1. Introduction
Vesicular trafficking disorders, which ensure that proteins and other molecules produced within the cell are delivered to the appropriate target, and genetic defects detected in autophagy processes that cause lysosomal degradation of damaged proteins, organelles, cellular material or macromolecules are among the new and most remarkable metabolic/neurometabolic disease groups. Although the pathophysiology of diseases caused by the interactions of these two pathways, which are very important for cellular homeostasis, has been revealed in various diseases, the elucidation of their role in the pathophysiology of newly defined neurometabolic diseases are one of the most recent research topics [
1,
2].
The process of modification in the endoplasmic reticulum (ER), the delivery of molecules such as proteins and lipids to other compartments and the plasma membrane, or the removal of various molecules from the plasma membrane and their subsequent transport to intracellular target compartments, is controlled by vesicular traffic. However, the return of membrane proteins from the Golgi apparatus to the ER and the return of various receptors to the Golgi or plasma membrane for use again (retrograde pathway) are also mediated by vesicular traffic. [
3,
4]. The Golgi apparatus is key to the structure of eukaryotic cells and is made of flattened, perforated membrane stacks, called cisternae. It is divided into three sections: cis, medial, and trans, where parallel cisternae are arranged in a stack. The cis-Golgi network (CGN), situated closest to the endoplasmic reticulum (ER), is responsible for receiving ER-derived transport vesicles and returning ER-resident proteins to the ER. [
5,
6]. The cisterna at the extreme position is continuous with a tubular, branching and reticulate compartment called the trans-Golgi (TGN). The TGN is involved in the final stage of sorting, packaging and delivery of most secretory proteins to their targets. Furthermore, cargo from early and late endosomes is retrogradely targeted to the TGN. When the endosome-mediated transport vesicle approaches the TGN, the transporter-Golgi interaction is mediated by tethering proteins, one of which is the Golgi-associated retrograde proteins (GARP) complex [
7,
8]. The GARP complex (a heterotetrameric binding complex) is composed of four subunits, namely VPS51, VPS52, VPS53, and VPS54. Defects in the GARP complex have been demonstrated to result in defects in both retrograde and anterograde trafficking. Studies in the “wobbler” mouse, considered a model for ALS, have shown that mutations in the gene encoding the VPS54 subunit lead to a severe reduction in VPS54 levels and disrupt GARP complex assembly, thus causing spinal muscle atrophy. More recently, however, mutation in another GARP complex subunit, VPS51, have been associated with a neurodevelopmental disorder that causes lysosomal enlargement and impaired CI-M6PR distribution, demonstrating the importance of this complex in cellular processes [
9].
Lysosomal hydrolases are similarly glycosylated in the endoplasmic reticulum and transported to the Golgi, where they are prepared for transduction by the addition of mannose-6-phosphate (M6P) in the trans-Golgi network (TGN). Here, it is recognized by M6P receptors (M6PR), packaged into the vesicle and delivered to the lysosome [
10,
11,
12]. Clathrin and COP coat proteins, GEF proteins, ARF, Sar1, SNAREs, VPS proteins, RAB proteins, microtubular structures, actin binding proteins, molecular motor proteins and cell membrane lipid regulators, which are necessary for the main stages of vesicular content (cargo) delivery, are encoded by more than a hundred genes and participate in the transport system [
13,
14] In the transport system in which all these proteins participate in the process, membrane-membrane interaction complexes are also involved in the delivery of lysosomal cargo through endosomes [
15]. In this transduction pathway, membran fusion complexes promote the attachment of endosome-lysosome and autophagosome-lysosome membranes mediated by Rab7. In this way, it also ensures the delivery of vesicle contents to the lysosome in the autophagic pathway [
16,
17,
18]. The GARP complex mediates membrane fusion here of recycling endosomes that retrograd transport M6P receptors to the trans-Golgi network for use again, which complete the transport of hydrolases to the lysosome [
19,
20].
In this article, we present two siblings with a novel homozygous variant in VPS51 gene (c.1511C>T; p.Thr504Met), exhibiting developmental delay, thin corpus callosum, severe ID, epilepsy, microcephaly, hearing loss and dysphagia. In VPS51 gene, a novel homozygous missense pathogenic variant c.1511C>T; p.Thr504Met is identified in two affectedsiblings by whole exome sequencing (WES) and confirmed by Sanger DNA sequencing.
Here, we aimed to determine the effects of the novel VPS51 gene mutation at the RNA and protein level in fibroblast cells derived from patients. It was also, investigated the target protein(s) associated with vesicular traffic, which has not been previously elucidated by comparative proteome analysis, the link between autophagic flow and protein defects that provide intermembrane binding and fusion involved in vesicular traffic.
Our study leads to a better understanding of the molecular pathways leading to disease pathophysiology in vesicular traffic disorders, at least in part, serving as a prerequisite for the development of putative therapies. We believe that this study confirms the key role of the GARP and EARP complexes in normal autophagic flux, vesicular traffic, organelle-organelle interaction and cellular homeostasis.
2. Results
DNA sequencing of the VPS51 gene in all family members was shown results consistent with an autosomal recessive pattern of inheritance. As a result of DNA segregation analyses of novel missense variant NM_013265.4: c.1511C>T; p.Thr504Met in VPS51 gene family members revealed that the affected siblings were homozygous, parents and healthy sibling were heterozygous carriers for this nucleotide change.
