Submitted:
14 May 2025
Posted:
15 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection
2.3. Laboratory Analysis
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The Bovine Milk Microbiota: Insights and Perspectives from -Omics Studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.F.; Teixeira, A.G.V.; Lima, F.S.; Ganda, E.K.; Higgins, C.H.; Oikonomou, G.; Bicalho, R.C. The Bovine Colostrum Microbiome and Its Association with Clinical Mastitis. J. Dairy Sci. 2017, 100, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; Godden, S.; Bey, R.; Rapnicki, P.; Fetrow, J.; Farnsworth, R.; Scanlon, M.; Arnold, Y.; Clow, L.; Mueller, K.; et al. Preventing Bacterial Contamination and Proliferation during the Harvest, Storage, and Feeding of Fresh Bovine Colostrum. J. Dairy Sci. 2005, 88, 2571–2578. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.G.; Silva, S.R.; Pereira, A.M.F.; Cerqueira, J.L.; Conceição, C. A Comprehensive Review of Bovine Colostrum Components and Selected Aspects Regarding Their Impact on Neonatal Calf Physiology. Animals 2024, 14, 1130. [Google Scholar] [CrossRef]
- Otte, J.-M.; Podolsky, D.K. Functional Modulation of Enterocytes by Gram-Positive and Gram-Negative Microorganisms. Am. J. Physiol. Liver Physiol. 2004, 286, G613–G626. [Google Scholar] [CrossRef]
- Malik, M.I.; Rashid, M.A.; Raboisson, D. Heat Treatment of Colostrum at 60°C Decreases Colostrum Immunoglobulins but Increases Serum Immunoglobulins and Serum Total Protein: A Meta-Analysis. J. Dairy Sci. 2022, 105, 3453–3467. [Google Scholar] [CrossRef]
- Barry, J.; Bokkers, E.A.M.; De Boer, I.J.M.; Kennedy, E. Pre-Weaning Management of Calves on Commercial Dairy Farms and Its Influence on Calf Welfare and Mortality. Animal 2020, 14, 2580–2587. [Google Scholar] [CrossRef]
- Abuelo, A.; Havrlant, P.; Wood, N.; Hernandez-Jover, M. An Investigation of Dairy Calf Management Practices, Colostrum Quality, Failure of Transfer of Passive Immunity, and Occurrence of Enteropathogens among Australian Dairy Farms. J. Dairy Sci. 2019, 102, 8352–8366. [Google Scholar] [CrossRef]
- Chase, C.C.L.; Hurley, D.J.; Reber, A.J. Neonatal Immune Development in the Calf and Its Impact on Vaccine Response. Vet. Clin. North Am. - Food Anim. Pract. 2008, 24, 87–104. [Google Scholar] [CrossRef]
- Heinrichs, J.; Jones, C. Composition and Hygiene of Colostrum on Modern Pennsylvania Dairy Farms. Available online: https://extension.psu.edu/composition-and-hygiene-of-colostrum-on-modern-pennsylvania-dairy-farms (accessed on 21 July 2024).
