Introduction
The escalating menace of cancer has prompted researchers to explore potential targets for therapeutic interventions in cancer drug delivery [
1]. Maintaining a delicate balance between cell proliferation and apoptosis is vital for cellular homeostasis, and any disruption in this equilibrium can lead to various pathological conditions, including cancer [
2]. Challenges in cancer therapeutics arise from issues such as delayed detection, indiscriminate systemic distribution, resistance to multiple drugs, and inadequate drug concentration at the tumor site [
3].
Breast cancer is a predominant global cause of mortality, with its incidence rapidly rising in India. According to Globocan data 2022, BC accounted for 11.6% of all cancer cases and 6.9% of all deaths [
4]. Despite the growing global research efforts, the pathogenesis of this disease remains largely elusive. Consequently, there is a pressing need to develop potential therapeutic strategies to overcome these obstacles. Furthermore, it has been recognized that targeting specific molecular processes involved in cancer development could offer a more precise treatment strategy.
Targeting genes associated with angiogenesis, cell motility, invasion, and metastasis using shRNA libraries presents a formidable challenge in cancer treatment [
5]. Numerous studies have reported successful utilization of RNA interference (RNAi) to inhibit cancer progression in both
in vitro and
in vivo models [
6]. Short hairpin RNA (shRNA) is a specialized RNA sequence forming a tight hairpin turn, capable of suppressing gene expression through RNA interference (RNAi). The contemporary RNAi model involves both initiation and effector stages. Initially, double-stranded RNAs undergo processing into 20-25 nucleotide (nt) small interfering RNAs (siRNAs) through the action of an RNase III-like enzyme called Dicer (initiation step). Subsequently, siRNAs assemble into endoribonuclease-containing complexes known as RNA-induced silencing complexes (RISCs), unwinding during the process. The siRNA strands then guide the RISCs to complementary RNA molecules, leading to cleavage and degradation of the targeted RNA (effector step). Cleavage of the targeted RNA occurs near the middle of the region bound by the siRNA strand [
6]. The activation of RISC involves ATP-dependent unwinding of the siRNA duplex. Consequently, mRNA degradation results in the silencing of the target gene [
6,
7].
shRNA libraries can be employed to introduce gene-specific shRNA into mammalian cells using lentiviral vectors. The primary objective of this study is to target key genes associated with angiogenesis and metastasis that are overexpressed in breast cancer by utilizing their respective shRNA libraries. Silencing these cancer-specific genes demonstrate a novel therapeutic approach for inhibiting tumor growth and angiogenesis in breast cancer. This strategy aims to enhance our comprehension of shRNA-based targeted delivery in cancer therapy, paving the way for subsequent preclinical and clinical investigations.
Protein phosphatase, Mg2+/Mn2+ dependent, 1D (PPM1D), also known as wild-type p53 inducible protein 1 (Wip1) phosphatase, belongs to the protein phosphatase 2C (PP2C) family of Ser/Thr protein phosphatases [
8]. It is emerging as an oncogene due to its inhibitory influence on several tumor suppressor pathways, including ATM, CHK2, p38 MAPK, and p53. PPM1D exhibits overexpression and/or mutations in various human primary cancers, particularly in breast tumors, correlating with tumor progression and poor prognosis [
9,
10].
Bulavin et al. were the first to investigate the association between PPM1D and tumors. Their study involved assessing PPM1D mRNA levels in various cell types including human embryonic fibroblasts, breast cancer cells, ovarian cancer cells, non-small cell lung cancer cells, renal cancer cells, and T lymphocyte leukemia cell lines. Significantly, they identified elevated level of PPM1D in BT474 and MCF7 cell lines. This research not only uncovered a pivotal link between PPM1D and cancer but also underscored the importance of breast cancer in PPM1D-related tumor research [
11]. Recent advancements in the field have unveiled the potential of anti-PPM1D-directed therapies to delay tumor onset or reduce tumor burden.
PPM1D, an oncogene have been observed in various cancers such as breast, prostate, ovarian, neuroblastoma, medulloblastoma, pancreatic adenocarcinoma, gastric carcinoma, papillary thyroid cancer, hepatocellular carcinoma, colorectal, and bladder transitional cell carcinomas [
4]. Notably, gain-of-function of PPM1D have been detected in primary tumors, reinforcing the concept that inhibiting PPM1D could play a crucial therapeutic role in suppressing tumor growth and evolution. Several chemical inhibitors, including orally active allosteric inhibitors of PPM1D, have been recently discovered [
4,
12,
13].
The PPM1D gene, located on chromosome 17q23 in humans, is composed of six exons [
8]. The PPM1D protein comprises three domains: the N-terminus, the phosphatase domain, and the C-terminus. Furthermore, the phosphatase domain of PPM1D shows evolutionary conservation with that of other members of the protein phosphatase 2C (PP2C) family of Ser/Thr phosphatases [
9]. Genomic abnormalities involving PPM1D may manifest as amplifications of chromosome 17q, particularly observed in ovarian and breast cancer cases [
6,
14].
Among the diverse human cancers affected by PPM1D, mammary gland cancers have been the subject of intensive study. Therefore, PPM1D, encoding PPM1D, a serine-threonine phosphatase previously identified as the driver oncogene of a 17q23 amplicon found in approximately 15% of human breast tumors, is likely to play a significant role in tumorigenesis. Given its therapeutic importance, we opted for PPM1D as a target and achieved stable downregulation using lentiviral shRNA [
15].
Materials and Methods
Cell culture- The breast cancer cell line, MDA-MB-231 was cultured in L-15 supplemented with 10% fetal bovine serum, 100 units penicillin and 100μg/ml streptomycin in a humidified atmosphere without CO 2 at 37°C.
Transduction- MDA-MB-231 breast cancer cells were transduced with gene specific recombinant lentiviral particle as per the standard instructions of the manufacturer. Cells were cultured for long-term in puromycin containing medium to select specific clones.
MTT Assay- The MTT assay was performed as per standard procedure. Briefly, MDA-MB-231 cells have been transduced with lentiviral shRNA specific to metastasis and angiogenesis genes. MTT (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide) was added to the wells and the cells were incubated for 3 h. Formazan crystals were dissolved with 200 µl isopropanol. Optical density of formazan solution was read using a spectrophotometer at 570 nm. The data was analyzed and represented as percent cell survival.
Wound migration assay- The wound migration assay was performed using MDA- MB-231 cells. Briefly, MDA-MB-231 cells (3x105 per well), both clones and un-transduced control cells, were grown as monolayer in a 12 well culture plate, wound with uniform size was made using sterile micro tip and cells were photographed at t=0 h and t=16 h by phase contrast microscope (Nikon, Melville, NY, USA). Wound closure was measured by Image-Pro plus software, analyzed statistically and represented in the form of bar graph.
Cell migration assay- The migration assay was performed using Transwell cell culture chambers (Corning, NY, USA) according to standard protocol. Briefly, MDA- MB-231 (1x105 ) cells (both clones and un-transduced) were seeded in upper part of Boyden chamber. 10 % FBS was used as chemoattractant in the lower chamber. After incubating at 37°C for 16 h, the cells that migrated to the lower surface of the membrane were fixed with methanol, stained with Giemsa and photographed under an inverted microscope (Nikon, Melville, NY, USA). Cells were counted from five different images for each well, analyzed statistically and represented in the form of bar graph.
RNA isolation, cDNA synthesis and qPCR analysis- Total RNA from stably transduced MDA-MB-231 cells were isolated by Trizol RNA isolation protocol and quantified spectrophotometrically. The isolated RNA was reverse transcribed, analyzed and quantified by using Real Time-SYBRgreen qPCR as per standard protocol.