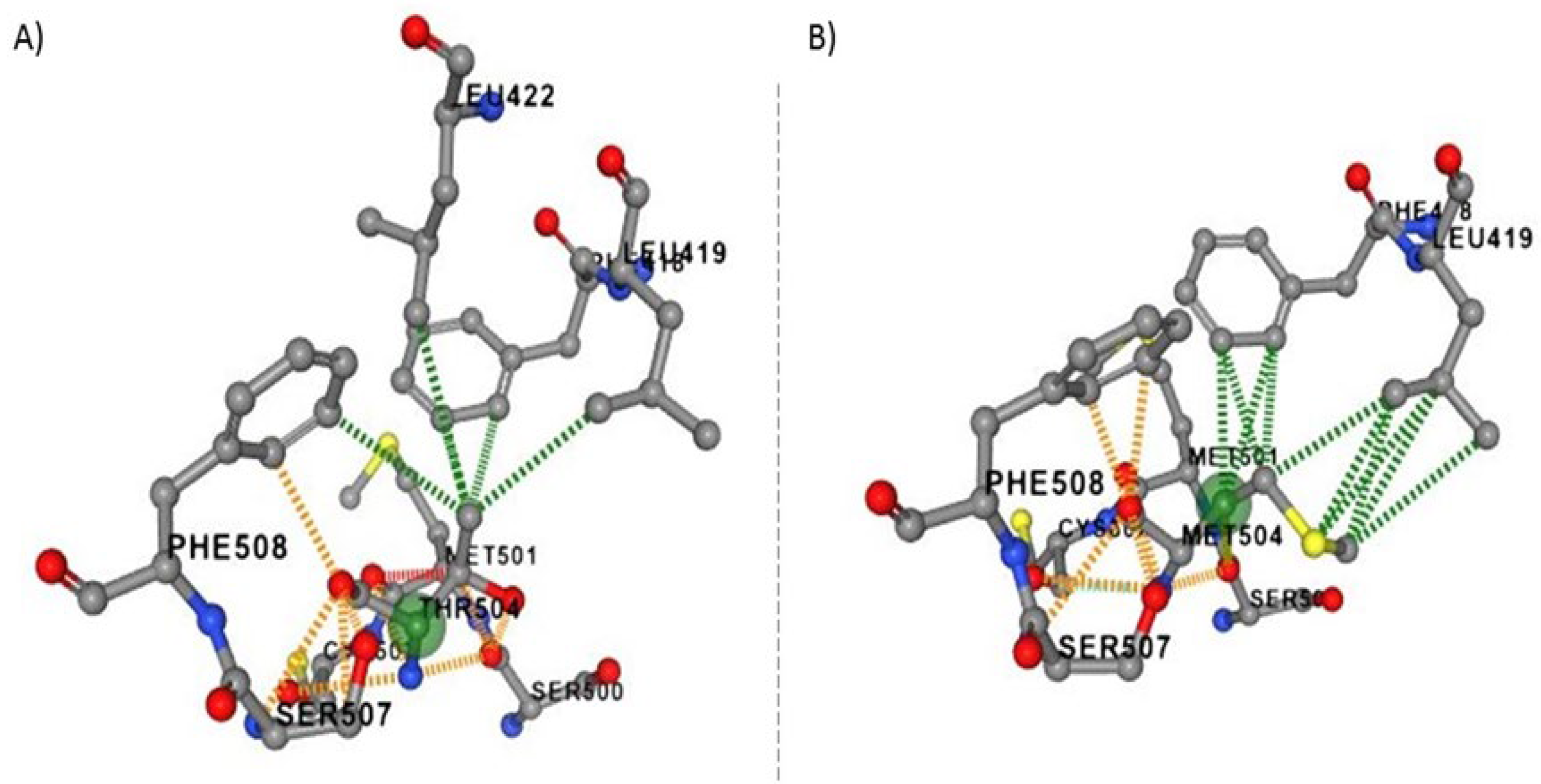
To predict the effect of p.Thr504Met the novel variant on VPS51 protein stability and dynamics, the wild-type and mutant-type predicted structural model generated by the DynaMut2 software using Alphafold database (structure: AF-Q9UID3-F1) as the template structure is shown in (
Figure 1A-B). Based on in silico analysis, the mutant protein is predicted to change the conformation and affect protein stabilization compared to the wild-type protein. The ΔΔG value of the c.1511C>T;p.Thr504Met variant (reported in this study) was calculated as -0,26 kcal/mol (destabilizing) (
Table 1). According to the ACMG guidelines, novel homozygous missense variant (NM_013265.4: c.1511C>T) in the VPS51 gene was classified as variant of uncertain significance (VUS). The potential pathogenic effects of this variant was predicted by in silico analysis program MutationTaster showed that the variant is assigned as disease causing pathogenic variation. The CADD analysis for this variant (CADD score:25.6) and SIFT4G analysis (SIFT4G score :0.01) suggest that it has damaging effect (
Table 1).
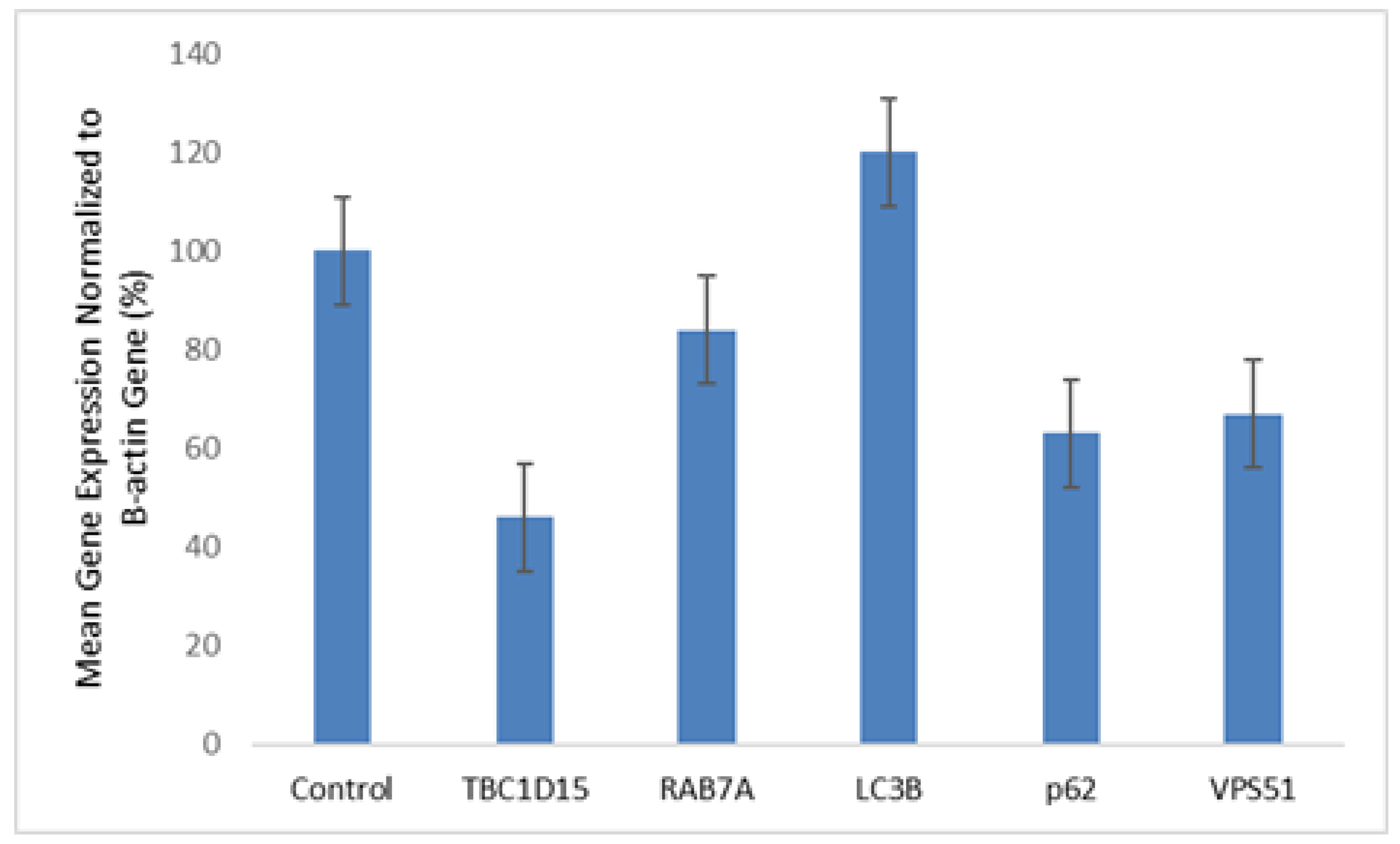
We investigated the effect of homozygous missense novel variant c.1511C>T;p.Thr504Met in VPS51 gene at the RNA/protein level and the relationship with autophagy. mRNA expression levels of disease-associated gene (VPS51), autophagosome-lysosome fusion related genes (RAB7A, TBC1D15) and autophagy pathway-related genes (LC3B, p62 )were detected in this patient. Quantitative RT-PCR analysis have shown that mRNA expression level of VPS51 (67%) was decreased in the patients compared to control group (100%). Increased mRNA expressions of LC3B (169%) and decreased mRNA expressions of p62 (63%), RAB7A(84%), TBC1D15 (46%) were observed compared to control group (100%) (
Figure 2).
