1. Introduction
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide, ranking as the third most commonly diagnosed cancer and the second leading cause of cancer-related deaths. Both genetic predispositions and environmental factors contribute to its development [
1,
2]. DNA repair is an essential pathway in CRC pathogenesis, which maintains genomic stability by managing replication errors. Critical alterations in the DNA mismatch repair (MMR) system, including mutations and polymorphisms in key genes such as
hMLH1 and
hMSH2, have been strongly implicated in CRC susceptibility [
3,
4,
5].
The
hMLH1 and
hMSH2 genes encode proteins that form part of the MMR complex, which corrects base mismatches and insertion-deletion loops generated during DNA replication. Dysfunction of these genes leads to microsatellite instability (MSI), a hallmark of hereditary nonpolyposis colorectal cancer (HNPCC) and a significant factor in sporadic CRC cases [
6,
7,
8].
Several single nucleotide polymorphisms (SNPs) in these genes have been reported to affect gene expression and protein function, potentially modifying CRC risk. One of the most extensively studied polymorphisms in the
hMLH1 gene is
-93G>A (rs1800734), located in the promoter region. This SNP has been associated with reduced
hMLH1 expression, which may impair MMR function and contribute to tumor development. Studies have shown that individuals carrying the A allele have a higher risk of developing CRC, particularly in the presence of other genetic or environmental factors [
9]. Similarly, the
hMSH2 1032G>A (rs4987188) polymorphism in exon 13 has been investigated for its potential role in cancer predisposition. Although its direct effect on protein function remains unclear, certain studies suggest that it may be linked to CRC susceptibility [
10,
11,
12]. Recent genome-wide association studies (GWAS) have identified multiple SNPs associated with CRC risk, including polymorphisms in DNA repair genes. However, findings have been inconsistent across different ethnic groups, necessitating further investigation. Given the crucial role of DNA repair mechanisms in maintaining genomic stability, identifying genetic variations that influence CRC susceptibility is essential for improving early detection and personalized treatment strategies [
13,
14].
This study aims to assess the association between hMLH1 -93G>A and hMSH2 1032G>A polymorphisms and CRC risk by analyzing their distribution in patients and healthy controls. By investigating these polymorphisms, we aim to deepen our understanding of genetic risk factors for colorectal cancer and explore their potential as useful biomarkers for early diagnosis and treatment planning.
2. Materials and Methods
2.1. Subjects
A total of 134 patients diagnosed with CRC were enrolled in the study from Azerbaijan Medical University and the M. A. Topchubashov Scientific Surgery Center. The study included sporadic CRC cases, excluding cases of inflammatory bowel disease and hereditary CRC syndromes. The histopathological data of the tumors, including tumor grade and stage, were specified in the pathology report. The study population included 137 healthy individuals with no family history of cancer who underwent routine colonoscopy screening. Upon obtaining informed consent, additional data - including age, smoking status, and alcohol consumption—were collected from both patients and control subjects. The study protocol was approved by the Ethics Committee of the Institute of Genetic Resources, and written informed consent was obtained from each patient.
2.2. Genotyping
Blood samples were collected from patients and controls, followed by DNA extraction, which was performed at the Human Genetics Laboratory of the Institute of Genetic Resources using the
Non-Enzymatic Salting Out Method [
15]. The extracted DNA samples were then stored at −20°C until required for the subsequent stage of the experiment. The quantitative and qualitative parameters of the DNA were measured using a NanoDrop™ 2000/2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). PCR-RFLP (restriction fragment length polymorphism) methods were used to determine
hMLH1 -93G>A (rs1800734) and
hMSH2 1032G>A (rs4987188) gene polymorphisms. The PCR reaction mixture was prepared in 20 µl according to the instructions. The PCR reaction mixture consists of 2 µl genomic DNA, 2 µl 10Xbuffer [10 mM Tris-HCl pH 8.0, 50 mM KCl], 2 µl MgCl2, 0.2 µl (20 mM) dNTP mixture, 0.5 µl (100 µM) primer and 0.2 µl (5 U/µl) Taq polymerase enzyme. The PCR reaction consisted of initial denaturation (5 min at 95°C), 35 cycles (30 sec at 95°C, 1 min at 57°C, 2 min at 72°C) and final extension (5 min at 72°C).
