2011, 8, P31. 22373414. [CrossRef]
- Balasubramanian, S., Matasci, M., Kadlecova, Z., Baldi, L., Hacker, D.L., Wurm, F.M. Rapid recombinant protein production from piggyBac transposon-mediated stable CHO cell pools. J Biotechnol 2015, 200, 61-69. 25758242. [CrossRef]
- Balasubramanian, S., Rajendra, Y., Baldi, L., Hacker, D.L., Wurm, F.M. Comparison of three transposons for the generation of highly productive recombinant CHO cell pools and cell lines. Biotechnol Bioeng 2016, 113, 1234-1243. 26616356. [CrossRef]
- Yusa, K., Zhou, L., Li, M.A., Bradley, A., Craig, N.L. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A 2011, 108, 1531-1536. 21205896. [CrossRef]
- Burnight, E.R., Staber, J.M., Korsakov, P., Li, X., Brett, B.T., Scheetz, T.E., Craig, N.L., McCray, P.B. Jr A Hyperactive Transposase Promotes Persistent Gene Transfer of a piggyBac DNA Transposon. Mol Ther Nucleic Acids 2012, 1, e50. 23344650. [CrossRef]
- Wen, W., Song, S., Han, Y., Chen, H., Liu, X., Qian, Q. An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications. Int J Mol Sci 2020, 21, 3064. 32357554. [CrossRef]
- Ivics, Z., Hackett, P.B., Plasterk, R.H., Izsvak, Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 1997, 91, 501-510. 9390559. [CrossRef]
- Izsvák, Z., Ivics, Z., Plasterk, R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol 2000, 302, 93-102. 10964563. [CrossRef]
- Geurts, A.M., Yang, Y., Clark, K.J., Liu, G., Cui, Z., Dupuy, A.J., Bell, J.B., Largaespada, D.A., Hackett, P.B. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 2003, 8, 108-117. 12842434. [CrossRef]
- Zayed, H., Izsvak, Z., Walisko, O., Ivics, Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther 2004, 9, 292-304. 14759813. [CrossRef]
- Mates, L., Chuah, M.K., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., Grzela, D.P., Schmitt, A., Becker, K., Matrai, J., Ma, L., Samara-Kuko, E., Gysemans, C., Pryputniewicz, D., Miskey, C., Fletcher, B., VandenDriessche, T., Ivics, Z., Izsvak, Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 2009, 41, 753-761. 19412179. [CrossRef]
- Voigt, F., Wiedemann, L., Zuliani, C., Querques, I., Sebe, A., Mátés, L., Izsvák, Z., Ivics, Z., Barabas, O. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun 2016, 7, 11126. 27025571. [CrossRef]
- Querques, I., Mades, A., Zuliani, C., Miskey, C., Alb, M., Grueso, E., Machwirth, M., Rausch, T., Einsele, H., Ivics, Z., Hudecek, M., Barabas, O. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat Biotechnol 2019, 37, 1502-1512. 31685959. [CrossRef]
- Wang, S., Gao, B., Miskey, C., Guan, Z., Sang, Y., Chen, C., Wang, X., Ivics, Z., Song, C. Passer, a highly active transposon from a fish genome, as a potential new robust genetic manipulation tool. Nucleic Acids Res 2023, 51, 1843-1858. 36688327. [CrossRef]
- Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y., Xu, T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 2005, 122, 473-483. 16096065. [CrossRef]
- Wang, W., Lin, C., Lu, D., Ning, Z., Cox, T., Melvin, D., Wang, X., Bradley, A., Liu, P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A 2008, 105, 9290-9295. 18579772. [CrossRef]
- O’Kane, C.J., Gehring, W.J. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A 1987, 84, 9123-9127. 2827169. [CrossRef]
- Bier, E., Vaessin, H., Shepherd, S., Lee, K., McCall, K., Barbel, S., Ackerman, L., Carretto, R., Uemura, T., Grell, E., et, al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev 1989, 3, 1273-1287. 2558049. [CrossRef]
- Bellen, H.J., O’Kane, C.J., Wilson, C., Grossniklaus, U., Pearson, R.K., Gehring, W.J. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev 1989, 3, 1288-1300. 2558050. [CrossRef]
- Wilson, C., Pearson, R.K., Bellen, H.J., O’Kane, C.J., Grossniklaus, U., Gehring, W.J. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev 1989, 3, 1301-1313. 2558051. [CrossRef]
- Osborne, B.I., Baker, B. Movers and shakers: maize transposons as tools for analyzing other plant genomes. Curr Opin Cell Biol 1995, 7, 406-413. 7662372. [CrossRef]