In vivo tumorigenicity- To study the effect of shRNAs on tumorigenicity, 1 x 106 of MDA-MB-231 cells stably transduced with PPM1D specific shRNA with equal volume of Matrigel were injected orthotopically into NOD-SCID mice. The mice were kept under pathogen-free condition as per Experimental Animal Facility guidelines to develop tumors. Tumor volumes were measured twice a week. After 6 weeks, mice were sacrificed and the tumors were dissected.
Result
Table 1.
Breast cancer metastasis and angiogenesis gene-specific shRNA Library (96 well plate format) details.
Table 1.
Breast cancer metastasis and angiogenesis gene-specific shRNA Library (96 well plate format) details.
Format of the
Library |
Metastasis |
Angiogenesis |
| Gene No |
Clone No |
Gene No |
Clone No |
| Lentiviral |
146 |
514 |
42 |
210 |
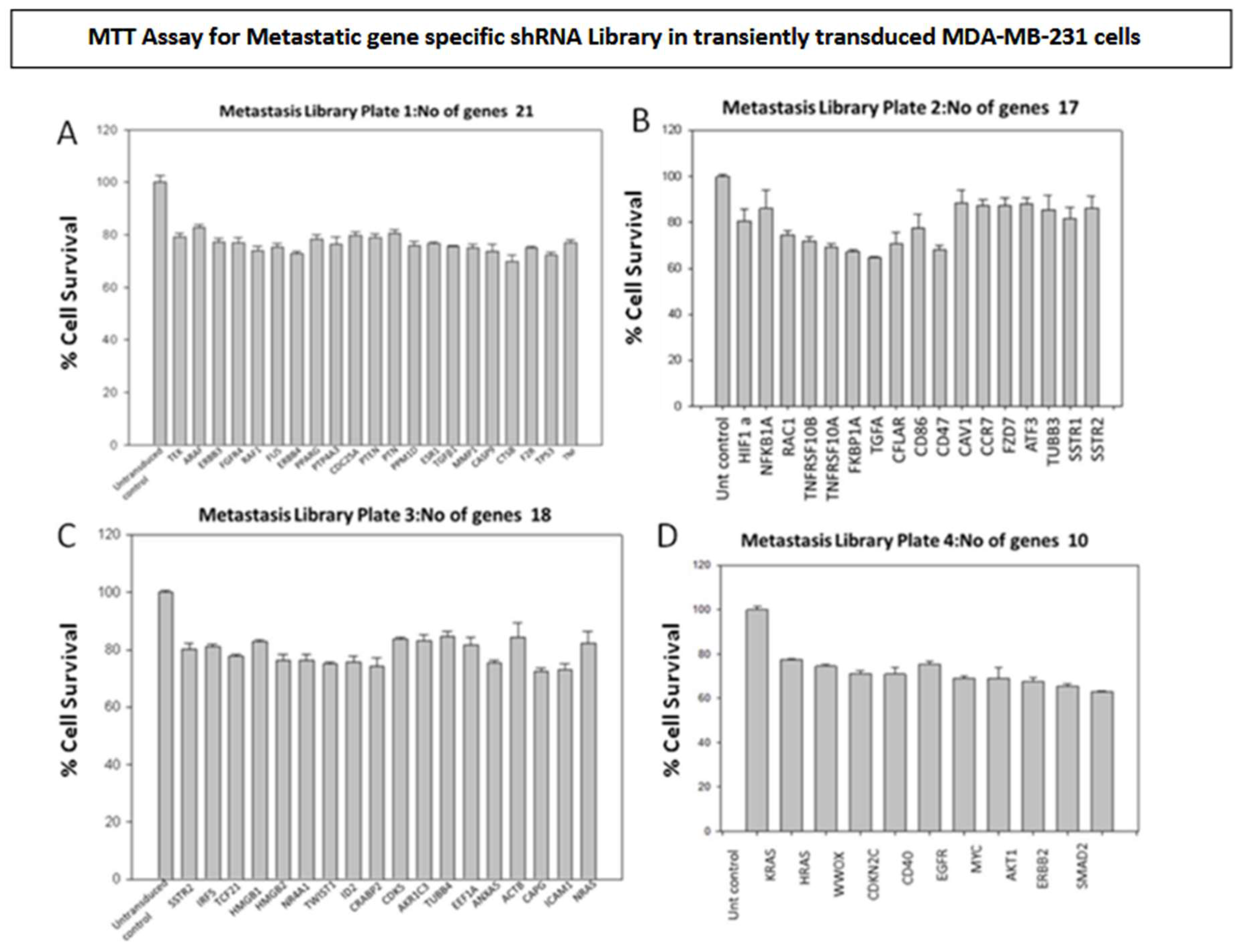
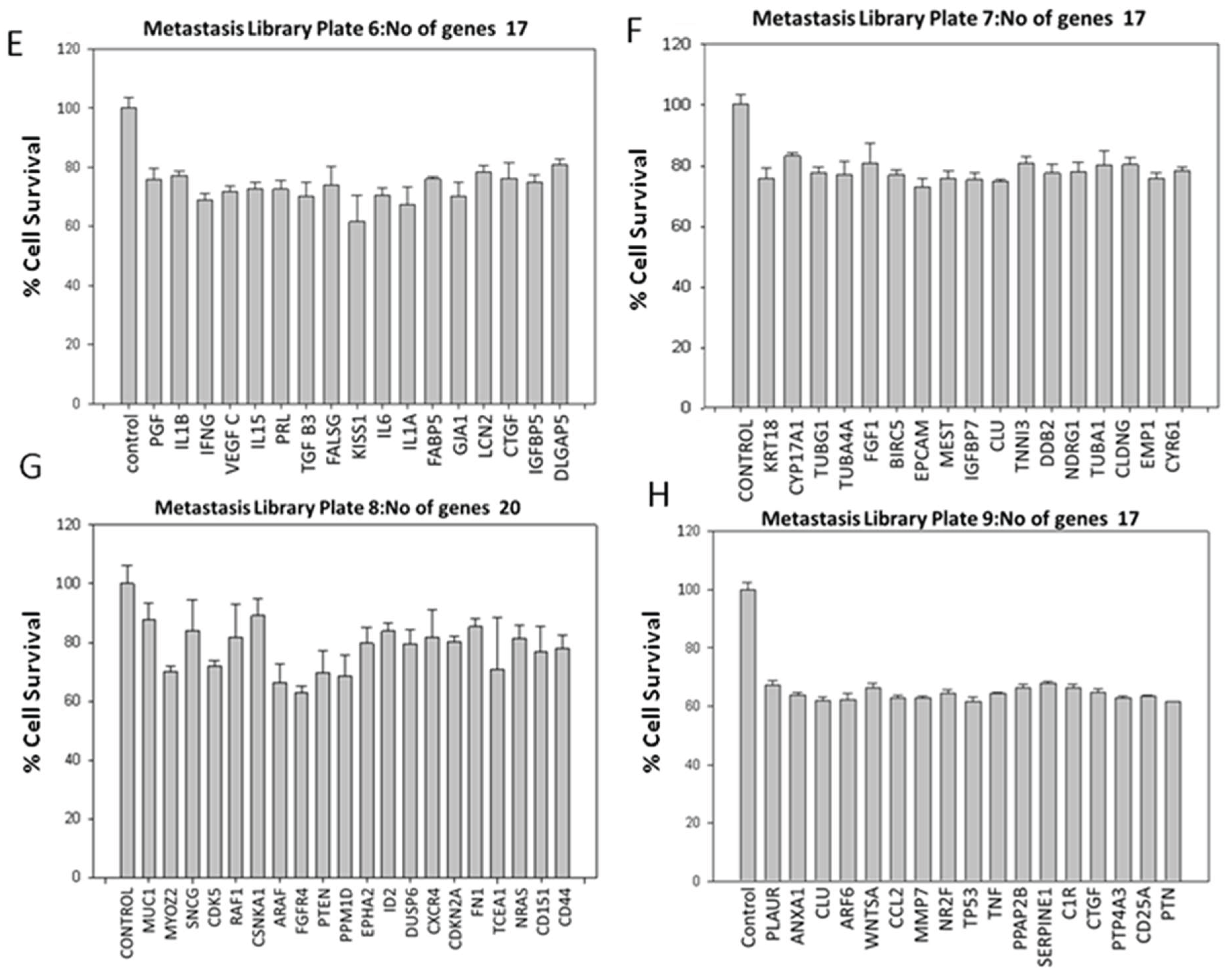
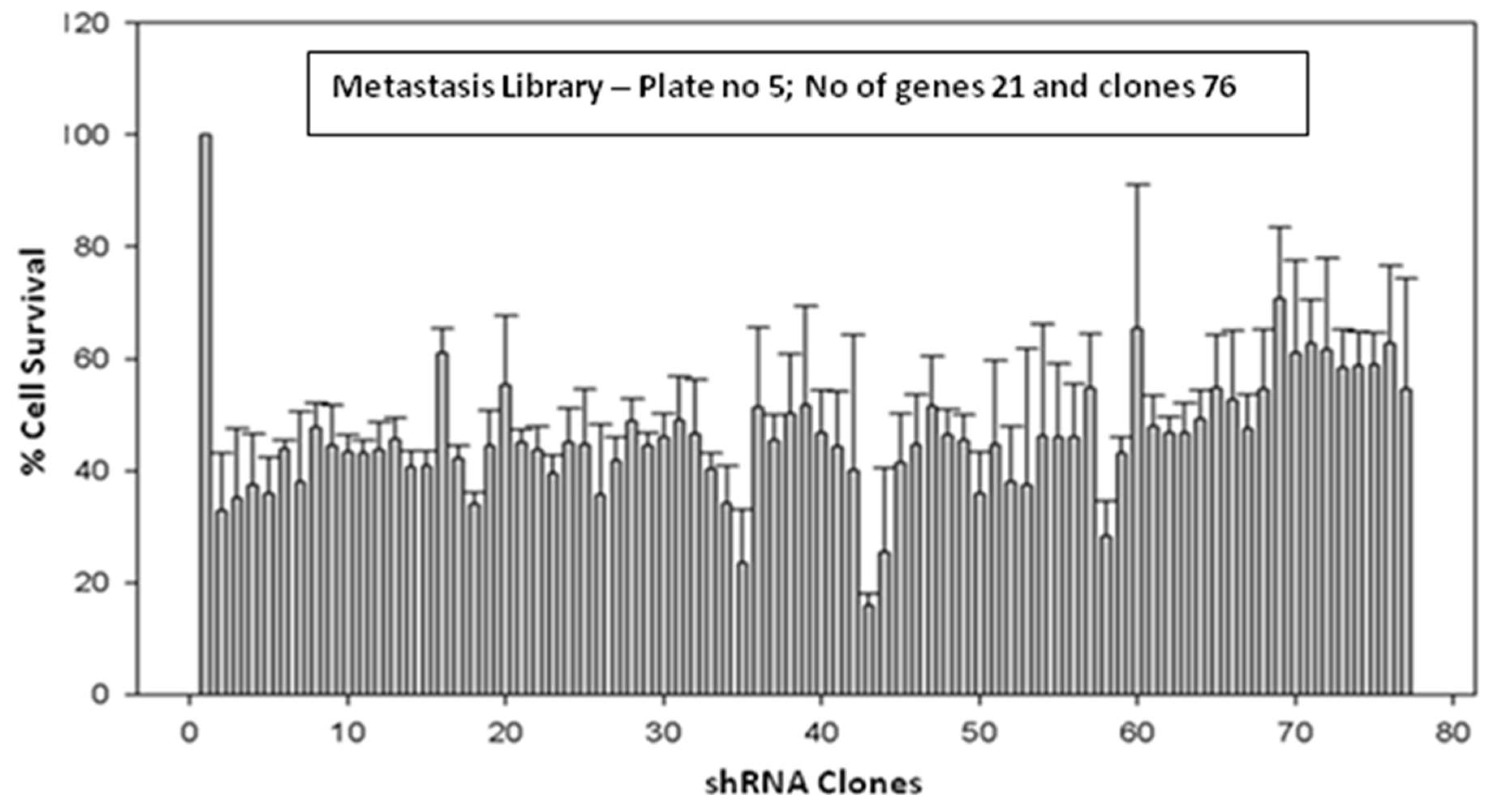
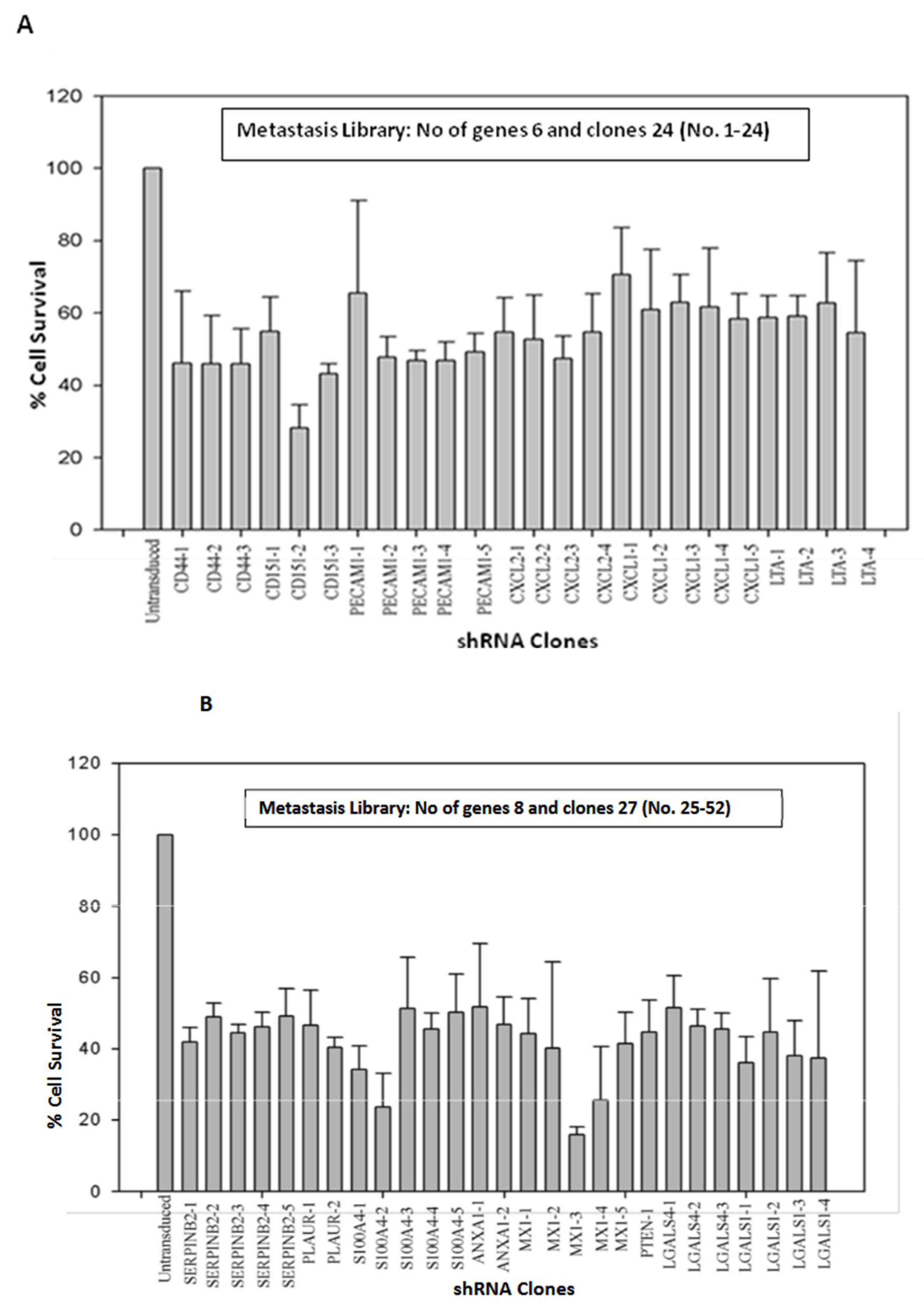
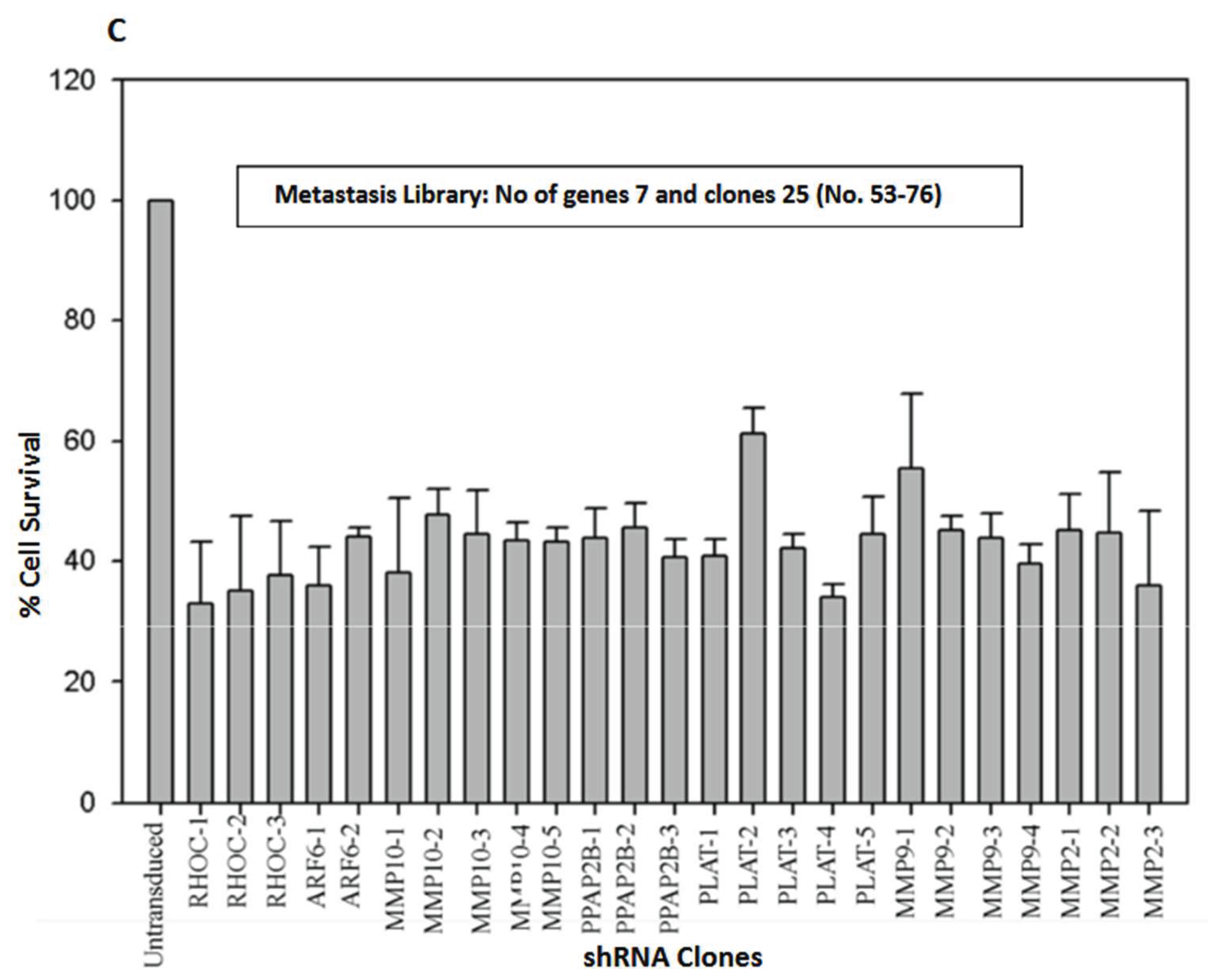
MDA-MB-231 breast cancer cells were transiently transduced by shRNA Library with Multiplicity of Infection of 0.5. The shRNA library screening for the transduced cells was conducted using the MTT assay on Plate number 5, featuring 21 genes and 76 clones (
Figure 1 and
Figure 2 A-C). The metastasis gene-specific library from Plate number 5 was selected for screening process. The cell growth of breast cancer metastasis specific shRNA transiently transduced MDA-MB-231 cells was analyzed by MTT assay and the findings indicated