Submitted:
14 August 2023
Posted:
15 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methodology
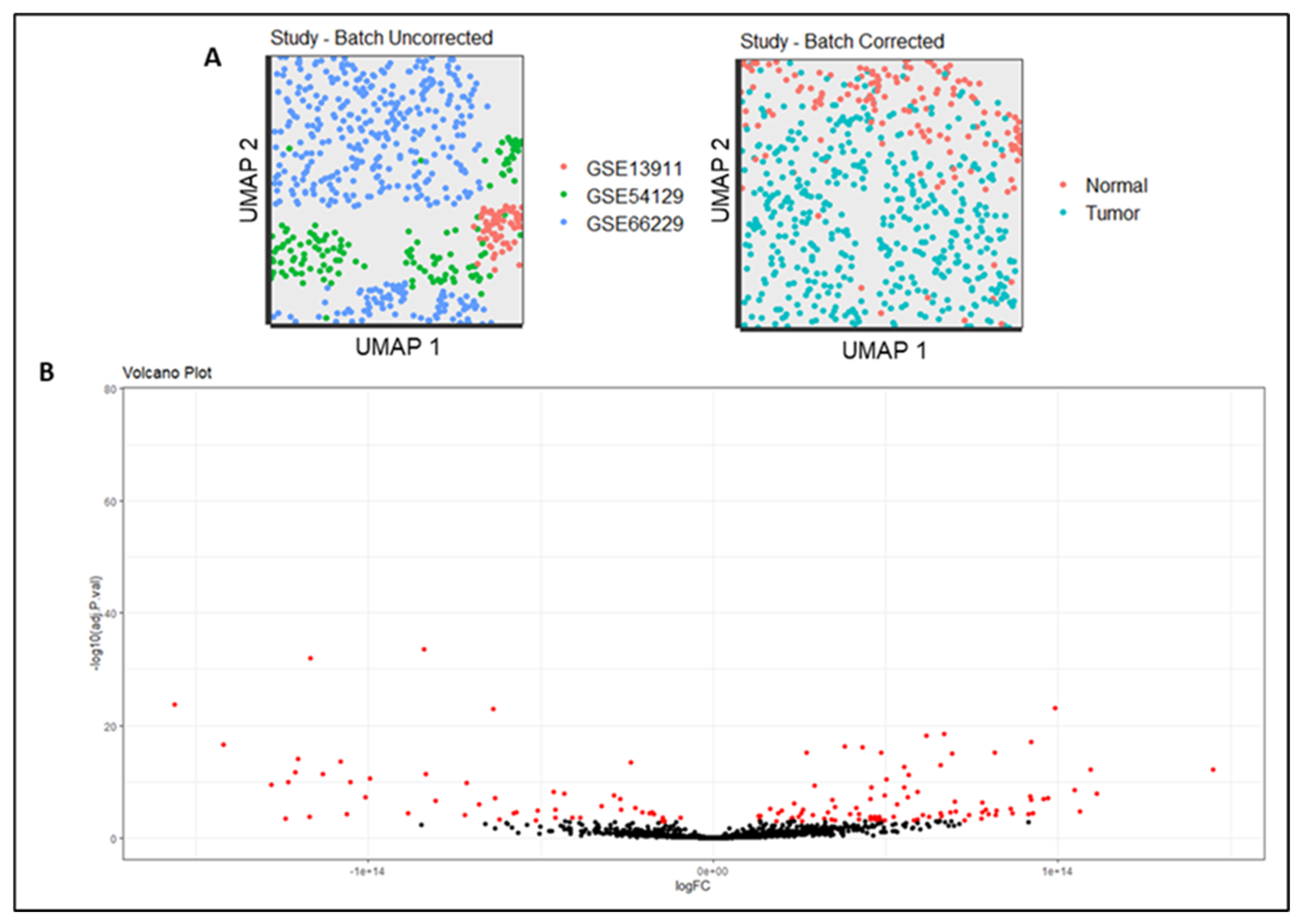
2.1. Selection of Microarray Transcriptome Studies
| Differentially Expressed Genes | |||||||
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| Gene Symbol | LogFC | t | Adjusted p-value | Gene Symbol | LogFC | t | Adjusted p-value |
| AJUBA | 1.44E+14 | 7.80 | 7.78E-13 | FBXL13 | -1.56E+14 | -11.18 | 2.01E-24 |