- Fraser, M.J., Smith, G.E., Summers, M.D. Acquisition of Host Cell DNA Sequences by Baculoviruses: Relationship Between Host DNA Insertions and FP Mutants of Autographa californica and Galleria mellonella Nuclear Polyhedrosis Viruses. J Virol 1983, 47, 287-300. 16789244. [CrossRef]
- Cary, L.C., Goebel, M., Corsaro, B.G., Wang, H.G., Rosen, E., Fraser, M.J. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 1989, 172, 156-169. 2549707. [CrossRef]
- Shinmyo, Y., Mito, T., Matsushita, T., Sarashina, I., Miyawaki, K., Ohuchi, H., Noji, S. piggyBac-mediated somatic transformation of the two-spotted cricket, Gryllus bimaculatus. Dev Growth Differ 2004, 46, 343-349. 15367202. [CrossRef]
- Bonin, C.P., Mann, R.S. A piggyBac transposon gene trap for the analysis of gene expression and function in Drosophila. Genetics 2004, 167, 1801-1811. 15342518. [CrossRef]
- Luo, G., Ivics, Z., Izsv叩k, Z., Bradley, A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A 1998, 95, 10769-10773. 9724779. [CrossRef]
- Koga, A., Suzuki, M., Inagaki, H., Bessho, Y., Hori, H. Transposable element in fish. Nature 1996, 383, 30. 8779712. [CrossRef]
- Horn, C., Offen, N., Nystedt, S., Hacker, U., Wimmer, E.A. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 2003, 163, 647-661. 12618403. [CrossRef]
- Dupuy, A.J., Fritz, S., Largaespada, D.A. Transposition and gene disruption in the male germline of the mouse. Genesis 2001, 30, 82-88. 11416868. [CrossRef]
- Fischer, S.E., Wienholds, E., Plasterk, R.H. Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci U S A 2001, 98, 6759-6764. 11381141. [CrossRef]
- Horie, K., Kuroiwa, A., Ikawa, M., Okabe, M., Kondoh, G., Matsuda, Y., Takeda, J. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc Natl Acad Sci U S A 2001, 98, 9191-9196. 11481482. [CrossRef]
- Carlson, C.M., Largaespada, D.A. Insertional mutagenesis in mice: new perspectives and tools. Nat Rev Genet 2005, 6, 568-580. 15995698. [CrossRef]
- Collier, L.S., Largaespada, D.A. Hopping around the tumor genome: transposons for cancer gene discovery. Cancer Res 2005, 65, 9607-9610. 16266976. [CrossRef]
- Cadiñanos, J., Bradley, A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res 2007, 35, e87. 17576687. [CrossRef]
- Collier, L.S., Adams, D.J., Hackett, C.S., Bendzick, L.E., Akagi, K., Davies, M.N., Diers, M.D., Rodriguez, F.J., Bender, A.M., Tieu, C., Matise, I., Dupuy, A.J., Copeland, N.G., Jenkins, N.A., Hodgson, J.G., Weiss, W.A., Jenkins, R.B., Largaespada, D.A. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res 2009, 69, 8429-8437. 19843846. [CrossRef]
- Copeland, N.G., Jenkins, N.A. Harnessing transposons for cancer gene discovery. Nat Rev Cancer 2010, 10, 696-706. 20844553. [CrossRef]
- Dupuy, A.J., Akagi, K., Largaespada, D.A., Copeland, N.G., Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 2005, 436, 221-226. 16015321. [CrossRef]
- Collier, L.S., Carlson, C.M., Ravimohan, S., Dupuy, A.J., Largaespada, D.A. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 2005, 436, 272-276. 16015333. [CrossRef]
- Keng, V.W., Villanueva, A., Chiang, D.Y., Dupuy, A.J., Ryan, B.J., Matise, I., Silverstein, K.A., Sarver, A., Starr, T.K., Akagi, K., Tessarollo, L., Collier, L.S., Powers, S., Lowe, S.W., Jenkins, N.A., Copeland, N.G., Llovet, J.M., Largaespada, D.A. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol 2009, 27, 264-274. 19234449. [CrossRef]
- Dupuy, A.J., Rogers, L.M., Kim, J., Nannapaneni, K., Starr, T.K., Liu, P., Largaespada, D.A., Scheetz, T.E., Jenkins, N.A., Copeland, N.G. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009, 69, 8150-8156. 19808965. [CrossRef]
- Rad, R., Rad, L., Wang, W., Cadinanos, J., Vassiliou, G., Rice, S., Campos, L.S., Yusa, K., Banerjee, R., Li, M.A., de, la Rosa J., Strong, A., Lu, D., Ellis, P., Conte, N., Yang, F.T., Liu, P., Bradley, A. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 2010, 330, 1104-1107. 20947725. [CrossRef]