a reduction in the cell growth of shRNA-transduced MDA-MB-231 cells in comparison to the untransduced MDA-MB-231 control cells (
Figure 1 and
Figure 2 A-C).
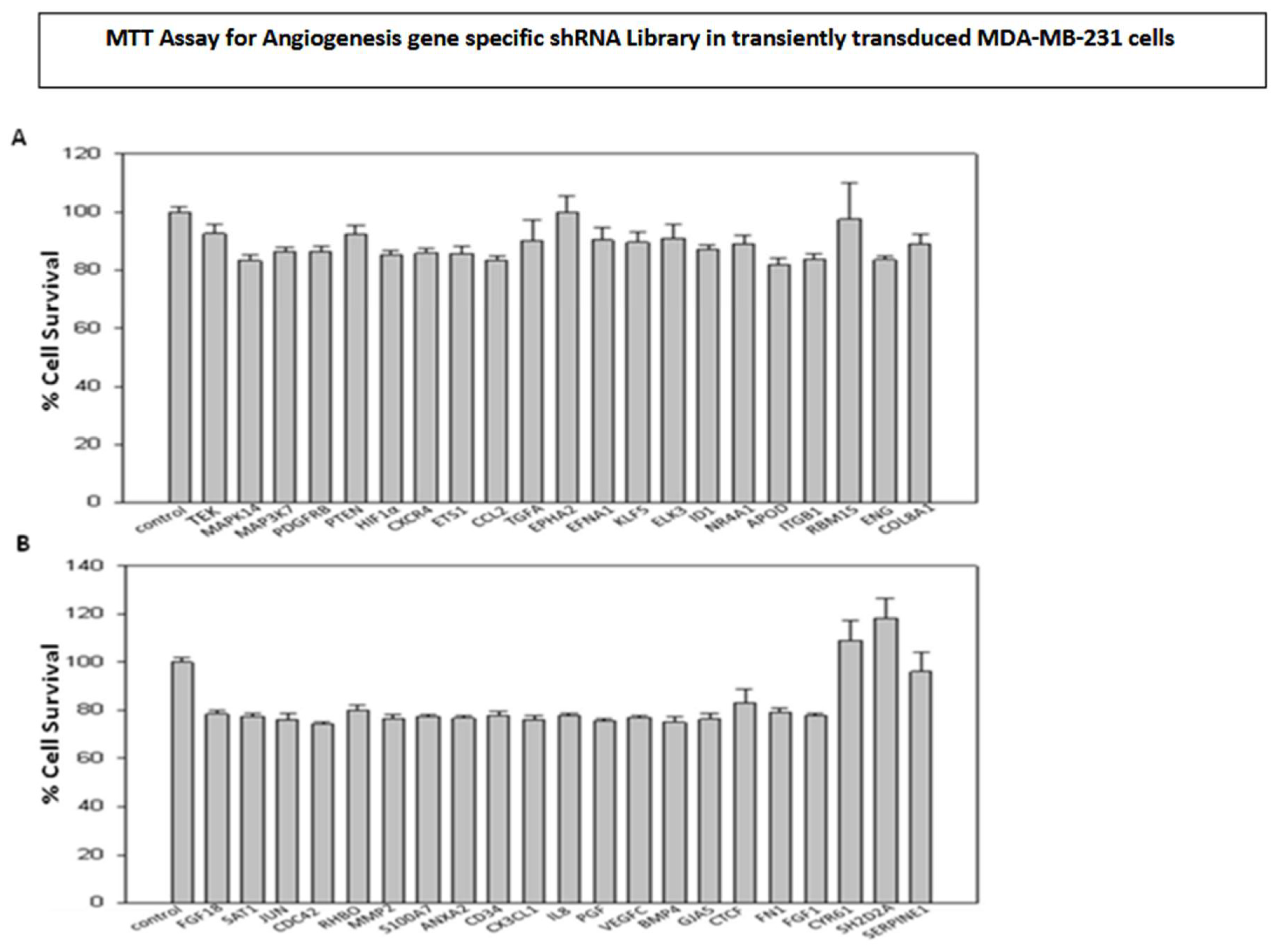
The majority of genes associated with angiogenesis and metastasis are recognized for their role in controlling cell proliferation and survival. Consequently, the shRNA library screening involved the transient transduction of MDA-MB-231 cells with lentiviral particles containing angiogenesis-specific shRNAs (42 genes and 210 clones) (
Figure 3) and metastasis-specific shRNAs (146 genes and 491 clones) (
Figure 1 and Figure 4). Based on the outcomes, genes that demonstrated a decrease in cell proliferation compared to the control were chosen for subsequent assays.