- Godden, S.M.; Lombard, J.E.; Woolums, A.R. Colostrum Management for Dairy Calves. Vet. Clin. North Am. - Food Anim. Pract. 2019, 35, 535–556. [Google Scholar] [CrossRef]
- Fecteau, G.; Baillargeon, P.; Higgins, R.; Paré, J.; Fortin, M. Bacterial Contamination of Colostrum Fed to Newborn Calves in Québec Dairy Herds. Can. Vet. J. 2002, 43, 523–527. [Google Scholar] [PubMed]
- McGuirk, S.M.; Collins, M. Managing the Production, Storage, and Delivery of Colostrum. Vet. Clin. North Am. - Food Anim. Pract. 2004, 20, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Šlosárková, S.; Pechová, A.; Staněk, S.; Fleischer, P.; Zouharová, M.; Nejedlá, E. Microbial Contamination of Harvested Colostrum on Czech Dairy Farms. J. Dairy Sci. 2021, 104, 11047–11058. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.J.; Beggs, D.S.; Murray, A.J.; Mansell, P.D.; Stevenson, M.A.; Pyman, M.F. Survey of Bovine Colostrum Quality and Hygiene on Northern Victorian Dairy Farms. J. Dairy Sci. 2016, 99, 8981–8990. [Google Scholar] [CrossRef]
- Godden, S.; McMartin, S.; Feirtag, J.; Stabel, J.; Bey, R.; Goyal, S.; Metzger, L.; Fetrow, J.; Wells, S.; Chester-Jones, H. Heat-Treatment of Bovine Colostrum. II: Effects of Heating Duration on Pathogen Viability and Immunoglobulin G. J. Dairy Sci. 2006, 89, 3476–3483. [Google Scholar] [CrossRef]
- Van Driessche, L.; Santschi, D.E.; Paquet, É.; Renaud, D.; Charbonneau, É.; Gauthier, M. Lou; Chancy, A.; Barbeau-Grégoire, N.; Buczinski, S. Hygiene Management Practices and Adenosine Triphosphate Luminometry of Feeding Equipment in Preweaning Calves on Dairy Farms in Quebec, Canada. J. Dairy Sci. 2023, 106, 8885–8896. [Google Scholar] [CrossRef]