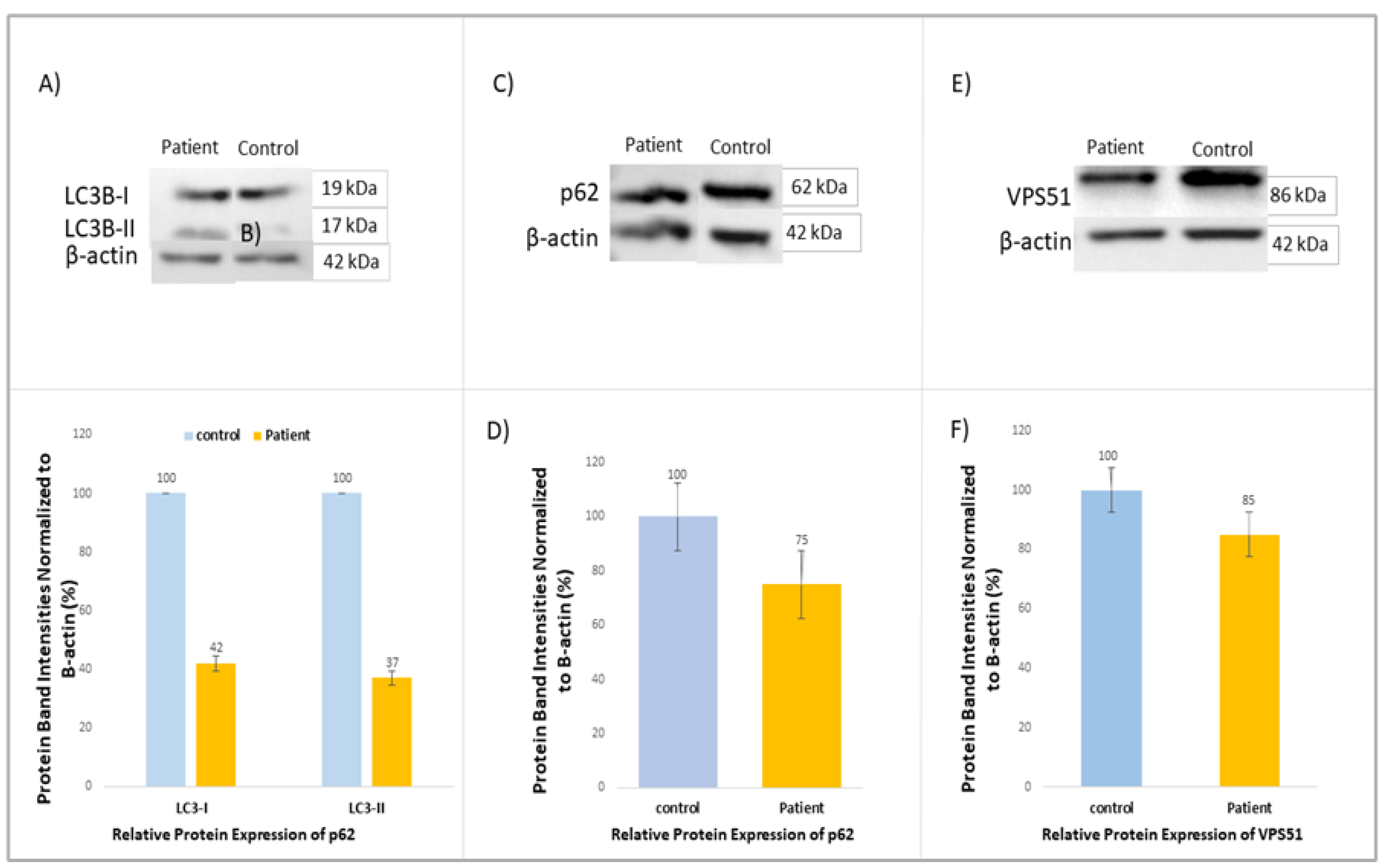
Western blot analysis revealed decreased protein expressions for LC3B, p62 (SQSTM1), and VPS51 in patients compared to the control (
Figure 3A-E).
2.1. Proteomic Profiling
The 1561 proteins identified in the three fractions were quantified using label-free quantification (LFQ) and converted into relative expression data in fibroblast cells from the patient and healthy individual (amplified from skin biopsy), resulting in expression profiles for all of the proteins. From the data, those identified by potential contamination, reverse effects and proteins identified only at a single site were removed and the analysis continued with 1498 proteins. LFQ intensities were filtered by Log2(x) transformation according to the valid value (minimum percentage of valid value is 70%) and those outside the normal distribution were removed. For the remaining 848 proteins, categorical annotation was performed by adding the UniProt Homo sapiens (UP000005640) annotation file. Statistically significant expression changes were determined by ANOVA analysis of variance (p<0.05), and then for the remaining 585 protein expressions, Posthoc Tukey’s test (FDR<0.05) was used to determine which sample was responsible for the expression change. For a total of 585 proteins, fold change between samples was determined and pathway analysis was performed with FC>1.5 (upregulated) and FC<-1.5 (downregulated) (
Table S1 and S2). Of the 322 proteins, 132 were upregulated and 190 were downregulated. These proteins were grouped according to whether they were upregulated or downregulated and pathway analysis was performed [
34,
35].