Figure 3.
(A & B): Effect of transiently transduced breast cancer angiogenesis gene specific shRNA library on survival of MDA-MB-231 cells.
Figure 3.
(A & B): Effect of transiently transduced breast cancer angiogenesis gene specific shRNA library on survival of MDA-MB-231 cells.
Figure 4.
(A-D): Effect of transiently transduced breast cancer metastasis gene-specific library on survival of MDA-MB-231 cells.
Figure 4.
(A-D): Effect of transiently transduced breast cancer metastasis gene-specific library on survival of MDA-MB-231 cells.
Figure 4.
(E-H): Effect of transiently transduced breast cancer metastasis gene specific library on survival of MDA-MB-231 cells.
Figure 4.
(E-H): Effect of transiently transduced breast cancer metastasis gene specific library on survival of MDA-MB-231 cells.
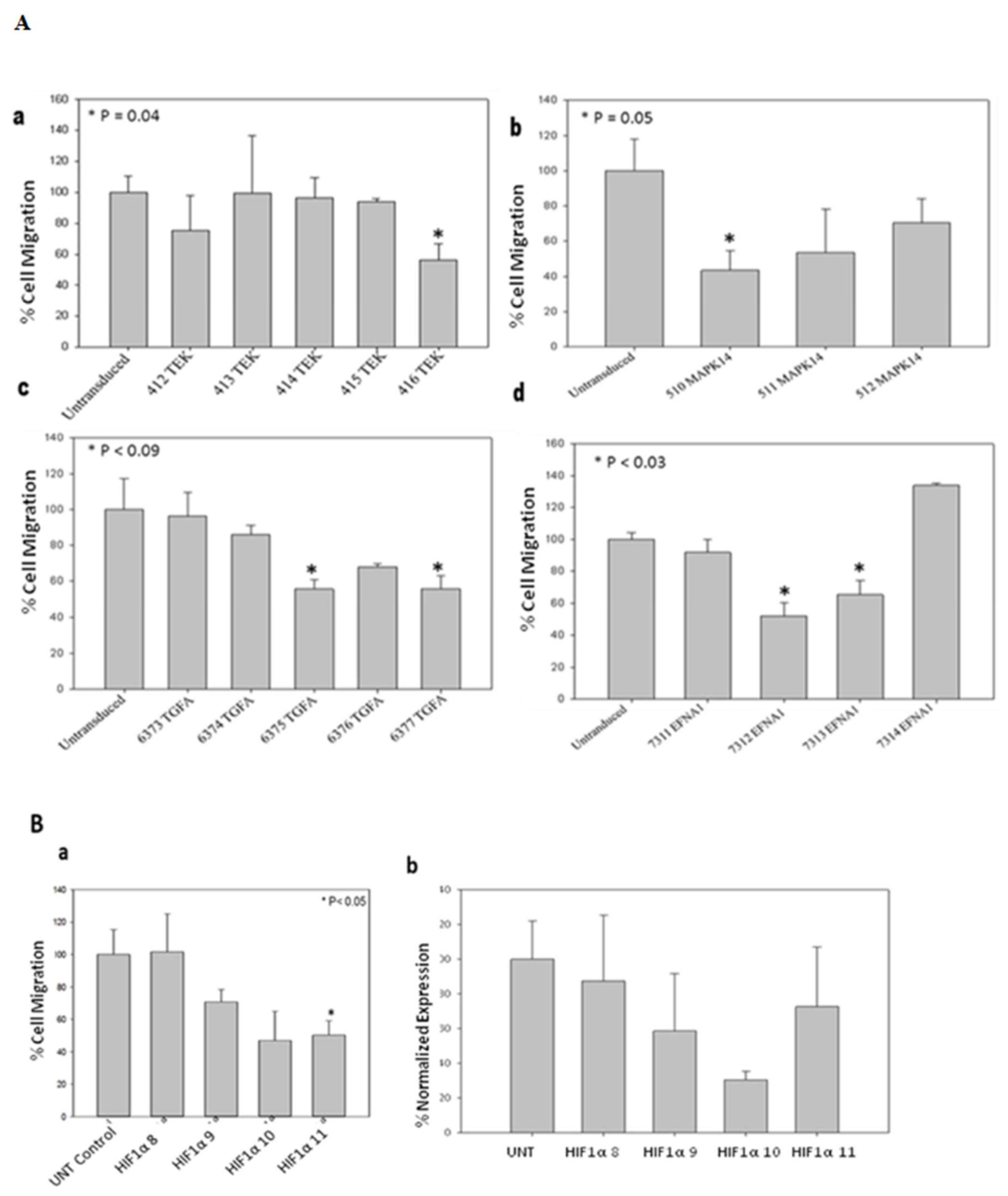
Based on the screening results of the angiogenesis gene-specific library, the TEK, MAPK14, TGFA, and EFNA1 genes exhibited the highest inhibition of cell proliferation when transiently transduced into MDA MB 231 breast cancer cells. Stable clones of these selected genes were established through puromycin selection in MDA MB 231 cells. Subsequently, a wound healing assay was conducted to assess the impact of the shRNA clones on cell migration (
Figure 5A). HIF1α was singled out for further investigations, considering its significance in tumor angiogenesis. To explore its role in both cell proliferation and migration, MDA-MB-231 cells were stably transduced with HIF1α-specific shRNA lentiviral particles. The gene expression of HIF1α in the stably transduced MDA-MB-231 cells was examined through RT-qPCR (
Figure 5B).