- Barry, J.; Bokkers, E.A.M.; Berry, D.P.; de Boer, I.J.M.; McClure, J.; Kennedy, E. Associations between Colostrum Management, Passive Immunity, Calf-Related Hygiene Practices, and Rates of Mortality in Preweaning Dairy Calves. J. Dairy Sci. 2019, 102, 10266–10276. [Google Scholar] [CrossRef]
- Hyde, R.M.; Green, M.J.; Hudson, C.; Down, P.M. Quantitative Analysis of Colostrum Bacteriology on British Dairy Farms. Front. Vet. Sci. 2020, 7, 1–12. [Google Scholar] [CrossRef]
- Guzman-Carazo, V.; Reyes-Vélez, J.; Elsohaby, I.; Olivera-Angel, M. Factors Associated with Microbiological Quality of Bovine Colostrum in Colombian Dairy Herds. Int. Dairy J. 2020, 105, 104670. [Google Scholar] [CrossRef]
- Donahue, M.; Godden, S.M.; Bey, R.; Wells, S.; Oakes, J.M.; Sreevatsan, S.; Stabel, J.; Fetrow, J. Heat Treatment of Colostrum on Commercial Dairy Farms Decreases Colostrum Microbial Counts While Maintaining Colostrum Immunoglobulin G Concentrations. J. Dairy Sci. 2012, 95, 2697–2702. [Google Scholar] [CrossRef]
- Raboisson, D.; Trillat, P.; Cahuzac, C. Failure of Passive Immune Transfer in Calves: A Meta-Analysis on the Consequences and Assessment of the Economic Impact. PLoS One 2016, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.; Silva, J.T.; Santos, F.H.R.; Bittar, C.M.M. Nutritional and Microbiological Quality of Bovine Colostrum Samples in Brazil. Rev. Bras. Zootec. 2017, 46, 72–79. [Google Scholar] [CrossRef]