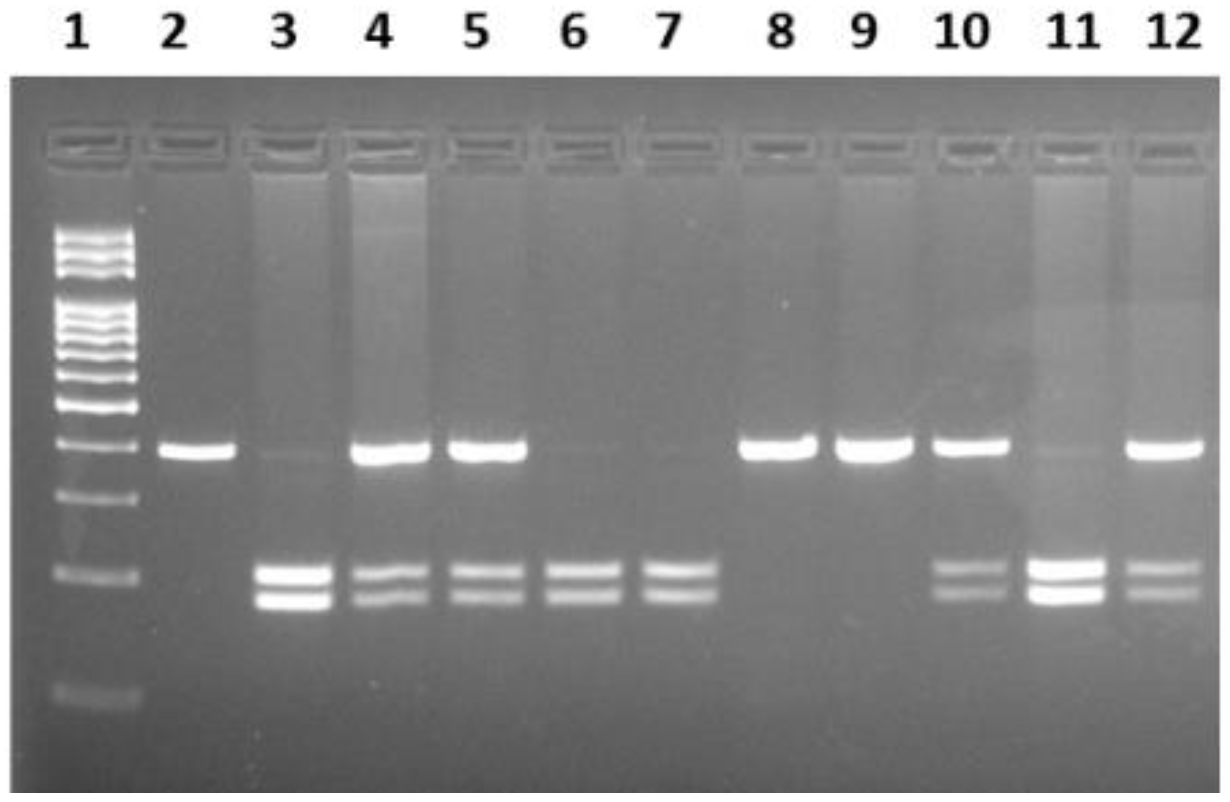
The primer sequences for determining the -93G>A polymorphism of the
hMLH1 gene are listed below: 5'-CCGAGCTCCTAAAAACGAAC-3'; reverse strand: 5'-CTGGCCGCTGGATAACTTC-3'. In the PCR reaction, the amplified 387 bp region of the gene was restricted with
PvuII (Thermo Scientific, USA) enzyme at 37°C for 5 hours. Samples were then electrophoresed on a 2% agarose gel. The homozygous AA genotype produced undigested fragments of 387bp, while the homozygous GG genotype yielded two digested fragments of sizes 207 bp and 180 bp. The AG heterozygous condition revealed three bands sized 387 bp, 207 bp and 180 bp (
Figure 1). For the
hMSH2 1032G>A (rs4987188) polymorphism, the following primers were used: sense 5’-GTTTTCACTAATGAGCTTGC-3' and antisense 5’-AGTGGTATAATCATGTGGGT-3'. In the PCR experiment, the amplified 252 bp fragment of the gene was subjected to restriction using the
HinfI enzyme (Thermo Scientific, USA) at 37°C for 5 hours. The wild-type GG genotype produced undigested fragments of 252 bp, whereas the heterozygous GA genotype yielded fragments of 252, 182, and 70 bp. The homozygous AA genotype produced fragments of 182 and 70 bp.