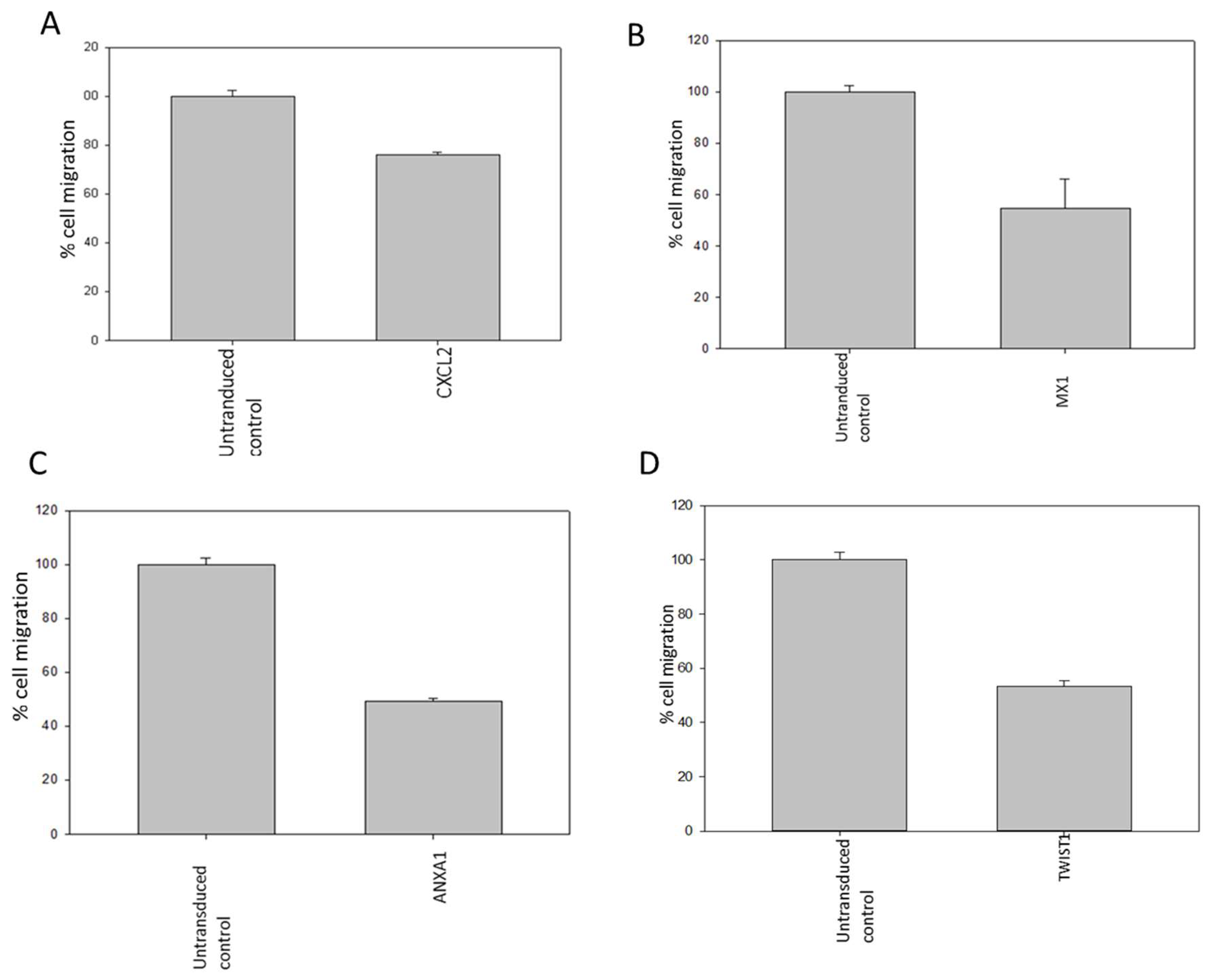
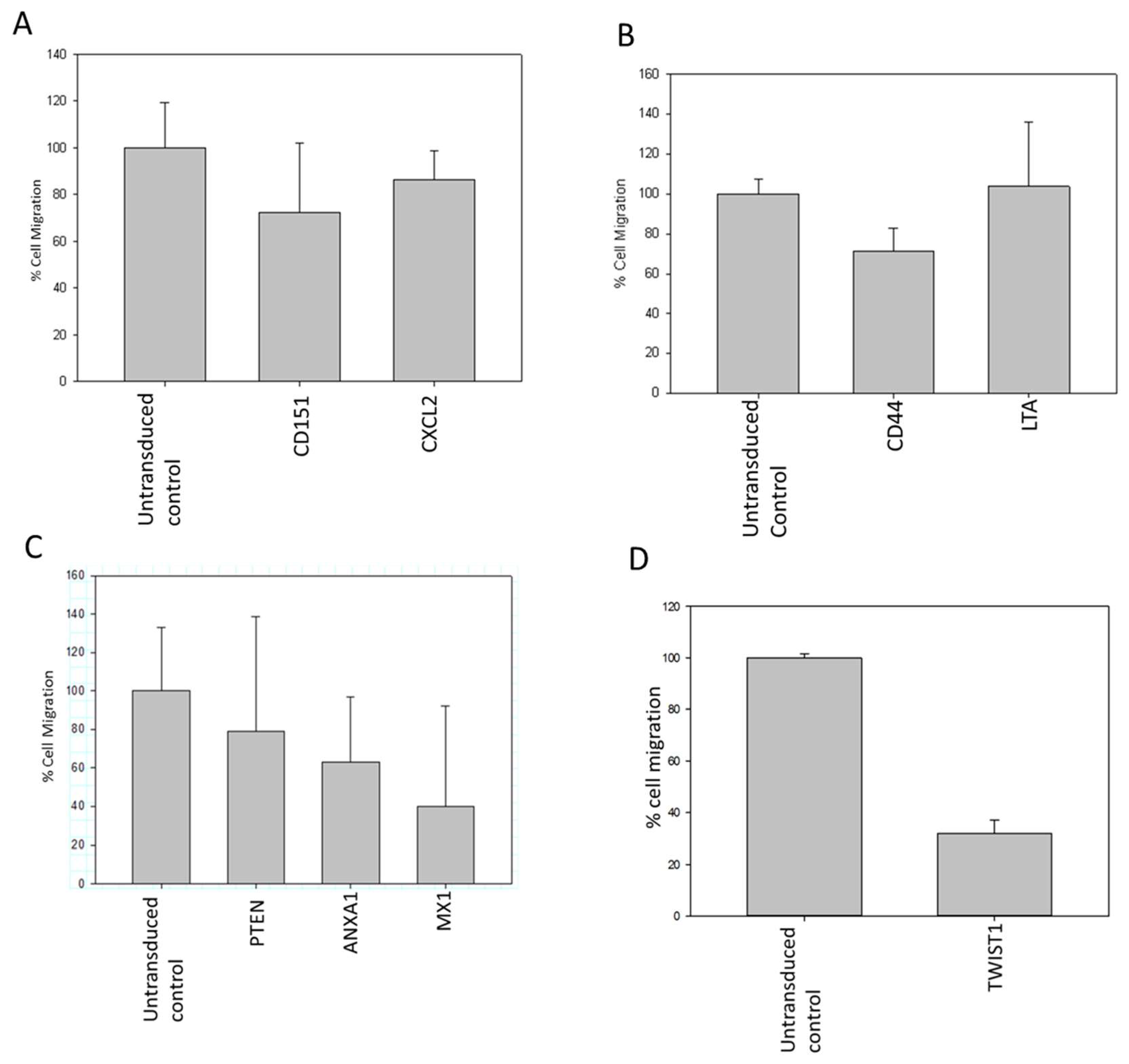
Following the outcomes of the metastasis gene-specific library screening, CXCL2, MX1, ANXA1, and TWIST1 genes, which exhibited diminished cell proliferation, were chosen for further investigation. Stable clones of these selected genes were established through puromycin selection in MDA MB 231 cells. To study the metastatic properties of the transduced cells, a cell migration assay using the Boyden chamber was conducted (
Figure 6). CD151, CXCL2, CD44, LTA, PTEN, ANXA1, MX1, and TWIST1, identified for their significance in breast cancer metastasis, were selected as metastasis-specific genes. Subsequently, a wound healing assay was performed to assess the impact of shRNA transduction on cell migration (
Figure 7).