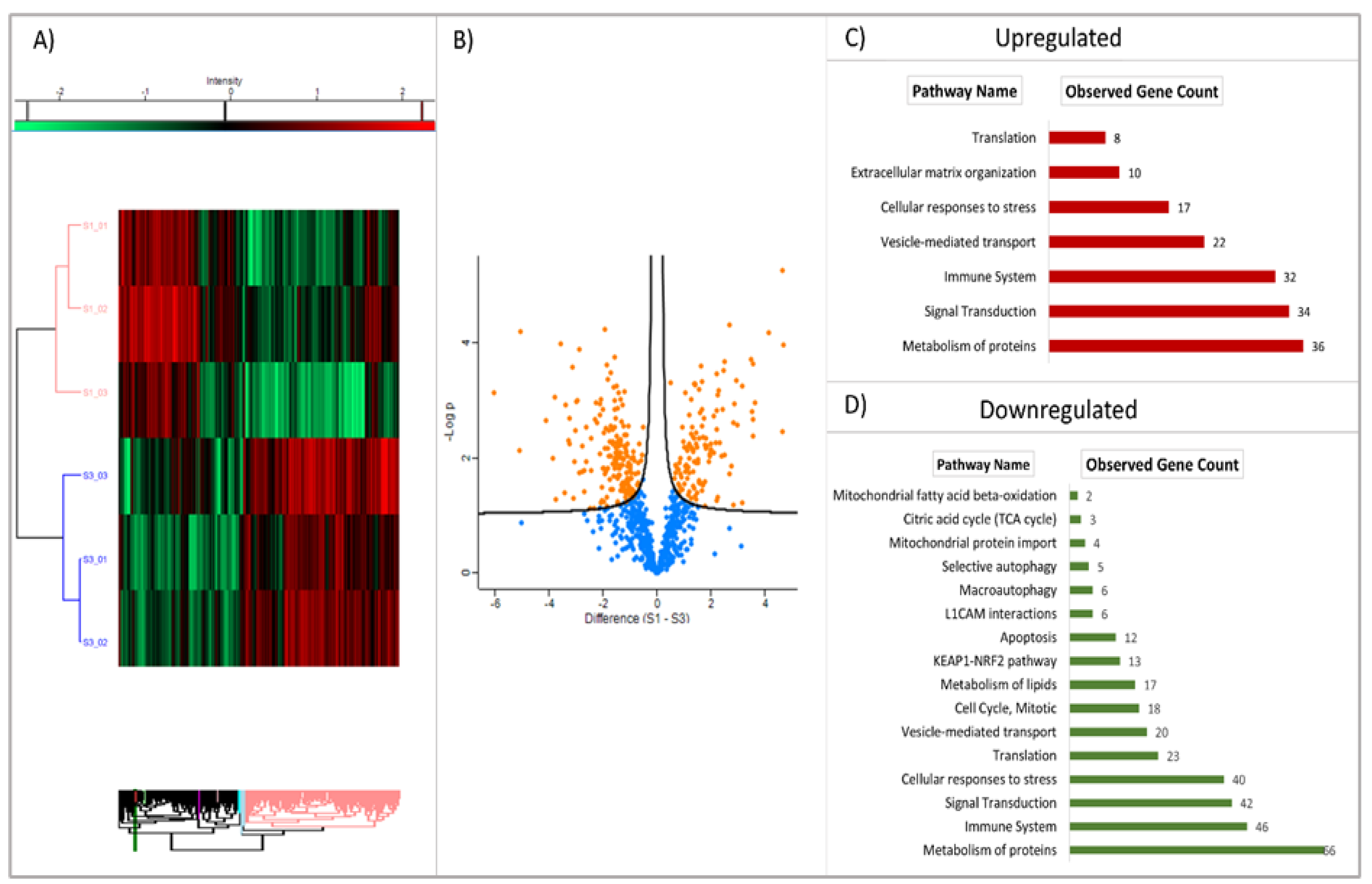
The heatmap, derived from the proteomic analysis (see
Figure 4A), exhibits a clear distinction between the patient and control cohorts. The volcano plot (see
Figure 4B) further elucidates the statistical significance of protein alterations (p<0.05). In both graphs, the separation is evident, with the Patient (S1) and Healthy Control (S3) group data distinctly separated. The dendrogram, which is a hierarchical cluster analysis, further demonstrates that the expression profiles of these two groups are different. In the patient group, some proteins demonstrated markedly increased expression (red), while others exhibited decreased expression (green). The volcano plot employs the Y-axis (-log10 p-value) to indicate statistical significance [
36], providing a quantitative metric. Positively-valued points (in orange) indicate proteins that are elevated in the patient group, whilst negatively-valued points (in blue) indicate proteins that are decreased in the patient group.
Proteins involved in vesicular traffic (22), protein metabolism (36), cellular response to stress (27), signal transduction (34) and immune system (32) pathways were found to be upregulated (
Figure 4C), while proteins associated with protein metabolism (66), cellular stress response (40), KEAP1-NRF2 pathway (33), translation (23), L1CAM interactions (6), autophagy (total 11), apoptosis (12), energy metabolism and mitotic activity pathways (total 27) were found to be downregulated (
Figure 4D). When the molecular functions of upregulated proteins are examined, it is seen that they are mostly grouped in protein binding, cell adhesion molecule binding and cytoskeletal protein binding (
Table S2). Downregulated proteins are mostly grouped in protein binding, catalytic activity, ion binding, hydrolase activity, enzyme binding, cell adhesion molecule binding and GTPase activity (
Table S3).
In order to comprehend the molecular adaptations that are induced by the VPS51 mutation, a proteomic analysis was conducted with the objective of identifying differentially expressed proteins.The results of the analysis revealed that there were significant changes in several key metabolic pathways, including those relating to energy metabolism, lipid metabolism, and oxidative stress responses. Increased gene expression and pathway activations were observed, with proteins involved in glycolysis and lipid synthesis showing notable upregulation, suggesting a shift in the mechanisms of cellular energy production. Among the proteins that were found to be expressed at elevated levels were ACLY (ATP Citrate Lyase), which facilitates the conversion of citrate to acetyl-CoA, thus providing precursors for fatty acid synthesis; FASN (Fatty Acid Synthase), which was found to be critically elevated, indicating enhanced lipogenesis in response to the VPS51 mutation; and PGK1 (Phosphoglycerate Kinase 1), a key glycolytic enzyme that is expressed at elevated levels to support ATP production under altered metabolic conditions. Finally, ATP5F1D (ATP Synthase Subunit D) has been identified as a key player in this process, with its upregulation suggesting a compensatory response to disrupted oxidative phosphorylation. Collectively, these findings underscore a metabolic reprogramming towards increased glycolysis and lipid synthesis, which may be linked to cellular energy demands and stress adaptation [
37].
Conversely, a number of key enzymes implicated in mitochondrial energy metabolism and β-oxidation demonstrated a decline in expression, suggesting a decrease in reliance on oxidative phosphorylation and lipid degradation. ACADM (Acyl-CoA Dehydrogenase Medium Chain) and DECR1 (2,4-Dienoyl-CoA Reductase 1) are particularly noteworthy, as their reduced expression indicates suppressed mitochondrial β-oxidation. Downregulation of IDH1 affects NADPH production, potentially impairing antioxidant defences and lipid metabolism. Reduced levels of CS (citrate synthase) and ACO2 (aconitase 2) suggest reduced flux through the TCA cycle. These changes imply a metabolic shift away from mitochondrial energy metabolism, which may contribute to the lipid accumulation observed in VPS51 mutant cells. Taken together, these findings suggest a dual metabolic adaptation - increased glycolysis and lipogenesis to support cellular energy requirements under stress. Reduced mitochondrial β-oxidation and TCA cycle activity, likely contributing to energy conservation and reduction of reactive oxygen species (ROS).
Supplementary tables illustrates the pathways affected by these changes, while
Figure 4C and
Figure 4D summarises the key up- and down-regulated proteins and their roles in cellular metabolism.