| GPNMB | 1.11E+14 | 6.20 | 1.53E-08 | PDILT | -1.14+14 | -9.20 | 2.98E-17 |
| CD80 | 1.09E+14 | 7.84 | 5.74E-13 | CCDC69 | -1.28E+14 | -6.83 | 3.98E-10 |
| ANLN | 1.06+14 | 4.82 | 1.61E-05 | PDZK1IP1 | -1.24E+14 | -4.03 | 4.02E-04 |
| ADGRG7 | 1.06E+14 | 4.75 | 2.22E-05 | SCIN | -1.23E+14 | -7.05 | 1.02E-10 |
| BICD1 | 1.04E+14 | 6.47 | 3.44E-09 | ITIH5 | -1.21E+14 | -7.66 | 1.98E-12 |
| KNL1 | 9.91E+13 | 11.01 | 8.37E-24 | NKX2-3 | -1.20E+14 | -8.46 | 7.58E-15 |
| ABCD3 | 9.71E+13 | 5.89 | 8.40E-08 | ITGB1 | -1.17E+14 | -4.27 | 1.59E-04 |
| CENPL | 9.54E+13 | 5.85 | 1.03E-07 | SIGLEC11 | 1.16E+14 | -13.16 | 1.33E-32 |
| PGTS2 | 9.26E+13 | 4.60 | 4.17E-05 | PTCHD1 | -1.13E+14 | -7.52 | 5.07E-12 |
2.2. Data Acquisition and Processing
2.3. Meta-Data Construction
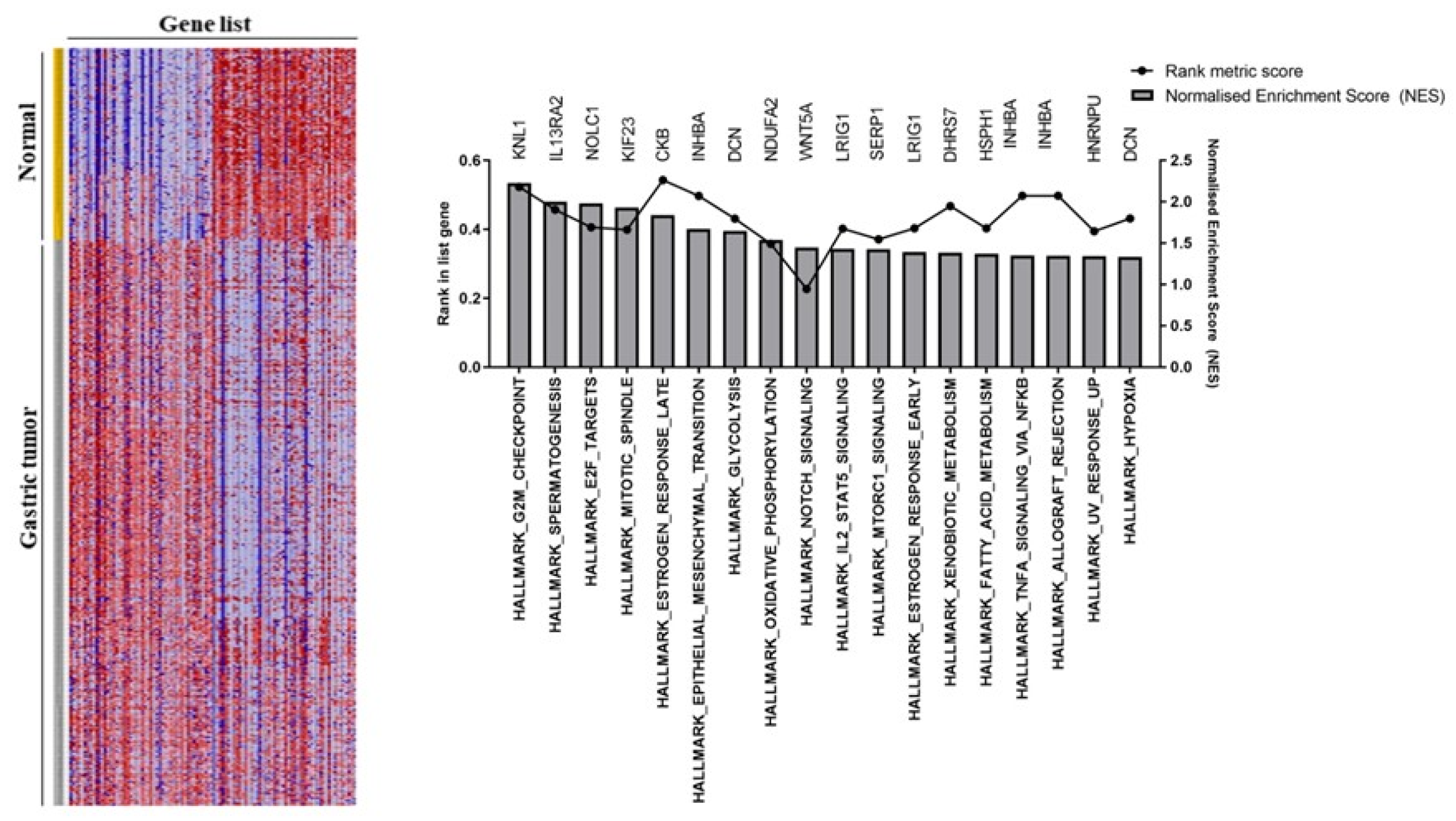
2.4. Gene Set Enrichment Analysis (GSEA)
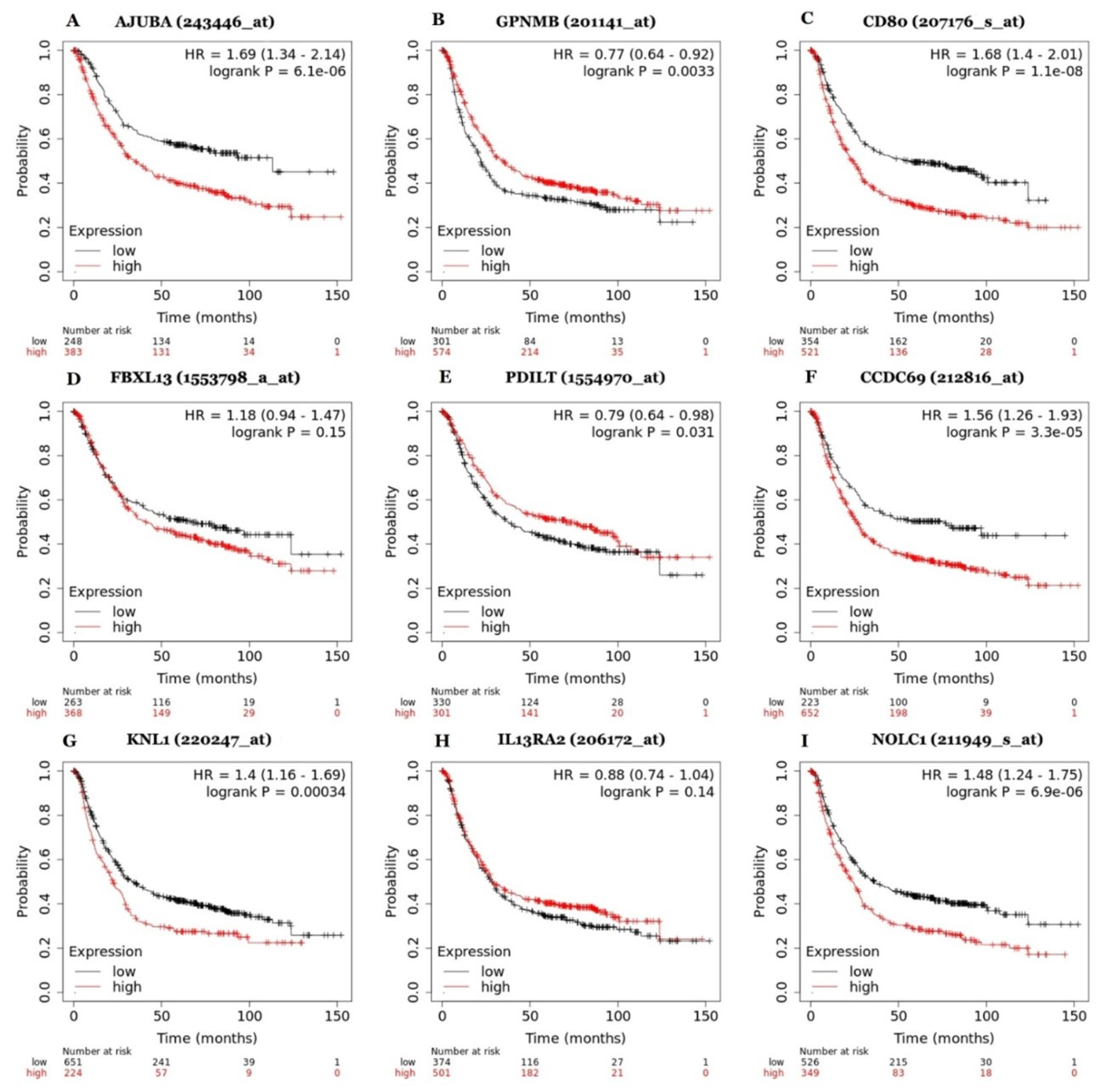
2.5. Survival Analysis
2.6. Primer Designer