PPM1D is frequently overexpressed and/or mutated in various human primary cancers. Additionally, elevated PPM1D expression has been linked to tumor progression and poor prognosis in several cancers, including breast. Recent advancements in the field have unveiled the potential of anti-PPM1D-directed therapies in delaying tumor onset or reducing the tumor burden. In line with PPM1D's oncogenic function, its amplification and/or overexpression have been observed in various tumors [
12].
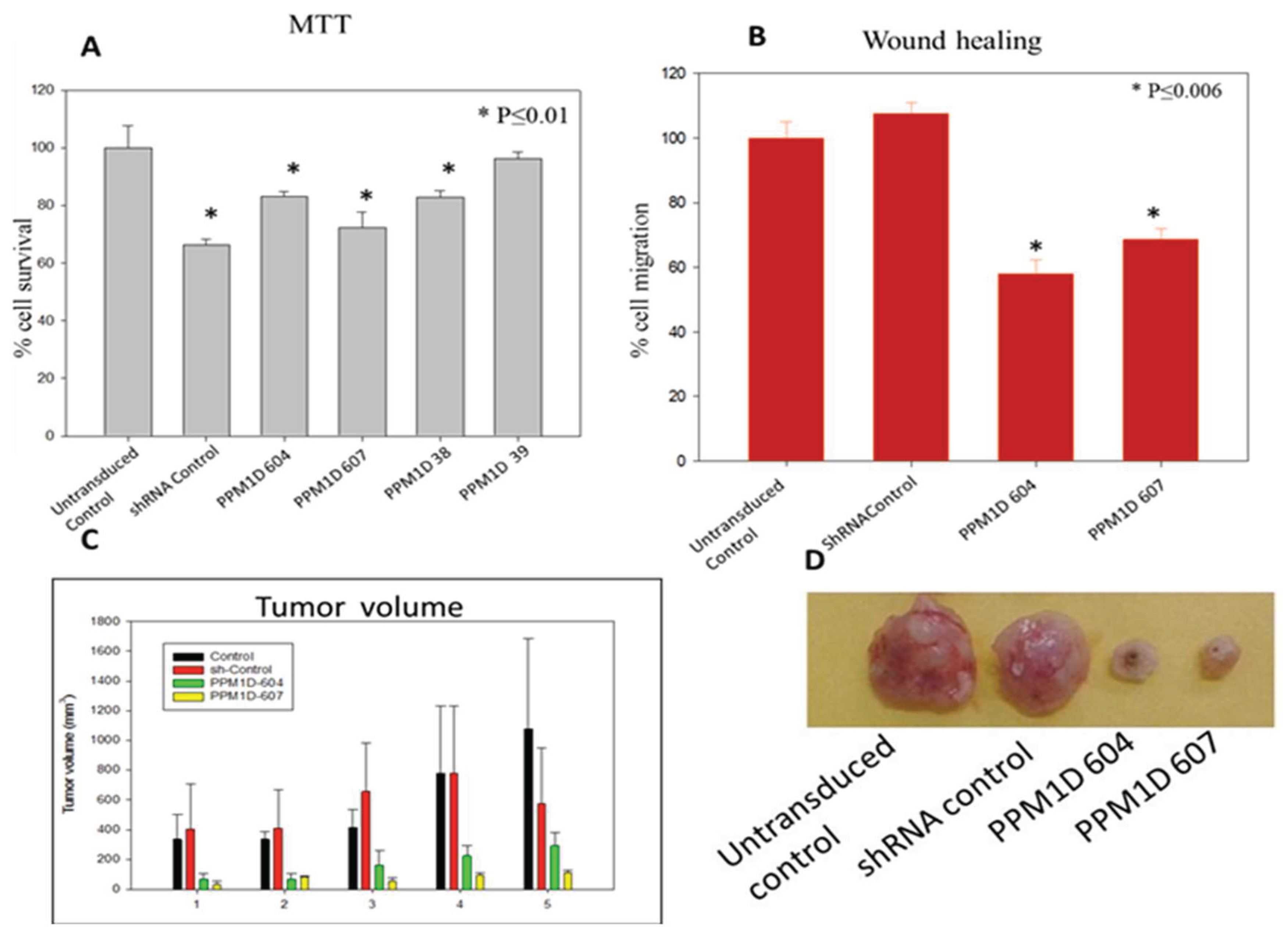
Given its therapeutic relevance, we have selected PPM1D as a target and achieved stable downregulation using lentiviral shRNA. The silencing of PPM1D in cancer cells resulted in decreased cell proliferation and migration, as confirmed by MTT and wound migration assays (Figure 8 A & B). To assess the impact of shRNAs on tumorigenicity, 1 x 106 of MDA-MB-231 cells stably transduced with PPM1D-specific shRNA were orthotopically injected with an equal volume of Matrigel into NOD-SCID mice. The mice were maintained under pathogen-free conditions in accordance with Experimental Animal Facility (EAF) guidelines for tumor development. Tumor volumes were measured twice a week, and after 6 weeks, mice were sacrificed, and the tumors were dissected. Tumorigenicity results revealed reduced tumor volumes in PPM1D knocked-down tumor xenografts compared to the control (Figure 8 C & D).
Discussion
The shRNA library targeting angiogenesis and metastasis genes was screened using an MTT assay to assess cell viability. MDA MB 231 breast cancer cells were transduced with lentiviral shRNAs specific to these genes. Subsequent analysis evaluated the impact of these shRNA clones on cancer cell proliferation and migration. Promising shRNA clones exhibiting significant effects were chosen for further investigation. To gauge the influence of selected clones on MDA-MB-231 breast cancer cells' proliferation and migration, MTT, wound healing, and Boyden chamber assays were conducted.
Silencing genes associated with angiogenesis, including TEK, MAPK14, TGFA, and EFNA1 resulted in reduced migration of breast cancer cells compared to un-transduced MDA-MB-231 cells. Interestingly, HIF1α clones demonstrated a substantial decrease in breast cancer cell migration relative to the un-transduced control, with its down-regulation confirmed through RT-qPCR analysis.
Similarly, silencing metastasis-related genes such as CXCL2, MX1, ANXA1, and TWIST1 led to reduced migration of breast cancer cells compared to controls. The wound migration assay revealed attenuated cell migration in MDA MB 231 cells treated with metastasis gene-specific shRNA clones, including CD151, CXCL2, CD44, LTA, PTEN, ANXA1, MX1, and TWIST1.
PPM1D phosphatase plays a crucial role as a negative regulator of the p53 pathway and DNA damage response. When overexpressed, PPM1D disrupts p53 function, promoting tumorigenesis, often in conjunction with the activation of other oncogenes. Amplification, overexpression, or mutation of PPM1D is closely associated with numerous human tumors. While several inhibitors of PPM1D have been identified, selectively inhibiting its phosphatase activity remains a significant challenge. The lack of specific small molecule inhibitors hampers the advancement of PPM1D as a viable pharmacological target for cancer treatment.
Targeting PPM1D in breast cancer cells resulted in decreased proliferation and migration, as evidenced by MTT and wound migration assays. Additionally, tumor volume was diminished in PPM1D knockdown tumor xenografts compared to controls, indicating a suppressive effect on tumor growth. Initially recognized as a gene regulated by p53, PPM1D has since been identified as amplified and more recently mutated in various human cancers, including breast. Recent advancements in this area suggest that selectively inhibiting PPM1D holds promise as an anti-cancer strategy to either delay tumor onset or diminish tumor burden. Continual investigation into PPM1D and its association with cancer is crucial, as it significantly impacts the understanding of cancer's development and potential treatment approaches in the management of breast cancer.
Author Contributions
G.C.K. conceptualized the entire study. P.G., M.B. & G.C.K. wrote the manuscript, analyzed the data and prepared the figures and tables. D.M., and G.C.K. edited the entire manuscript. All authors have read and agreed to publish the manuscript.