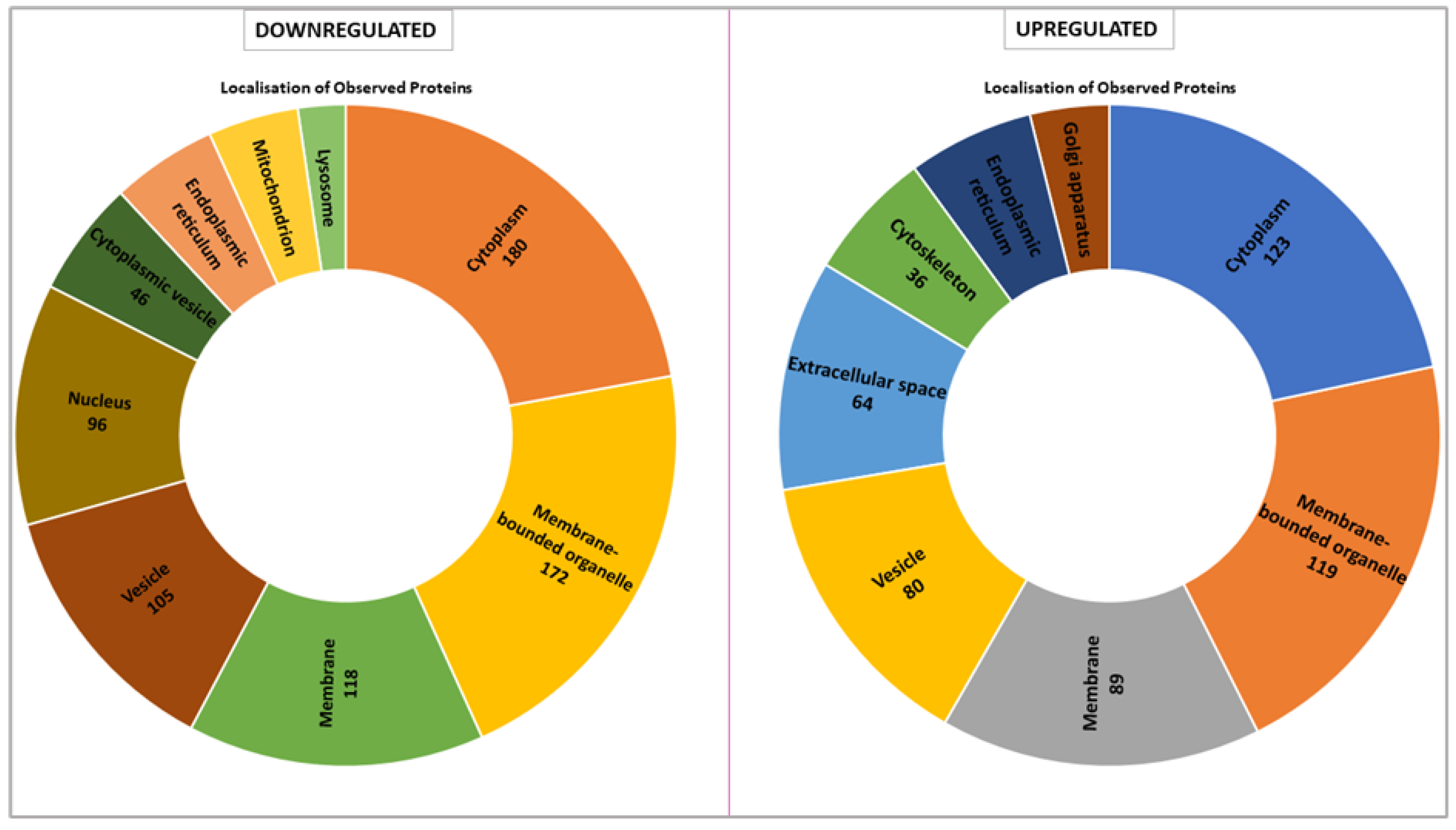
The proteomic analysis reveals distinct localization patterns of upregulated and downregulated proteins in VPS51-mutant cells, with the majority of downregulated proteins being localized to the cytoplasm (180 proteins) and membrane-bounded organelles (172 proteins). Other major localizations include membrane-associated proteins (118 proteins), vesicle-related proteins (105 proteins), and nuclear proteins (96 proteins). In contrast, fewer proteins are localized to the cytoplasmic vesicle, endoplasmic reticulum, mitochondria, and lysosome compartments. Conversely, the majority of up-regulated proteins are predominantly localized to the cytoplasm (123 proteins) and membrane-bounded organelles (119 proteins). Notable increases are observed in membrane-associated proteins (89 proteins), vesicle-associated proteins (80 proteins), and proteins localised to the extracellular space (64 proteins). Proteins associated with the Golgi apparatus and cytoskeleton are also enriched, but represent smaller proportions (
Figure 5).
When we evaluated the intracellular location of proteins with expression differences between control and patient by proteomic analysis, we found that they were mostly membrane-bound organelles. Collectively and mostly they were grouped in the cell membrane, vesicle, cytoplasm and endoplasmic reticulum (
Figure 5). In contrast, upregulated proteins were found in the Golgi, extracellular matrix and cytoskeleton. The proteins localized in the Golgi were mostly proteins that stimulate the formation of Golgi stacks and ribbons and are involved in the transport and localization of molecules in both anterograde and retrograde pathways [
38,
39]. Upregulation of proteins involved in both structural and functional processes of the Golgi suggests a Golgi stress response. Similarly, some of the proteins localized in the extracellular matrix are involved in the cellular stress response (molecular functions have been shown in
Table S4). Lysosome and mitochondria highlighted in downregulated proteins (
Figure 5). In terms of molecular function, many proteins with lysosomal hydrolase activity are downregulated (
Table S3), indicating that the lysosome cannot fulfill its normal function. However, the downregulation of proteins involved in mitochondrial pathways is not surprising considering the crosstalk between mitochondria and lysosomes. It is known that the function of these two organelles is coordinated and loss of function in one leads to secondary impairment in the other [
40]. Our proteomic data support that the defect in lysosomal hydrolase delivery caused by the VPS51 mutation disrupts the mitochondria-lysosome crosstalk, including the contact site. Taken together, increased RAB7A levels and decreased p62 and LC3 levels suggest proteasomal dysfunction and aggrephagy [
41,
42].
2.2. VPS51 Mutation Affects M-L Contact Sites
We performed mitochondrial and lysosomal colabeling and staining in patient and control fibroblasts using Lysotracker Red DND-99 and Mitotracker Green FM. After the staining of the target organelles, live cell images were captured using a Zeiss LSM980 confocal microscope at ×63 objectives (
Figure 6A-B). Twenty cell images from each group were included in the analysis and analyzed separately for mitochondria and lysosomes by using the Fiji-ImageJ program. Then, the distance and the number of surface connections between the labeled organelles in each cell were measured in terms of size and number. Statistical significance of patient and control data was tested by Student’s t-test and Mann-Whitney U test according to the distribution patterns.