| Ligand-ID | Target | Chemical formula | Mol. Wt. | 2D-Structure |
|---|---|---|---|---|
| 1. MCULE-2386589557-0-6 | AJUBA | C20H16N2O2 | 316.352 | 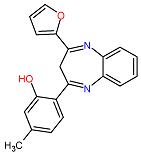 |
| 2. MCULE-7343047040-0-1 | FBXL13 | C19H21N3O3 | 339.388 | 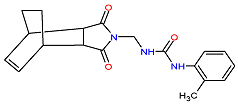 |
| 3. MCULE-5230409338-0-3 | CCDC69 | C22H22FN3OS | 395.495 | 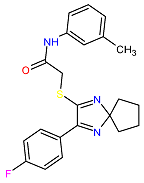 |
| 4. MCULE-9178344200-0-1 | CD80 | C17H20N2O3S | 332.419 | 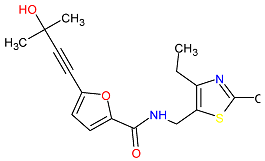 |
| 5. MCULE-5881513100-0-29 | NOLC1 | C20H19ClFN5 | 383.848 | 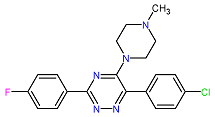 |
2.7. Cell Culture
2.8. Gene Expression by qRT-PCR
2.8.1. RNA Extraction
2.8.2. Conversion of mRNA to cDNA
2.8.3. Real-Time Quantitative PCR (qRT-PCR)
2.10. Virtual Screening
2.11. In Silico Toxicity Prediction
2.12. Prediction of ADME Parameters
2.13. Molecular Docking (MD) Assays
2.14. Protein-Protein Interaction Network
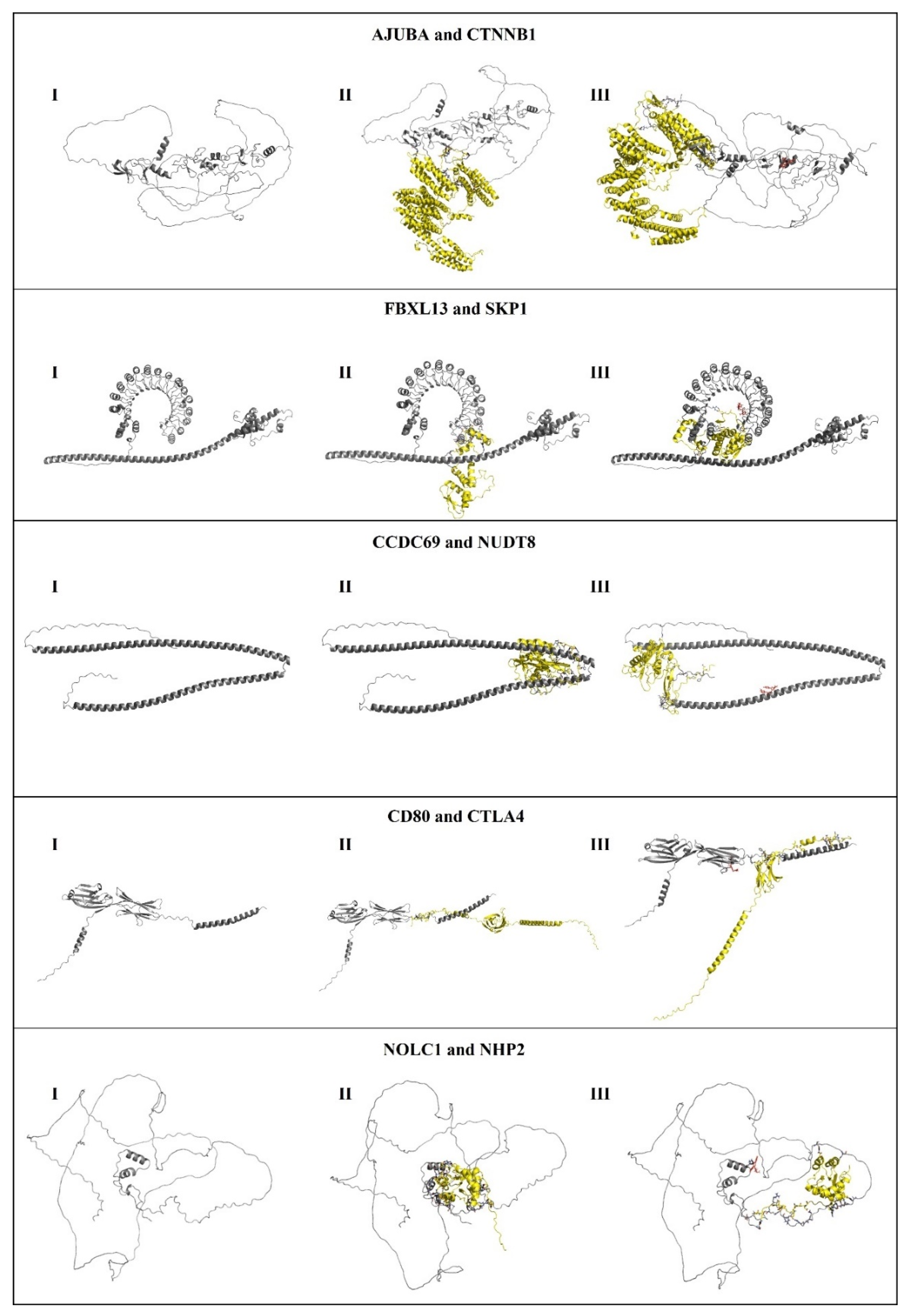