- Starr, T.K., Allaei, R., Silverstein, K.A., Staggs, R.A., Sarver, A.L., Bergemann, T.L., Gupta, M., O’Sullivan, M.G., Matise, I., Dupuy, A.J., Collier, L.S., Powers, S., Oberg, A.L., Asmann, Y.W., Thibodeau, S.N., Tessarollo, L., Copeland, N.G., Jenkins, N.A., Cormier, R.T., Largaespada, D.A. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009, 323, 1747-1750. 19251594. [CrossRef]
- Takeda, H., Wei, Z., Koso, H., Rust, A.G., Yew, C.C., Mann, M.B., Ward, J.M., Adams, D.J., Copeland, N.G., Jenkins, N.A. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet 2015, 47, 142-150. 25559195. [CrossRef]
- Wang, W., Bradley, A., Huang, Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res 2009, 19, 667-673. 19233961. [CrossRef]
- Koso, H., Takeda, H., Yew, C.C., Ward, J.M., Nariai, N., Ueno, K., Nagasaki, M., Watanabe, S., Rust, A.G., Adams, D.J., Copeland, N.G., Jenkins, N.A. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci U S A 2012, 109, E2998-3007. 23045694. [CrossRef]
- Miyakura, H., Fukuda, M., Enomoto, H., Ishikawa, K., Watanabe, S., Semba, K. A screening system for identifying interacting proteins using biomolecular fluorescence complementation and transposon gene trap. PLoS One 2021, 16, e0251240. 33989302. [CrossRef]
- Ishikawa, K., Kobayashi, Y., Wakabayashi, Y., Watanabe, S., Semba, K. A highly sensitive trap vector system for isolating reporter cells and identification of responsive genes. Biol Methods Protoc 2018, 3, bpy003. 32161797. [CrossRef]
- Ishikawa, K., Tamamura, S., Semba, K., Watanabe, S. Establishment of reporter cells that respond to glucocorticoids by a transposon-mediated promoter-trapping system. Eur J Pharm Sci 2021, 162, 105819. 33775826. [CrossRef]
- Medico, E., Gambarotta, G., Gentile, A., Comoglio, P.M., Soriano, P. A gene trap vector system for identifying transcriptionally responsive genes. Nat Biotechnol 2001, 19, 579-582. 11385465. [CrossRef]
- Tanaka, T.S., Davey, R.E., Lan, Q., Zandstra, P.W., Stanford, W.L. Development of a gene-trap vector with a highly sensitive fluorescent protein reporter system for expression profiling. Genesis 2008, 46, 347-356. 18615730. [CrossRef]
- Asakawa, K., Suster, M.L., Mizusawa, K., Nagayoshi, S., Kotani, T., Urasaki, A., Kishimoto, Y., Hibi, M., Kawakami, K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A 2008, 105, 1255-1260. 18202183. [CrossRef]
- Asakawa, K., Kawakami, K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ 2008, 50, 391-399. 18482403. [CrossRef]
- Ogura, E., Okuda, Y., Kondoh, H., Kamachi, Y. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn 2009, 238, 641-655. 19191223. [CrossRef]
- Liu, Z., Okano, A., Sanada, E., Futamura, Y., Nogawa, T., Ishikawa, K., Semba, K., Li, J., Li, X., Osada, H., Watanabe, N. Identification of microbial metabolites that accelerate the ubiquitin-dependent degradation of c-Myc. Oncol Res 2023, 31, 655-666. 37547761. [CrossRef]
- Liu, Z., Ishikawa, K., Sanada, E., Semba, K., Li, J., Li, X., Osada, H., Watanabe, N. Identification of antimycin A as a c-Myc degradation accelerator via high-throughput screening. J Biol Chem 2023, 299, 105083. 37495110. [CrossRef]
- Wakabayashi, Y., Shimono, A., Terauchi, Y., Zeng, C., Hamada, M., Semba, K., Watanabe, S., Ishikawa, K. Identification of a novel RNA transcript TISPL upregulated by stressors that stimulate ATF4. Gene 2024, 917, 148464. 38615981. [CrossRef]
- Ishikawa, K., Tamamura, S., Takahashi, N., Takagi, M., Semba, K., Watanabe, S. Isolation of Reporter Cells That Respond to Vitamin A and/or D Using a piggyBac Transposon Promoter-Trapping Vector System. Int J Mol Sci 2022, 23, 9366. 36012634. [CrossRef]
- Kasahara, Y., Tamamura, S., Hiyama, G., Takagi, M., Nakamichi, K., Doi, Y., Semba, K., Watanabe, S., Ishikawa, K. Tyrosine Kinase Inhibitor Profiling Using Multiple Forskolin-Responsive Reporter Cells. Int J Mol Sci 2023, 24, 13863. 37762164. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