A randomly selected 10% study group was retested with PCR-RFLP, and the results were found to be 100% consistent.
2.3. Statistical Analysis
Comparisons of genotype and allele distributions between colorectal cancer patients and control subjects were conducted using the chi-square (χ
2) test or Fisher's exact test when appropriate. For contingency tables larger than 2×2, Fisher's exact test was performed using the Social Science Statistics platform
http://www.socscistatistics.com/tests/chisquare2/Default2.aspx). To assess the relationship between genetic variants and CRC risk, odds ratios (ORs) with 95% confidence intervals (CIs) were calculated through binary logistic regression. Genetic association was evaluated under both dominant and recessive models. All tests were two-tailed, with statistical significance defined as P<0.Data analysis was carried out using SPSS software, version 22 (IBM Corp., Chicago, IL, USA).
3. Results
Among the patients, 76 (56.7%) were male, and 58 (43.3%) were female, while the control group comprised 64 (43.3%) males and 73 (56.7%) females. The patients' ages ranged from 25 to 85 years, whereas the control group ranged from 32 to 82 years. The mean ages of the patient and control groups were 60±10.2 and 60±11.3 years, respectively. Tumor grade was observed to be G1 in 11 patients (8.2%), G2 in 90 patients (67.1%), and G3 in 33 patients (24.7%).
Table 1.
Clinical and demographic characteristics of the study groups.
Table 1.
Clinical and demographic characteristics of the study groups.
| |
Patients
N=134 (%) |
Controls
N=137 (%) |
P value |
Gender
Male
Female |
76 (56.7)
58 (43.3) |
64 (43.3)
73 (56.7) |
0.256 |
Age
Average
Mean |
25-85
60±10.2 |
32-82
60±11.3 |
|
Grade
G1
G2
G3 |
11 (8.2)
90 (67.1)
33 (24.7) |
|
|
Stage
T I
T II
T III
T IV |
3 (2.1)
12 (9.3)
109 (81.5)
10 (7.1) |
|
|
Smoking
Smokers
Non-smokers
Unknown |
44 (34.3)
82 (60)
8 (5.7) |
55 (33.5)
98 (59.8)
11 (6.7) |
0.942
|
Alcohol consumption
User
Non-user
Unknown |
36 (32.1)
90 (62.2)
8 (5.7) |
59 (36)
93 (56.6)
12 (7.4) |
0.410 |
The total number of patients diagnosed with each tumor stage was as follows: T1 (2.1%), T2 (9.3%), T3 (81.5%), and T4 (7.1%). The patient survey revealed that 34.3% of patients were smokers, while 60% were non-smokers. In the control group, these percentages were 33.5% and 59.8%, respectively. It should be noted that smoking data was unavailable for some individuals. Among the patients, 36 (32.1%) consumed alcohol, 90 (62.2%) did not, and for 8 (5.7%), no information was available. In the control group, these values were recorded as 59 (36%), 93 (56.6%), and 5 (3.3%), respectively. A comparison of gender, age, smoking, and alcohol consumption between the study groups revealed no statistically significant relationship (P>0.05).
The genotype and allele frequencies of the hMLH1 gene -93G>A polymorphism were calculated and statistically analyzed for both groups (
Table 2). The frequency of the GG genotype in the patient group was 15.7%, heterozygous GA was 32.1%, and mutant AA was 52.2%. In contrast, in the control group, the frequencies of GG, GA, and mutant AA genotypes were 16.8%, 45.2%, and 38.0%, respectively. No significant association was found between disease risk and the heterozygous GA (OR=0.76; 95% CI=0.37–1.54; P=0.446) or homozygous mutant AA (OR=1.47; 95% CI=0.74–2.95; P=0.270) genotypes. In the dominant model (GG vs. GA+AA), no statistically significant association with disease risk was observed (OR=0.92; 95% CI=0.48–1.76; P=0.803). However, in the recessive model (GG+GA vs. AA), a statistically significant association with disease risk was identified (OR=0.56; 95% CI=0.35–0.91; P=0.018). Additionally, the G allele was found in 53.1% of patients and 56.1% of the control group, while the mutant A allele was observed in 46.9% of patients and 43.9% of the control group. Statistical analyses revealed no significant association between the mutant A allele and CRC risk (P=0.748).