- McAloon, C.G.; Doherty, M.L.; Donlon, J.; Lorenz, I.; Meade, J.; O’Grady, L.; Whyte, P. Microbiological Contamination of Colostrum on Irish Dairy Farms. Vet. Rec. 2016, 178, 474. [Google Scholar] [CrossRef] [PubMed]
- Morrill, K.M.; Conrad, E.; Lago, A.; Campbell, J.; Quigley, J.; Tyler, H. Nationwide Evaluation of Quality and Composition of Colostrum on Dairy Farms in the United States. J. Dairy Sci. 2012, 95, 3997–4005. [Google Scholar] [CrossRef]
- Cummins, C.; Berry, D.P.; Murphy, J.P.; Lorenz, I.; Kennedy, E. The Effect of Colostrum Storage Conditions on Dairy Heifer Calf Serum Immunoglobulin G Concentration and Preweaning Health and Growth Rate. J. Dairy Sci. 2017, 100, 525–535. [Google Scholar] [CrossRef]
- Elizondo-Salazar, J.A.; Jayarao, B.M.; Heinrichs, A.J. Effect of Heat Treatment of Bovine Colostrum on Bacterial Counts, Viscosity, and Immunoglobulin G Concentration. J. Dairy Sci. 2010, 93, 961–967. [Google Scholar] [CrossRef]
- Mann, S.; Curone, G.; Chandler, T.L.; Moroni, P.; Cha, J.; Bhawal, R.; Zhang, S. Heat Treatment of Bovine Colostrum: I. Effects on Bacterial and Somatic Cell Counts, Immunoglobulin, Insulin, and IGF-I Concentrations, as Well as the Colostrum Proteome. J. Dairy Sci. 2020, 103, 9368–9383. [Google Scholar] [CrossRef]
- Pyörälä, S.; Taponen, S. Coagulase-Negative Staphylococci—Emerging Mastitis Pathogens. Vet. Microbiol. 2009, 134, 3–8. [Google Scholar] [CrossRef]