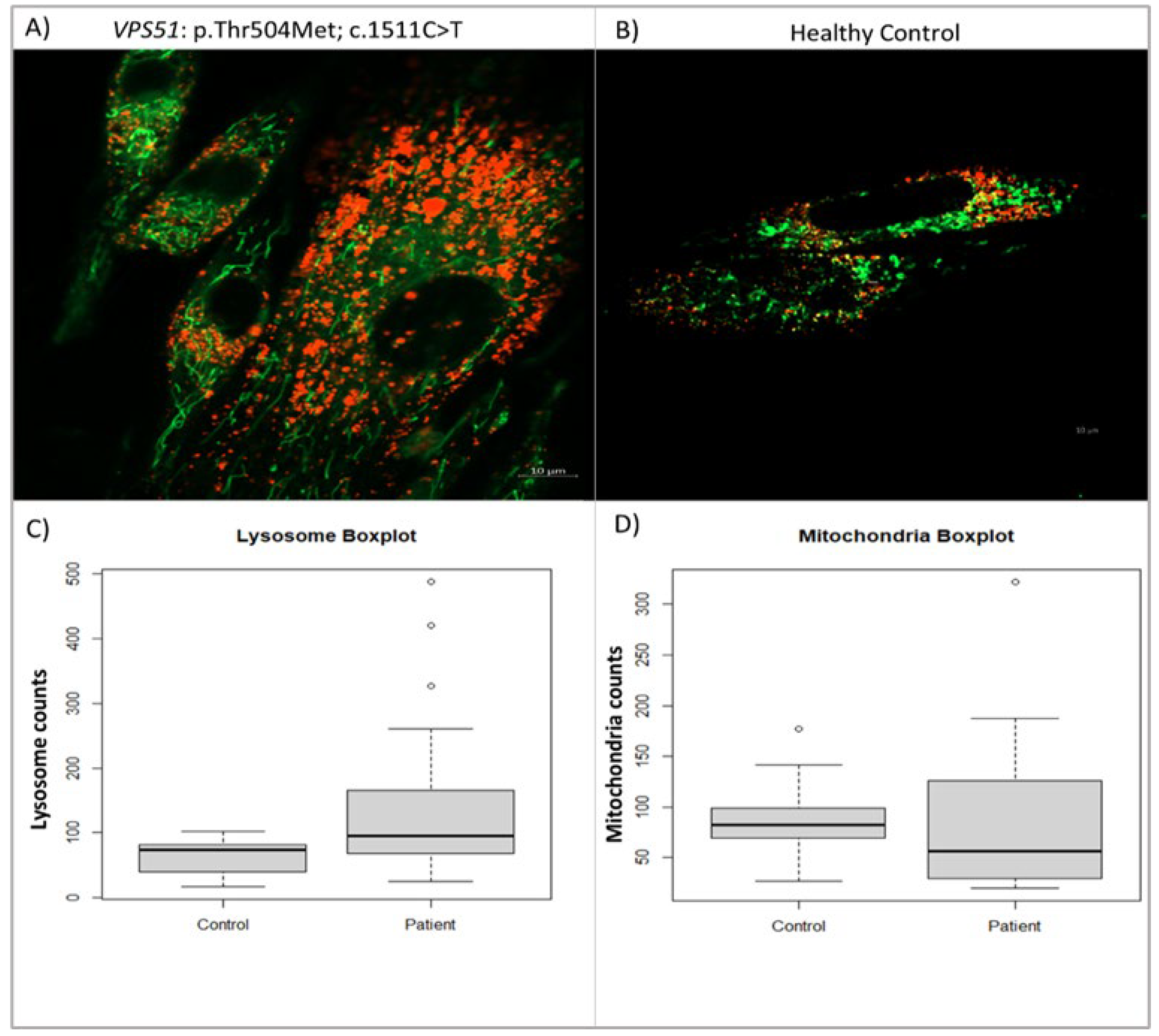
In this study comparing lysosomes in patient and control fibroblasts, it was found that there was a significant increase in the number of lysosomes in patient cells. The study measured the number of lysosomes and mitochondria separately in both groups and used the Shapiro-Wilk test to determine the normal distribution of the results. The difference between patients and controls was determined using a Student T-test, which revealed a 136.9% increase in lysosome numbers in patient cells compared to the control group (
Figure 6C). Additionally, Cohen’s d test confirmed that there was a large difference in lysosome data between patients and controls. Overall, this study suggests that lysosome numbers are significantly increased in patients with the VPS51 mutation (n= 20, p-value: 0.0066).
We also compared the number of mitochondria in patient cells with the VPS51 mutation to control cells and found that there was no significant increase in the number of mitochondria in patient cells. The significance of the difference was determined using Student’s t-test, which showed that the slight difference in the number of mitochondria in the patient cells compared to the control was not significant (
Figure 6D).
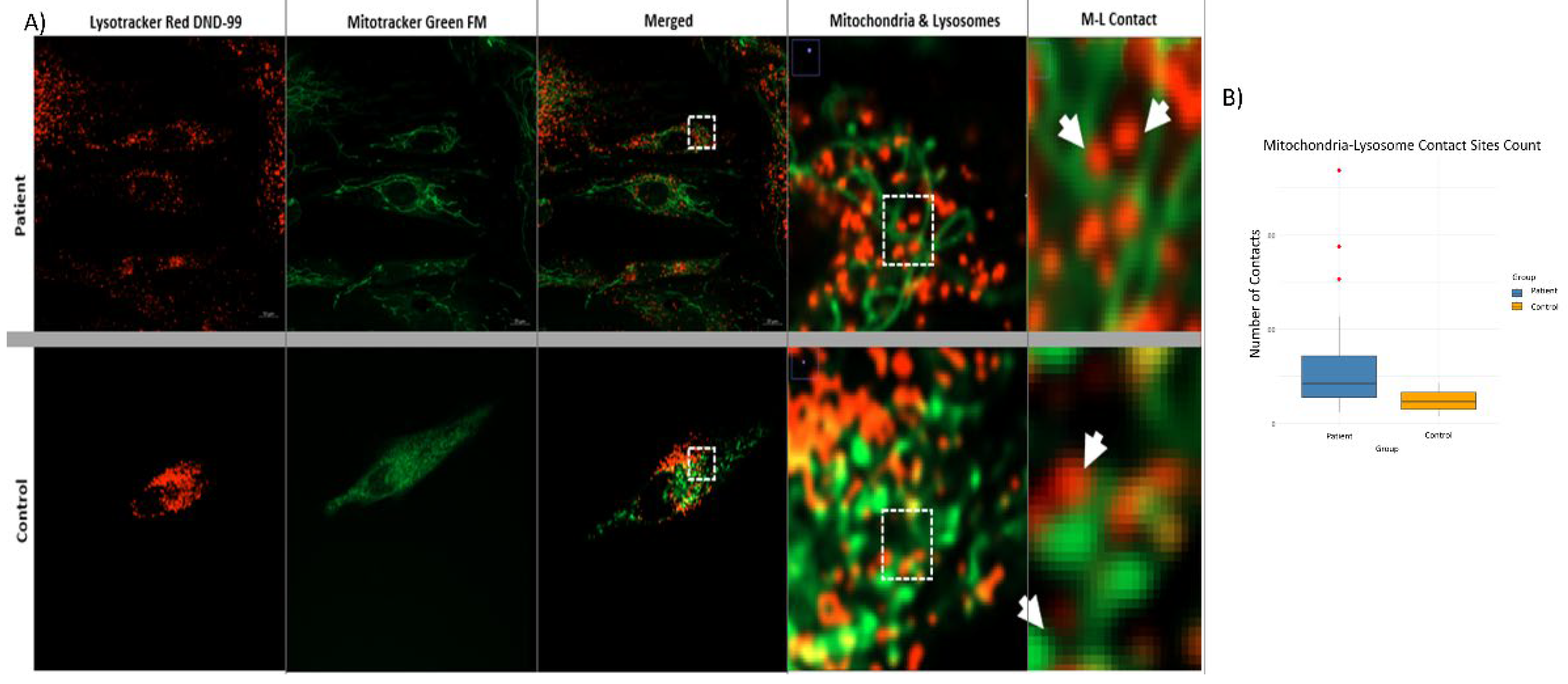
Confocal microscope images obtained from patient and control fibrobasts (n = 20) were examined to investigate the potential impact of mutations in the membrane tethering complex subunits on the mitochondria-lysosome contact sites in the cell (see
Figure 7A). The DiAna plugin of the Fiji-ImageJ program was utilized to determine the organelles in contact between the mitochondria and lysosomes within the cells, with a maximum distance of 10 nm. It is possible that there may be differences in size, number of organelles, or structural differences between cells. Therefore, the contact numbers obtained were normalized to the number of mitochondria and lysosomes in each cell. This approach was adopted to mitigate the impact of these variations on the cellular level. Statistical significance tests were conducted, similar to the approach employed for mitochondria and lysosome numbers. The results of the Shapiro-Wilk test suggested that the data of both groups might be following a normal distribution. Therefore, it was decided to use Student’s t-test to determine whether there was a difference between the number of contacts. The analysis revealed that the mean of the control group was notably lower than the mean of the patient group, and the normalized number of M-L contacts in the patient group increased by approximately 72.8% compared to the control group (
Figure 7B). This variation was found to be statistically significant and not merely random (p= 1.4807 × 10
-8).