The genotype frequencies of the hMLH1 gene based on gender were compared between both study groups (
Table 3). The heterozygous GA genotype was found in 32.9% of male patients, while it was more common in healthy males (43.8%). The homozygous mutant AA genotype was more frequently observed in male patients (48.7%) compared to healthy males (40.6%). In female patients, the heterozygous GA genotype (31%) was less common than in controls (46.6%), while the mutant AA genotype was more prevalent in female patients (56.9%) compared to healthy females (35.6%). However, despite these observations, when comparing the two groups based on gender, no statistically significant correlation was found between genotype and disease risk (P>0.05).
Table 4 demonstrates that the study groups were compared according to their genotypes, with the average age taken into account. In healthy individuals younger than 60 years, the heterozygous GA genotype (38.2%) was more common than in patients, while the mutant AA genotype was more prevalent in patients (54.2%) than in healthy individuals. In contrast, in individuals older than 60 years, the heterozygous GA genotype was more prevalent in the control group (47.4%), while the mutant AA genotype was more frequent in patients (51.1%). However, no statistically significant correlation was observed between these parameters (P>0.05).
Additionally, we calculated the distribution of genotypes in patients based on smoking and alcohol consumption (
Table 5). The heterozygous GA genotype was found more frequently in non-smoking patients (32.9%) compared to smokers (29.5%). In contrast, the mutant AA genotype was more prevalent in smokers (54.5%) than in non-smokers (52.5%). No statistically significant association was observed between genotypes and CRC risk in both smoking and non-smoking patients. Similarly, in patients who do not consume alcohol, the heterozygous GA genotype (33.3%) was more common, whereas in alcohol-consuming patients, the mutant AA genotype (61.1%) was observed at a higher frequency. No statistically significant difference was found between genotypes and disease risk with regard to alcohol consumption (P>0.05).
In our study, we also analyzed how the hMLH1 gene -93G>A polymorphism varies with the stage and grade of the tumor (
Table 6). The heterozygous GA genotype was primarily observed in grade G3 (54.5%), while the mutant AA genotype was more commonly found in grade G2 (20%). No statistically significant difference was found between tumor grade and hMLH1 gene -93G>A genotypes. Regarding the distribution of genotypes across cancer stages, the heterozygous GA genotype (46.8%) was most frequently observed in stage T3, while the homozygous AA genotype was more prevalent in stage T1 (33.3%). A positive statistical correlation was found between tumor stages and genotype distribution (P<0.05).
An evaluation of the genotype and allele frequencies of the
hMSH2 gene 1032G>A polymorphism was conducted, and the corresponding risk values were determined (
Table 7). It was observed that this polymorphism is uncommon among patients, with the heterozygous GA genotype present in 17.2% of patients and 9.5% of the control group. The homozygous mutant AA genotype was present in only one patient, and in the control group, only two individuals had the AA genotype. Nevertheless, no statistically significant difference was observed between genotype and allele frequencies and cancer risk (P>0.05).
4. Discussion
Deficiencies in DNA repair mechanisms are a hallmark of many cancers. While mutations in DNA repair genes are commonly found in malignant tumors, only a few studies have highlighted the critical role these genes play in determining disease prognosis and shaping responses to chemotherapy and radiotherapy [
16]. The contribution of MMR gene polymorphisms to CRC risk has been widely acknowledged, but the picture remains incomplete, namely in genetically understudied populations. This study aimed to evaluate two well-characterized polymorphisms,
hMLH1 -93G>A and
hMSH2 1032G>A, in a cohort of Azerbaijani patients and controls, contributing to a broader understanding of CRC genetics.
While inherited mutations and promoter methylation of
hMLH1 and
hMSH2 are well-established contributors to Lynch syndrome (hereditary nonpolyposis colorectal cancer, HNPCC), this study focused on patients with sporadic CRC. Although SNPs in these genes do not directly cause cancer, they are recognized as potential risk modifiers in certain individuals, particularly in the absence of a strong family history [
17].