- Trujillo, A.J.; Castro, N.; Quevedo, J.M.; Argüello, A.; Capote, J.; Guamis, B. Effect of Heat and High-Pressure Treatments on Microbiological Quality and Immunoglobulin G Stability of Caprine Colostrum. J. Dairy Sci. 2007, 90, 833–839. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Chen, Y.; Liang, G.; Goonewardene, L.A.; Guan, L.L. Heat-Treated Colostrum Feeding Promotes Beneficial Bacteria Colonization in the Small Intestine of Neonatal Calves. J. Dairy Sci. 2015, 98, 8044–8053. [Google Scholar] [CrossRef]
- Patangia, D. V.; Grimaud, G.; Linehan, K.; Ross, R.P.; Stanton, C. Microbiota and Resistome Analysis of Colostrum and Milk from Dairy Cows Treated with and without Dry Cow Therapies. Antibiotics 2023, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Adeniyi, B.A.; Adetoye, A.; Ayeni, F.A. Antibacterial Activities of Lactic Acid Bacteria Isolated from Cow Faeces against Potential Enteric Pathogens. Afr. Health Sci. 2015, 15, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tang, G.; Guo, W.; Lei, J.; Yao, J.; Xu, X. Detection of the Core Bacteria in Colostrum and Their Association with the Rectal Microbiota and with Milk Composition in Two Dairy Cow Farms. Animals 2021, 11, 3363. [Google Scholar] [CrossRef] [PubMed]
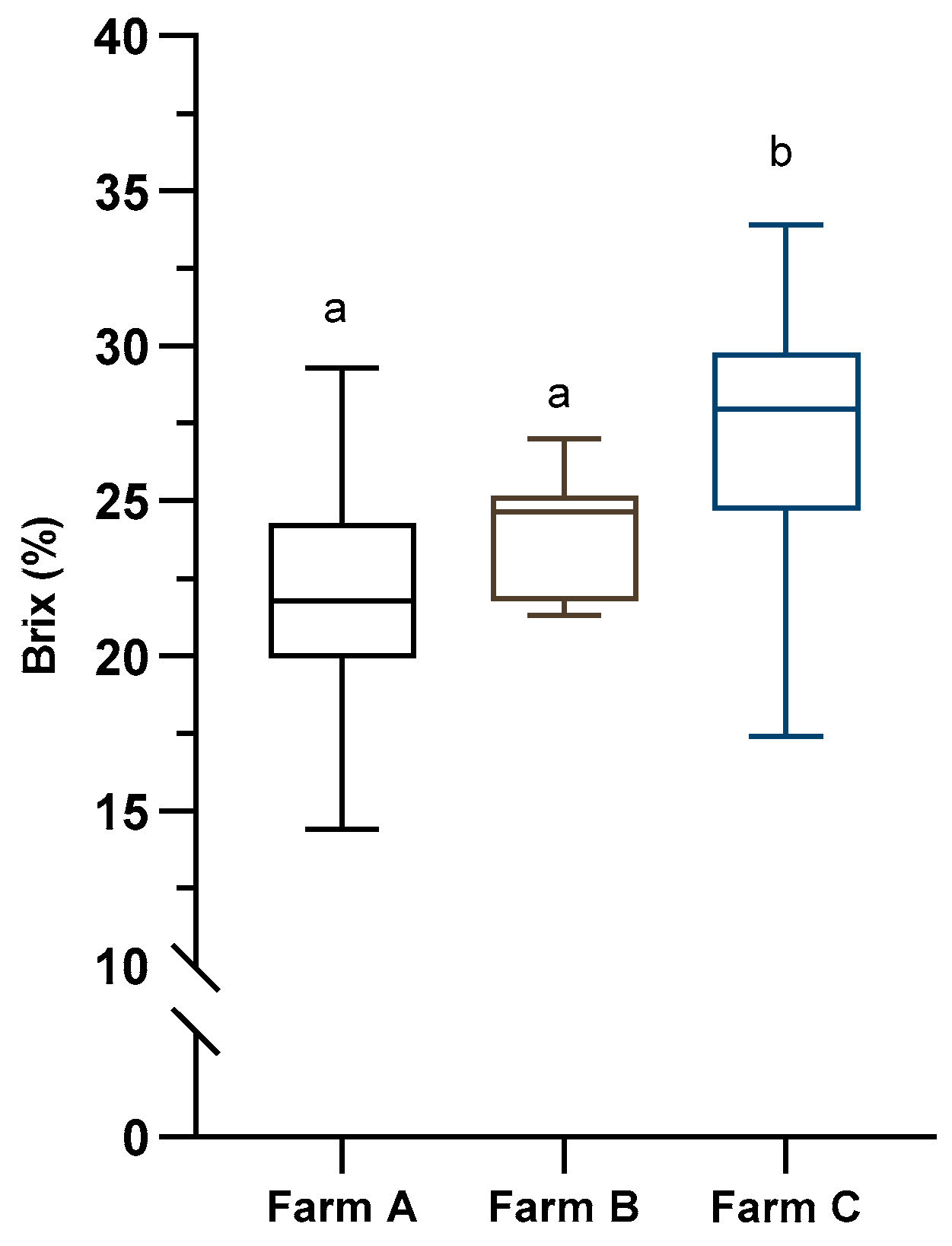
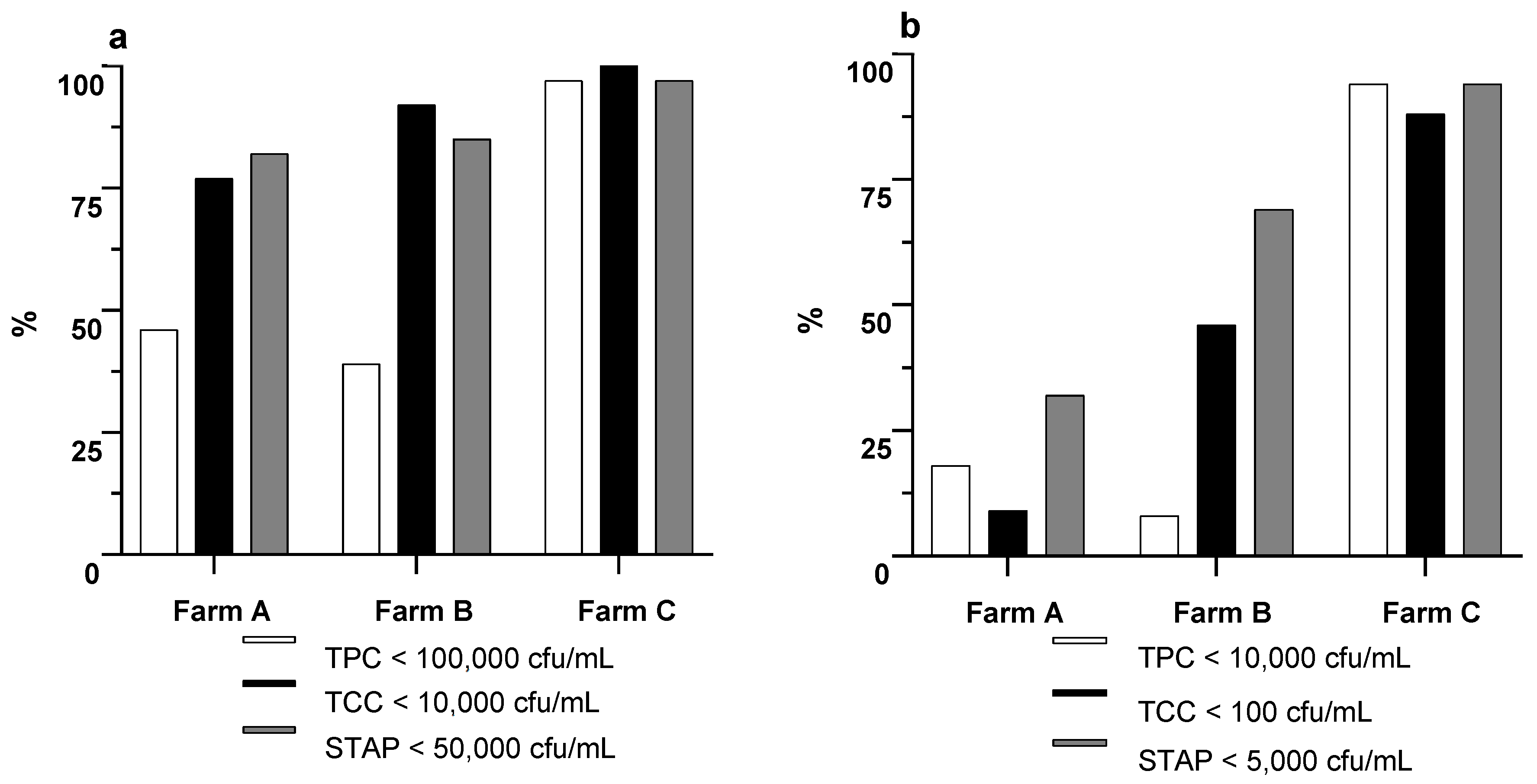
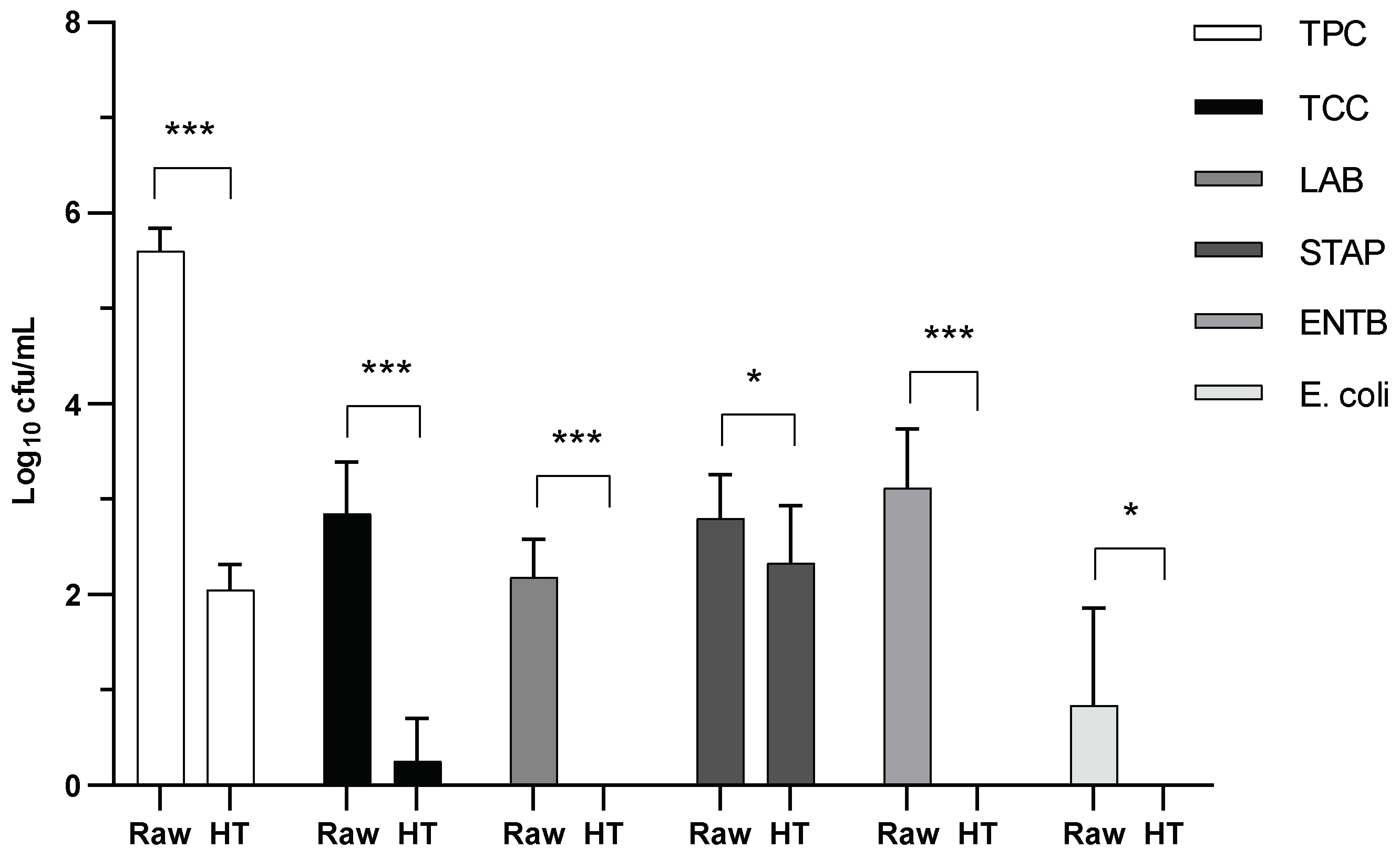
| Farm | Cows in milking | Average lactation nº | Milk production 305 d | Milk fat 305 d | Milk protein 305 d |
|---|---|---|---|---|---|
| A | 21 | 2.7 | 8813 | 4.46 | 3.43 |
| B | 484 | 2.6 | 13634 | 4.2 | 3.31 |
| C | 783 | 2.1 | 12118 | 4.11 | 3.22 |
| Farm | Cleaning of feeding apparatus | ||||