3. Discussion
Disorders associated with vesicular trafficking and genetic defects in autophagy processes are among the new and most remarkable groups of neurometabolic diseases, and therefore elucidation of their pathophysiology is of great importance.
Recently, Gershlick et al. revealed compound heterozygous mutations (c.1468C>T/p.Asp745Thrfs*93 and c.2232delC/p.Arg490Cys) in the gene encoding the common GARP/EARP subunit VPS51 in a 6-year-old patient with severe global developmental delay, microcephaly, hypotonia, epilepsy, cortical visual impairment, pontocerebellar abnormalities, failure to thrive, liver dysfunction, lower extremity edema and dysmorphic features were identified by exome sequencing. This study showed that the frameshift mutation caused by the c.2232delC mutation produces a longer but highly unstable protein, whereas the c.1468C>T mutation produces a stable protein that associates less efficiently with other GARP/EARP subunits. The levels of fully assembled GARP and EARP complexes were reduced in skin fibroblasts from the patient. The distribution of the cation-independent mannose 6 phosphate receptor was altered and some of the patient’s fibroblasts exhibited swelling in lysosomes. With these findings, a new genetic locus for a neurodevelopmental disorder has been identified, emphasizing the critical importance of GARP/EARP function in cellular and organismal physiology [
9].
Similarly, whole exome sequencing in two siblings with brain malformations consisting of cerebellar atrophy in the oldest sibling and hypoplastic corpus callosum in the younger sibling affected with delayed psychomotor development, speech deficit, severe intellectual disability and postnatal microcephaly, revealed a homozygous intragenic deletion in VPS51 (c.1419_1421del; p.Phe474del), but no further functional studies were performed [
43].
The novel homozygous missense mutation (c.1511C>T; p.Thr504Met) in the VPS51 gene detected by exome sequencing in siblings with microcephaly, thin corpus callosum, hearing loss, developmental delay, epilepsy and swallowing disorder reported in this article supports the pathogenic effect of VPS51 variants previously reported in the literature by Gershlick et al. and Uwineza et al. In this study, both mRNA expression and VPS51 protein expression were found to decrease as a result of examining the effect of the novel mutation detected in the VPS51 gene at the RNA/protein level using patient fibroblast cells. VPS51 protein expression were decreased in patients compared to the control, as previously shown in the literature [
9].
Since the GARP complex contributes to pre-autophagosomal structure and the lysosome is the final stop of autophagy, we also studied key autophagy markers known as p62 (SQSTM1) and LC3B. p62 mRNA expression levels was decreased, while LC3B mRNA expression was increased in the patients. We also investigated mRNA expression level for RAB7, which mediates membrane fusion, and found that total RAB7A mRNA expression levels were not highly reduced. However, there was a halving of TBC1D15 mRNA expression level, which mediates membrane separation by hydrolyzing RAB7-GTP at the mitochondria-lysosome membrane contact [
44].
In contrast to the increased levels of p62 and LC3 protein levels in the previous study in Saccharomyces cerevisiae by Perez-Victoria et al. our study showed a decrease in p62 and LC3 protein levels in patient with VPS51 gene defect encoding the GARP complex subunit [
45]. Although p62 mRNA expression level decreased, LC3 mRNA expression level increased and autophagy seemed to be increased, the expression level of both proteins decreased. The decrease in the total protein levels of LC3 and p62 in GARP-deficient cells may be due to the induction but not progression of the formation of autophagic structures. Proteomic analysis also showed that p62 (SQSTM1) protein expression was decreased in the patient sample compared to the control. To the best of our knowledge, this is the first study in which autophagic markers were examined in patients fibroblasts carrying VPS51 gene mutation.
Furthermore, this study aimed to identify target protein(s) and associated pathways associated with vesicular traffic, which have not been previously elucidated by comparative proteome analysis.
Proteomic analysis results revealed at least a 1.5-fold difference in vesicular traffic-related proteins between patients and control samples; LRP1, TUBA4A, LMAN1, TMED2, RAB10, SEC23A, RAB1B, TUBB6, HSP90AA1, AP2A1, ACTR1A, SURF4, OPTN, TXNDC5, YWHAQ, TFRC, ARF1, RAB31, RAB18, MYO1C were downregulated and GOLGA5, COL1A1, DCTN3, CAPZA1, ARCN1, USO1, RAB7A, PRKAG1, SPTBN1, CAPZA2, CTTN, PICALM, SEC31A, PAFAH1B1, MCFD2, AGFG1, ARPC4, GDI1, CAPZB, PAFAH1B3, AP2S1, ACTB were upregulated. Most of these proteins were vesicle coat protein, membrane protein and RAB GTPase proteins. In addition, proteins detected in pathways related to mitochondrial function such as TCA cycle, pentose phosphate pathway, fatty acid beta oxidation and pyruvate metabolism were found to be decreased in patients compared to control. In a study conducted in Parkinson’s patients linked to GBA1 (β-glucocerebrosidase), a lysosomal hydrolase, it was reported that there was no significant change in RAB7 total protein levels but TBC1D15 (RAB7-GAP) levels changed. However, further studies have shown that the RAB7-GTP/total RAB7 ratio increased. In our study, no information was obtained about TBC1D15 levels at the protein level, but RAB7 protein levels were found to increase [
46]. The fact that TBC1D15 did not show a significant change in proteomics, despite the increase in Rab7 and membrane contact sites, may be related to the fact that the function of this protein is provided by activity modulation or local dynamics rather than a quantitative change. In addition, the cell may have favoured alternative mechanisms to regulate the increased activities of Rab7. Further studies on the specific post-translational modifications and localisation of TBC1D15 may help to better understand these results. However, in the light of the present data, it appears that lysosomal and mitochondrial activity are impaired and also affects intermembrane junctions.
These changes reflect a coordinated cellular response to the VPS51 mutation, with metabolic reprogramming in favour of survival over efficiency. The observed lipid accumulation may result from suppressed β-oxidation, while increased glycolysis and FASN activity may help to counteract energy deficits [
37]. In support of this finding, changes in protein localisation suggest that cellular compartments are reorganised in response to the VPS51 mutation. Down-regulation of lysosomal, mitochondrial and vesicle-associated proteins may indicate impaired trafficking and organelle dynamics. In contrast, up-regulation of proteins associated with the extracellular space, membrane and Golgi apparatus may reflect compensatory mechanisms to maintain cellular homeostasis under stress conditions.
Decreased p62 induces activation of the autophagy pathway through the Nrf2/Keap1/p62 pathway.According to the results of proteome analysis, it is observed that the KEAP1/NRF2 pathway is downregulated in the patient. The decrease in the levels of proteins involved in this pathway and p62 protein expression suggests activation of the Nrf2/Keap1/p62 pathway [
47]. Decreased antioxidant proteins (CAT, PRDX2, TXN) may indicate that cells are vulnerable to oxidative stress. Both the reduction of p62 and the reduction of proteins in the NRF2/KEAP1 pathway lead to a more severe weakening of cellular defence mechanisms and cause autophagy mechanisms to collapse. This results in susceptibility of the cell to toxic accumulation, inability to control oxidative stress and increased mitochondrial dysfunction [
48,
49]. The data obtained as a result of proteomic analysis in fibroblast cells should be interpreted taking into account the minimum sample size. However, we believe that this study, which addresses many different perspectives and is the first of its kind in the EARP/GARP complex, is necessary and valuable for both research, clinical and relevant patient communities.