In this study, we found that the genotypic and allelic frequencies of hMLH1 -93G>A (rs1800734) and hMSH2 1032G>A (rs4987188) do not significantly contribute to the risk of CRC. Despite this, the hMLH1 -93G>A polymorphism was more frequently observed in patients, whereas the hMSH2 1032G>A polymorphism occurred at a lower frequency. Notably, a statistical link was identified between the hMLH1 -93G>A polymorphism and disease risk when analyzed under a recessive model.
An analysis conducted in China, which included 12 studies with 4,128 cancer cases and 4,678 control subjects, showed that the
hMLH1 -93G>A polymorphism is not associated with an increased risk of cancer development [
12]. Wang and colleagues obtained similar results in their study on CRC patients from northeastern China. Researchers found no link between
hMSH2 gene polymorphisms and cancer risk [
18]. Consistent with our findings, two different studies conducted in a Chinese population found no significant association between the -93G>A polymorphism and susceptibility to CRC [
19,
20]. Moreover, a large meta-analysis conducted in 2015 investigated the SNPs of
hMLH1 (rs1800734, rs1799977, and rs63750448), and no association was found between rs1800734 and CRC risk in both overall and subgroup analyses [
21].
Nevertheless, several studies have reported that this polymorphism increases the risk of CRC. Recent studies in Chinese and Turkish populations showed that
hMLH1 -93G>A polymorphism is associated with CRC risk under both dominant and recessive models, leading to the suggestion that variants in the
hMLH1 gene could potentially act as predictors of the disease [
18,
22]. In addition, our results revealed a statistically significant association only under the recessive model, which partially aligns with findings from Nizam and colleagues, who reported a stronger risk under the heterozygous model in sporadic CRC cases in the Malaysian population, highlighting possible differences in genetic impact across populations and inheritance patterns [
23].
In a cohort of 1,518 CRC patients, homozygosity for the
MLH1 -93A variant was significantly associated with a threefold increased risk of CRC, accompanied by loss of MLH1 protein expression, as assessed by immunohistochemical analysis [
24]. According to a comprehensive meta-analysis published in 2012, the
MLH1 -93G>A polymorphism may play a role in increasing an individual's susceptibility to CRC and could serve as a potential risk factor for microsatellite instability-positive CRC [
11].
A meta-analysis of 38 case-control studies found that the -93G>A and 655A>G polymorphisms in the
hMLH1 gene are associated with a higher risk of CRC in Asian populations, while no such link was identified in Caucasians [
10]. In the first study conducted in the Lower Northeastern Region of Thailand, it was found that the hMLH1 -93G>A polymorphism is associated with an increased risk of CRC [
25].
In the study conducted by Mik and his colleagues, a correlation was identified between the -93G>A
hMLH1 polymorphism and an increased risk of CRC. However, no association was found between
MSH2 Gly322Asp genotypes and the risk of the disease. Similarly, Liu et al. classified Gly322Asp as a benign polymorphism without clear segregation in hereditary cancer families [
26]. These findings are in concordance with our results; this variant was less frequent among CRC patients and showed no significant association with disease risk. Interestingly, the
MSH2 Gly322Asp polymorphism was proposed as a potential marker for patients at risk of disease recurrence [
17].
The given polymorphism has been investigated in relation to various cancers, although available studies remain limited, and findings are inconsistent. While few studies have specifically evaluated this variant in endometrial cancer, Romanowicz and the authors demonstrated that the Gly322Asp significantly increased the risk of disease, particularly among carriers of the Asp/Asp genotype. Interestingly, Smolarz et al. also reported that this polymorphism may have a protective effect against triple-negative breast cancer, highlighting that the impact of Gly322Asp might vary depending on cancer type and genetic background. Taken together, while our findings support the lack of a strong association with CRC risk, the available evidence suggests that the clinical significance of this variant may differ across tumor types, warranting further investigation [
27,
28].