Funding
This work was supported by Department of Biotechnology, Govt. of India (Project No. BT/PR4569/AGR/36/711/2012) and Science and Engineering Research Board (SERB), Govt. of India, (Project No. JCB/2023/000011) to G.C.K.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Chehelgerdi, M., Allela, O. Q. B., Pecho, R. D. C., Jayasankar, N., Rao, D. P., Thamaraikani, T., Vasanthan, M., Viktor, P., Lakshmaiya, N., Saadh, M. J., Amajd, A., Abo-Zaid, M. A., Castillo-Acobo, R. Y., Ismail, A. H., Amin, A. H., & Akhavan-Sigari, R. (2023). Progressing nanotechnology to improve targeted cancer treatment: overcoming hurdles in its clinical implementation. Molecular cancer, 22(1), 169. [CrossRef]
- Singh, R., Letai, A., & Sarosiek, K. (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nature reviews. Molecular cell biology, 20(3), 175–193.
- Zargar, A., Chang, S., Kothari, A., Snijders, A. M., Mao, J. H., Wang, J., Hernández, A. C., Keasling, J. D., & Bivona, T. G. (2020). Overcoming the challenges of cancer drug resistance through bacterial-mediated therapy. Chronic diseases and translational medicine, 5(4), 258–266c. [CrossRef]
- Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., & Jemal, A. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 74(3), 229–263.
- Stein EV, Price DK, Figg WD: shRNA technology, investigating Ras-dependent cancer. Cancer Biol Ther 2009, 8:1798–1799.
- Joshua A Bauer, Fei Ye, Clayton B Marshall, Brian D Lehmann, Christopher S Pendleton, Yu Shyr, Carlos L Arteaga and Jennifer A Pietenpol: RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Research 2010, 12: R41.
- Chris B. Moore, Elizabeth H. Guthrie, Max Tze-Han Huang, and Debra J. Taxman: Short Hairpin RNA (shRNA): Design, Delivery, and Assessment of Gene Knockdown. Methods Mol Biol. 2010; 629: 141–158.
- Wenhong Denga,b, Jieqing Lib, Kimberly Dorrahb, Denise Jimenez-Tapiab, Brando Arriagab, Qiongyu Haob, Wei Caob, Zhaoxia Gaoc,d, Jay Vadgamab, Yong Wub: The role of PPM1D in cancer and advances in studies of its inhibitors. Biomedicine & Pharmacotherapy 125 (2020) 109956.
- Chen Zhang, Yuanzhuo Chen, Mingsong Wang, Xianzhen Chen, Yongxin Li, E Song, Xiaoqing Liu, Sekwon Kim and Hu Peng: PPM1D silencing by RNA interference inhibits the proliferation of lung cancer cells. World Journal of Surgical Oncology 2014, 12:258.
- D.V. Bulavin, O.N. Demidov, S. Saito, P. Kauraniemi, C. Phillips, S.A. Amundson,C. Ambrosino, G. Sauter, A.R. Nebreda, C.W. Anderson, A. Kallioniemi, A.J. Fornace Jr., E. Appella: Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat. Genet. 31 (2) (2002) 210–215.
- Linda Zhang, Joanne I. Hsu, and Margaret A. Goodell: PPM1D in Solid and Hematologic Malignancies: Friend and Foe? Mol Cancer Res 2022;20:1365–78.
- Choi J, Appella E, Donehower LA.: The structure and expression of the murine wildtype p53-induced phosphatase 1 (Wip1) gene. Genomics 2000;64:298–306.
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, et al. Wip1: A novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A 1997;94: 6048–53.
- Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, et al.: Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet 2002;31:133–4.
- C.S. Sinclair, M. Rowley, A. Naderi, F.J. Couch: The 17q23 amplicon and breast cancer. Breast Cancer Res. Treat. 78 (3) (2003) 313–322.
- T.S. Peng, Y.H. He, T. Nie, X.D. Hu, H.Y. Lu, J. Yi, Y.F. Shuai, M. Luo: PPM1D is aprognostic marker and therapeutic target in colorectal cancer, Exp. Ther. Med. 8 (2) (2014) 430–434.
Figure 1.
Effect of transiently transduced breast cancer metastasis gene-specific shRNA library (Plate no 5; No of genes 21 and clones 76) on survival of MDA-MB-231 cells.
Figure 1.
Effect of transiently transduced breast cancer metastasis gene-specific shRNA library (Plate no 5; No of genes 21 and clones 76) on survival of MDA-MB-231 cells.
Figure 2.
A. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes CD44, CD151, PECAM1, CXCL2, CXCL1, LTA on survival of MDA-MB-231 cell. B. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes SERPINB2, PLAUR, S100A4, ANXA1, MX1, PTEN, LGALS4, LGALS1 on survival of MDA-MB-231 cells. C. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes RHOC, ARF6, MMP10, PPAP2B, PLAT, MMP9, MMP2 on survival of MDA-MB-231 cells.
Figure 2.
A. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes CD44, CD151, PECAM1, CXCL2, CXCL1, LTA on survival of MDA-MB-231 cell. B. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes SERPINB2, PLAUR, S100A4, ANXA1, MX1, PTEN, LGALS4, LGALS1 on survival of MDA-MB-231 cells. C. Effect of transient transduction with different shRNA lentivectors targeting metastasis specific genes RHOC, ARF6, MMP10, PPAP2B, PLAT, MMP9, MMP2 on survival of MDA-MB-231 cells.
Figure 5.
(A) Effect of stable transduction with different shRNA Lentivirus based targeting angiogenesis specific genes, TEK, MAPK14, TGFA and EFNA1 on migration of MDA-MB-231 cells. (B) Downregulation of HIF1α and its effect on MDA-MB-231 cell migration. (B) a. Wound healing assay to check migratory potential of HIF1α clones. (B) b. Normalized Expression of HIF1α mRNA in stably transduced MDA-MB-231 cells by real time PCR.
Figure 5.
(A) Effect of stable transduction with different shRNA Lentivirus based targeting angiogenesis specific genes, TEK, MAPK14, TGFA and EFNA1 on migration of MDA-MB-231 cells. (B) Downregulation of HIF1α and its effect on MDA-MB-231 cell migration. (B) a. Wound healing assay to check migratory potential of HIF1α clones. (B) b. Normalized Expression of HIF1α mRNA in stably transduced MDA-MB-231 cells by real time PCR.
Figure 6.
(A-D): Reduction in migratory property of MDA-MB-231 cells transduced with CXCL2, MX1, ANXA1 and TWIST1 shRNA as compared to control cells detected by Boyden chamber assay.
Figure 6.
(A-D): Reduction in migratory property of MDA-MB-231 cells transduced with CXCL2, MX1, ANXA1 and TWIST1 shRNA as compared to control cells detected by Boyden chamber assay.
Figure 7.
(A-D): Reduction in wound healing property of MDA-MB-231 cells transduced with CD151, CXCL2, CD44, LTA, PTEN, ANXA1, MX1 and TWIST1 shRNA as compared to control cells detected by scratch assay.
Figure 7.
(A-D): Reduction in wound healing property of MDA-MB-231 cells transduced with CD151, CXCL2, CD44, LTA, PTEN, ANXA1, MX1 and TWIST1 shRNA as compared to control cells detected by scratch assay.
Figure 8.
(A-D): Survival of MDA-MB-231 cells after stable transduction with PPM1D shRNA clones (A). Effect of stable transduction with PPM1D shRNA clones on MDA-MB-231 cell migration by wound healing assay (B). Tumor volume was reduced in PPM1D knocked down tumor xenografts when compared to control (C, D).
Figure 8.
(A-D): Survival of MDA-MB-231 cells after stable transduction with PPM1D shRNA clones (A). Effect of stable transduction with PPM1D shRNA clones on MDA-MB-231 cell migration by wound healing assay (B). Tumor volume was reduced in PPM1D knocked down tumor xenografts when compared to control (C, D).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).