In our study, intracellular vesicular traffic, immune system, signal transduction, L1CAM interaction and lysosomal activity pathways were found to be affected, similar to a previous study conducted with mucolipidosis, a lysosomal storage disease [
50].
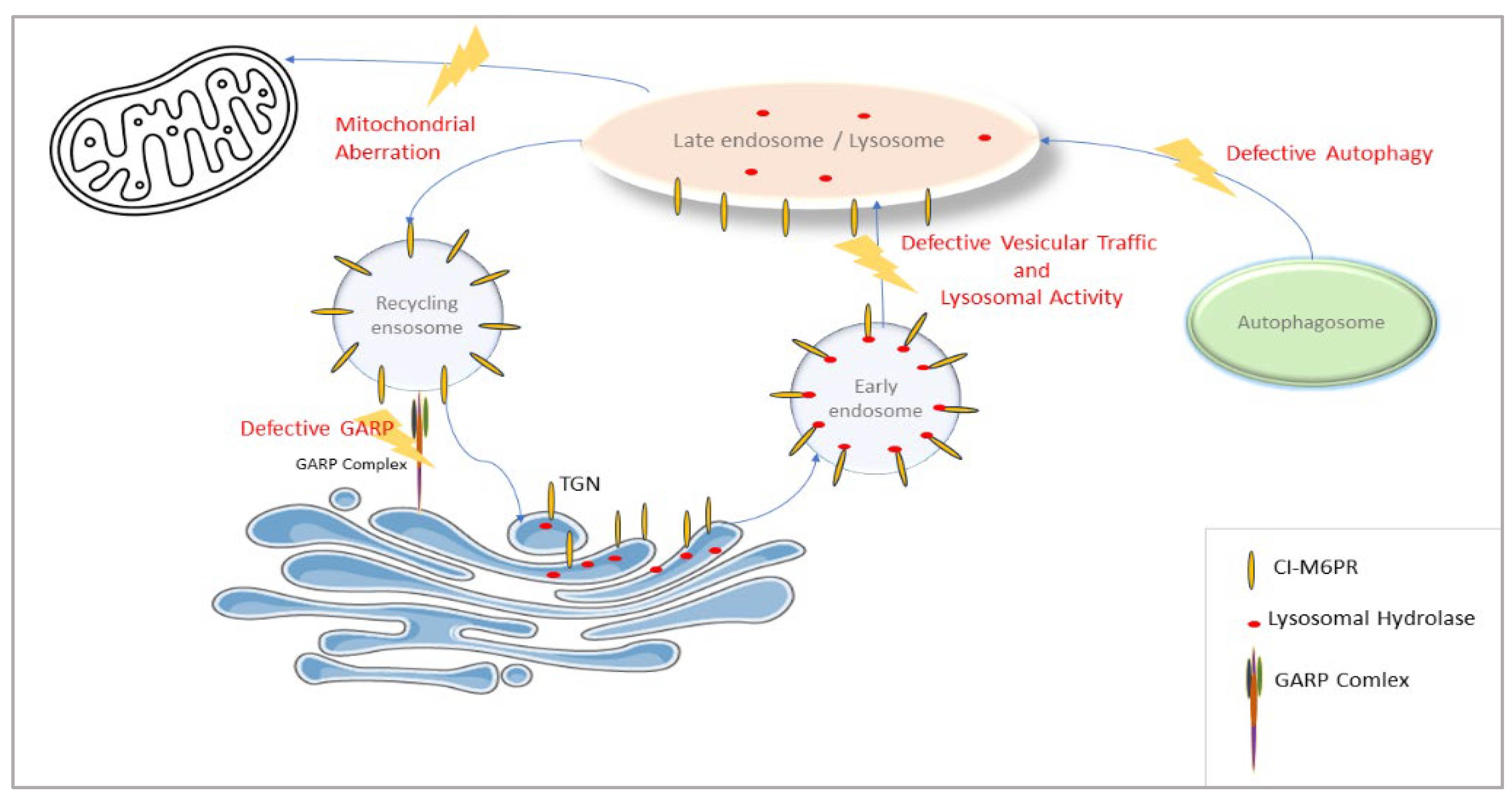
VPS51 is involved in the regulation of a wide range of lysosomal processes, as it encodes the GARP complex subunit for the retrieval of endosomes carrying receptors that transport lysosomal hydrolases back to trans-Golgi and the EARP complex subunit for the return of transferrin receptors from the cell membrane. The effect of reducing the activity of this gene may be more far-reaching than that seen, for example, in a specific lysosomal storage disease. Thus, the VPS51 defect leads to more far-reaching alterations in lysosomal function (supported in this study by changes in the levels of proteins in numerous lysosomal and vesicular pathways along with defective autophagy) (
Figure 8).
The findings indicate that the VPS51 mutation enhances communication between mitochondria and lysosomes, yet concomitantly leads to defects in the retromer complex and endosomal recycling mechanisms. Further functional studies are required to elucidate the mechanisms of lysosomal adaptation and mitophagy. The study of vesicular traffic and autophagy genes revealed that vesicular transmission is impaired by the VPS51 mutation, prompting increased vesicular traffic to regulate the accumulation of proteins and reduce cell stress. Among these genes, VPS29, SNX6 and RAB7A have been identified as particularly significant. The VPS29 is a component of the retromer complex, which is involved in the process of retrograde transport to new organelles, especially in endosomes. RAB7A is a Rab GTPase protein that plays an important role in endosomal transport pathways and regulates vesicular transport and fusion processes to direct the retromer complex to the correct location. SNX6 (Sorting Nexin 6) is a sorting nexin protein bound to the retromer complex and is involved in cargo selection.The combined function of Retromer, Nexin and RAB7A ensures the proper functioning of transduction between endosomes and lysosomes in the cell and the return of receptors to the Golgi [
51,
52]. Studies detailing the intracellular roles of Rab7A and VPS29 will provide a more comprehensive understanding of the mechanistic effects of the VPS51 mutation. Although further studies are being continued since this is the first article in this context with membrane tethering protein, the data obtained are mostly consistent with the studies conducted in the literature with lysosomal storage and lysosomal storage-like diseases [
50,
53]. In addition, findings that can also be evaluated as Golgipathies were obtained (Golgi-stress response) [
54].
In the context of energy metabolism, the proteins involved are of particular significance, including the enzymes that are interconnected in pathways such as the TCA cycle, glycolysis and lipogenesis (e.g., ACLY, FASN, PGK1, IDH1 and ACO2). Further studies on these enzymes may provide valuable insights into the complex structure of disease pathophysiology. It is hypothesized that the cell may be experiencing a combined vesicular trafficking disorder, with a switch in energy production affecting pathways such as oxidative phosphorylation and glycolysis. In the event of oxidative phosphorylation being ineffective, alternative pathways such as glycolysis and lipogenesis may be favored.