Furthermore, our research stratified the research groups according to age, gender, smoking and alcohol consumption, tumor grade and stage. Among these variables, a statistically significant association was found between the
hMLH1 -93G>A polymorphism and CRC stages. In contrast to these findings, a study has reported an association between the -93G>A polymorphism and smoking [
29]. In another study, the association of increased age at diagnosis and tumor characteristics with MSI tumors linked to the
MLH1 -93 G>A polymorphism suggests that this polymorphism may influence colorectal carcinogenesis at a later stage, promoting the progression of CIMP+ tumors via the MSI pathway [
30]. Whiffin et al. provided further evidence supporting the role of the
MLH1 -93A polymorphism as a low-penetrance susceptibility variant for CRC. In the same study, the rs1800734 polymorphism was found to be associated with age and sex in MSI-H CRC cases, which is more frequent in older individuals and females. No association was found with tumour site, stage, grade, or family history of CRC [
31]. In contrast, Zhang and colleagues reported comparable findings, showing a positive association between tumor stage and
hMLH1 -93 G>A polymorphism [
20].
The given investigation possesses several limitations. The relatively small sample size limits the ability to detect weak associations. Furthermore, the study did not incorporate molecular analyses such as methylation profiling or protein expression, which could have provided deeper insights into genotype–phenotype correlations. Lastly, although adjustments were made for demographic and major clinical variables, the inclusion of more comprehensive environmental data would enhance the understanding of gene-environment interactions in this population.
5. Conclusions
Our study investigated two common polymorphisms, hMLH1 -93G>A and hMSH2 1032G>A, in patients with sporadic CRC from the Azerbaijani population. Although the overall distribution of genotypes and alleles of the hMLH1 -93G>A polymorphism did not differ significantly between patients and controls, a statistically significant association was observed under the recessive model with a decreased risk of CRC. This finding supports previous evidence indicating that low-penetrance variants may contribute to colorectal cancer susceptibility, particularly when inherited under specific genetic models.
In contrast, the hMSH2 1032G>A polymorphism did not show a significant association with disease risk in this cohort. By focusing on a less-studied population, this study contributes to a broader understanding of genetic risk variation in colorectal cancer.
Further studies with larger sample sizes and functional assays will be essential to elucidate the biological relevance of these SNPs and to evaluate their potential impact on CRC risk stratification and early detection strategies.
Author Contributions
Conceptualization, B.B. and N.K.; methodology, B.B.; software, Z.S.; validation, N.K., R.K. and R.S.; formal analysis, O.I.; investigation, B.B.; resources, N.M.; data curation, H.V. and H.A.; writing—original draft preparation, B.B.; writing—review and editing, N.K.; visualization, O.I.; supervision, N.B. All authors have reviewed and approved the final version of the manuscript for publication.
Funding
This study was funded by the Science Development Foundation under the President of the Republic of Azerbaijan within the framework of Grant No. AEF-MCG-2023-1(43)-13/08/3-M-08.
Institutional Review Board Statement
The study was carried out in full compliance with the principles outlined in the Declaration of Helsinki and received ethical approval from the Ethics Committee of the Genetic Resources Institute under the Ministry of Science and Education (Protocol No. 68-12/258 approval date: 20 March 2023).
Informed Consent Statement
All experimental procedures in this study were carried out at the M. A. Topchubashov Scientific Surgery Center, the Educational-Surgical Clinic of Azerbaijan Medical University, and the Genetic Resources Institute. Prior to participation, all individuals received detailed information regarding the study protocol and voluntarily provided their written informed consent. Furthermore, written consent for publication was obtained from all participants.
Data Availability Statement
The entirety of the data employed in this research is comprehensively presented in the tables included in this article.