| HT1 | Hot water | Detergent | Disinfectant | Automated washing | |
| A | No | 40 – 50 °C | A | Yes | No |
| B | Yes | 40 – 50 °C | B | No | No |
| C | Yes | 60 °C | No | Yes | Yes |
| Counts | Mean | Median | SD | Minimum | First Q | Third Q | Maximum | Met (%) |
|---|---|---|---|---|---|---|---|---|
| TPC | 398,936.57 | 16,800.00 | 1,547,386.70 | 40.00 | 475.00 | 206,500.00 | 13,200,000.00 | 69.9a; 53.7b |
| TCC | 11,706.44 | 70.00 | 75,103.84 | < dl | < dl | 2,130.00 | 637,260.00 | 91.21a; 54.4b |
| LAB | 64,629.56 | 580.00 | 236,961.96 | < dl | 35.00 | 19,050.00 | 1,580,000.00 | |
| STAP | 17,720.67 | 1,460.00 | 50,892.69 | < dl | 350.00 | 14,750.00 | 350,000.00 | 89.7a; 69.1b |
| ENTB | 12,958.83 | 40.00 | 44,750.21 | < dl | < dl | 730.00 | 252,000.00 | |
| E. coli | 482.06 | < dl | 2,117.68 | < dl | < dl | 145.00 | 17,100.00 |
| Counts | Farm A | Farm B | Farm C | P-value |
|---|---|---|---|---|
| TPC1 | 5.13 (0.11) a | 5.26 (0.15) a | 2.80 (0.09) b | <0.001 |
| TCC1 | 3.47 (0.15) a | 2.12 (0.19) b | 0.71 (0.12) c | <0.001 |
| LAB1 | 4.31 (0.17) a | 3.18 (0.21) b | 1.41 (1.14) c | <0.001 |
| STAP1 | 4.02 (0.12) a | 3.55 (0.16) a | 2.46 (0.10) b | 0.002 |
| ENTB2 | 2.89 (1.60 – 5.40) a | 2.20 (< dl – 5.23) a | < dl (< dl – 3.13) b | <0.001 |
| E. coli2 | 2.18 (0.00 – 3.74) a | 0.00 (< dl – 4.23) b | < dl (< dl – 3.34) b | <0.001 |
| pH1 | 6.29 (0.02) a | 6.27 (0.03) ab | 6.22 (0.01) b | <0.001 |
| aw1 | 92.76 (0.13) a | 92.67 (0.17) a | 93.34 (0.09) c | 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