Acknowledgments
We would like to thank the Science Development Foundation under the President of the Republic of Azerbaijan for supporting this research.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CRC |
Colorectal cancer |
| SNP |
Single nucleotide polymorphism |
| MMR |
Mismatch repair |
| GWAS |
Genome-wide association studies |
| PCR |
Polymerase chain reaction |
| RFLP |
Restriction fragment length polymorphism |
| OR |
Odds ratios |
| CI |
Confidence interval |
| HNPCC |
Hereditary nonpolyposis colorectal cancer |
References
- Matsuda, T.; Fujimoto, A.; Igarashi, Y. Colorectal Cancer: Epidemiology, Risk Factors, and Public Health Strategies. Digestion 2025, 106, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef]
- Chung, D. C.; Rustgi, A. K. The Hereditary Nonpolyposis Colorectal Cancer Syndrome: Genetics and Clinical Implications. Annals of Internal Medicine 2003, 138, 560–570. [Google Scholar] [CrossRef]
- De’ Angelis, G. L.; Bottarelli, L.; Azzoni, C.; De’ Angelis, N.; Leandro, G.; Di Mario, F.; et al. Microsatellite instability in colorectal cancer. Acta bio-medica: Atenei Parmensis 2018, 89, 97–101. [Google Scholar] [CrossRef]
- dos Santos, I. B.; da Costa, A. C. A.; Gellen, L. P. A.; Sales, L. L. S.; Monte, N.; de Moraes, F. C. A.; et al. Identification of genomic variants associated with colorectal cancer heredity in indigenous populations of the Amazon. Scientific Reports 2025, 15, 14616. [Google Scholar] [CrossRef] [PubMed]
- Söreide, K.; Janssen, E. A. M.; Söiland, H.; Körner, H.; Baak, J. P. A. Microsatellite instability in colorectal cancer. British Journal of Surgery 2006, 93, 395–406. [Google Scholar] [CrossRef]
- Boland, C. R.; Goel, A. Microsatellite Instability in Colorectal Cancer. Gastroenterology 2010, 138, 2073–2087e3. [Google Scholar] [CrossRef] [PubMed]
- Vilar, E.; Gruber, S. B. Microsatellite instability in colorectal cancer—the stable evidence. Nature Reviews Clinical Oncology 2010, 7, 153–162. [Google Scholar] [CrossRef]
- Pan, X.-M.; Yang, W.-Z.; Xu, G.-H.; Bai, P.; Qin, H.-J.; Zhang, L.-S.; et al. The association between MLH1 -93 G>A polymorphism of DNA mismatch repair and cancer susceptibility: a meta-analysis. Mutagenesis 2011, 26, 667–673. [Google Scholar] [CrossRef]
- Zare, M.; Jafari-Nedooshan, J.; Jafari, M.; Neamatzadeh, H.; Abolbaghaei, S. M.; Foroughi, E.; et al. Relevance of hMLH1 -93G>A, 655A>G and 1151T>A polymorphisms with colorectal cancer susceptibility: a meta-analysis based on 38 case-control studies. Revista da Associação Médica Brasileira 2018, 64, 942–951. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Sima, L.; Shi, L.; Wang, Z.; Ni, C.; et al. Association between MLH1 -93G>A Polymorphism and Risk of Colorectal Cancer. PLoS ONE 2012, 7, e50449. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xu, X.; Chen, H.; Wang, J.; Xiong, W.; Xu, Y.; et al. The hMLH1 promoter polymorphisms and cancer susceptibility in Asian populations: A meta-analysis. Gene 2013, 523, 199–204. [Google Scholar] [CrossRef]
- Law, P. J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nature Communications 2019, 10, 2154. [Google Scholar] [CrossRef]
- Pardini, B.; Corrado, A.; Paolicchi, E.; Cugliari, G.; Berndt, S. I.; Bezieau, S.; et al. DNA repair and cancer in colon and rectum: Novel players in genetic susceptibility. International Journal of Cancer 2020, 146, 363–372. [Google Scholar] [CrossRef]
- Koshy, L.; Anju, A. L.; Harikrishnan, S.; Kutty, V. R.; Jissa, V. T.; Kurikesu, I.; et al. Evaluating genomic DNA extraction methods from human whole blood using endpoint and real-time PCR assays. Molecular Biology Reports 2017, 44, 97–108. [Google Scholar] [CrossRef]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Frontiers in Genetics 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Mik, M.; Dziki, L.; Malinowska, K.; Trzcinski, R.; Majsterek, I.; Dziki, A. Polymorphism of MSH2 Gly322Asp and MLH1 –93G>A in non-familial colon cancer – a case-controlled study. Archives of Medical Science 2017, 6, 1295–1302. [Google Scholar] [CrossRef]
- Wang, Y.; Li, G.; Hu, F.; Bi, H.; Wu, Z.; Zhao, X.; et al. The prognostic significance of polymorphisms in hMLH1/hMSH2 for colorectal cancer. Medical Oncology 2014, 31, 975. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Hu, F.; Yuan, F.; Fan, J.; Yu, Z.; Wu, Z.; et al. Intronic and promoter polymorphisms of hMLH1/hMSH2 and colorectal cancer risk in Heilongjiang Province of China. Journal of Cancer Research and Clinical Oncology 2015, 141, 1393–1404. [Google Scholar] [CrossRef]
- Zhang, L.-L.; Tang, X.-J.; Wang, X.-Y.; Zhu, Y.-W.; Peng, X.-B.; Gong, L. A promoter polymorphism in the hMLH1 gene (−93G/A) associated with sporadic colorectal cancer. Oncology Letters 2016, 12, 4035–4040. [Google Scholar] [CrossRef]
- Chen, H.; Shen, Z.; Hu, Y.; Xiao, Q.; Bei, D.; Shen, X.; et al. Association between MutL homolog 1 polymorphisms and the risk of colorectal cancer: a meta-analysis. Journal of Cancer Research and Clinical Oncology 2015, 141, 2147–2158. [Google Scholar] [CrossRef]
- Oztas, E.; Cilingir, U.; Akyuz, A.; Yanar, H. T.; Ozhan, G. MLH1 -93G>A and I219V polymorphisms are susceptible to increased risk of sporadic colorectal cancer in a Turkish population. Istanbul Journal of Pharmacy 2017, 47, 63–67. [Google Scholar] [CrossRef]
- Nizam, Z. M.; Abdul Aziz, A. A.; Kaur, G.; Abu Hassan, M. R.; Mohd Sidek, A. S.; Lee, Y. Y.; et al. Contribution of the MLH1 -93G>A Promoter Polymorphism in Modulating Susceptibility Risk in Malaysian Colorectal Cancer Patients. Asian Pacific Journal of Cancer Prevention 2013, 14, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Allan, J. M.; Shorto, J.; Adlard, J.; Bury, J.; Coggins, R.; George, R.; et al. MLH1 −93G>A promoter polymorphism and risk of mismatch repair deficient colorectal cancer. International Journal of Cancer 2008, 123, 2456–2459. [Google Scholar] [CrossRef] [PubMed]
- Pongsavee, M.; Wisuwan, K.; Pongsavee, K. MLH1 rs1800734 Pathogenic Variant among Patients with Colorectal Cancer in the Lower Northeastern Region of Thailand. Asian Pacific Journal of Cancer Prevention 2023, 24, 2911–2916. [Google Scholar] [CrossRef]
- Liu T; Stathopoulos P; Lindblom P; Rubio C; Wasteson Arve B; Iselius L; et al. Correspondence. European Journal of Cancer 1998, 34(12), 1981. [CrossRef]
- Romanowicz, H.; Bieńkiewicz, J.; Szaflik, T.; Malinowski, J.; Smolarz, B. Association between Gly322Asp polymorphism of hMSH2 (1032G>A, rs4987188) and endometrial cancer. International Journal of Clinical and Experimental Pathology 2017, 10, 2199–2204. [Google Scholar]
- Smolarz, B.; Makowska, M.; Samulak, D.; Michalska, M. M.; Romanowicz, H. Gly322Asp and Asn127Ser single nucleotide polymorphisms (SNPs) of hMSH2 mismatch repair gene and the risk of triple-negative breast cancer in Polish women. Familial Cancer 2015, 14, 81–88. [Google Scholar] [CrossRef]
- Campbell, P. T.; Curtin, K.; Ulrich, C. M.; Samowitz, W. S.; Bigler, J.; Velicer, C. M.; et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut 2009, 58, 661–667. [Google Scholar] [CrossRef]
- Samowitz, W. S.; Curtin, K.; Wolff, R. K.; Albertsen, H.; Sweeney, C.; Caan, B. J.; et al. The MLH1 −93 G>A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes, Chromosomes and Cancer 2008, 47, 835–844. [Google Scholar] [CrossRef]
- Whiffin, N.; Broderick, P.; Lubbe, S. J.; Pittman, A. M.; Penegar, S.; Chandler, I.; et al. MLH1-93G > A is a risk factor for MSI colorectal cancer. Carcinogenesis 2011, 32, 1157–1161. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).