2.15. Re-Docking
3. Results
3.1. Differentially Gene Wxpression in Gastric Tumor Meta-Dataset
3.2. Gastric Tumor Group Involved in Multiple Cancer Progression Pathways
3.3. Prognostic Value of Differentially Expressed Genes Involved in Gastric Cancer Progression
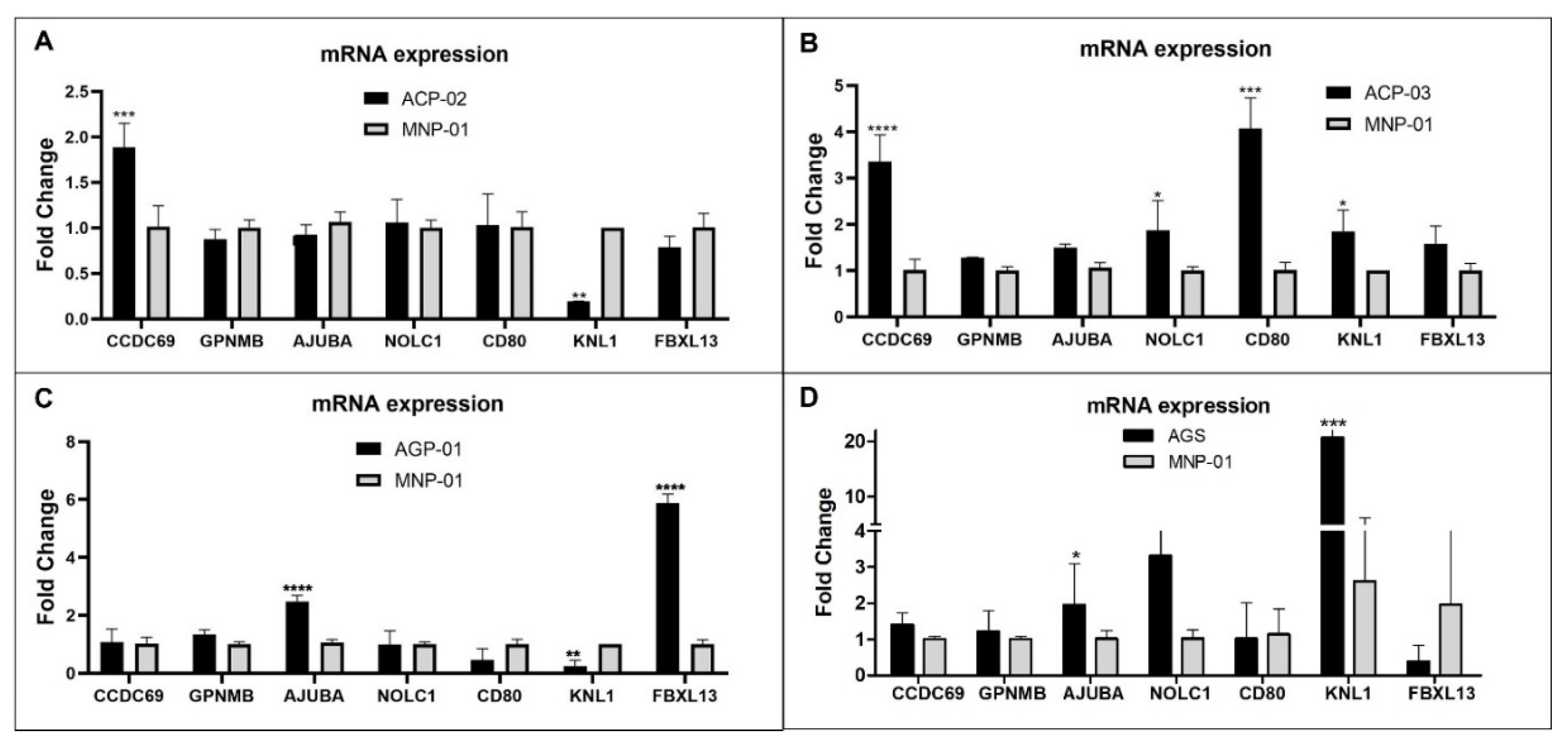
3.4. Validation of Relevant Genes in Gastric Tumor Cell Lines Compared to Normal Gastric Cell Lines
3.5. In Silico High-Throughput Virtual Screening
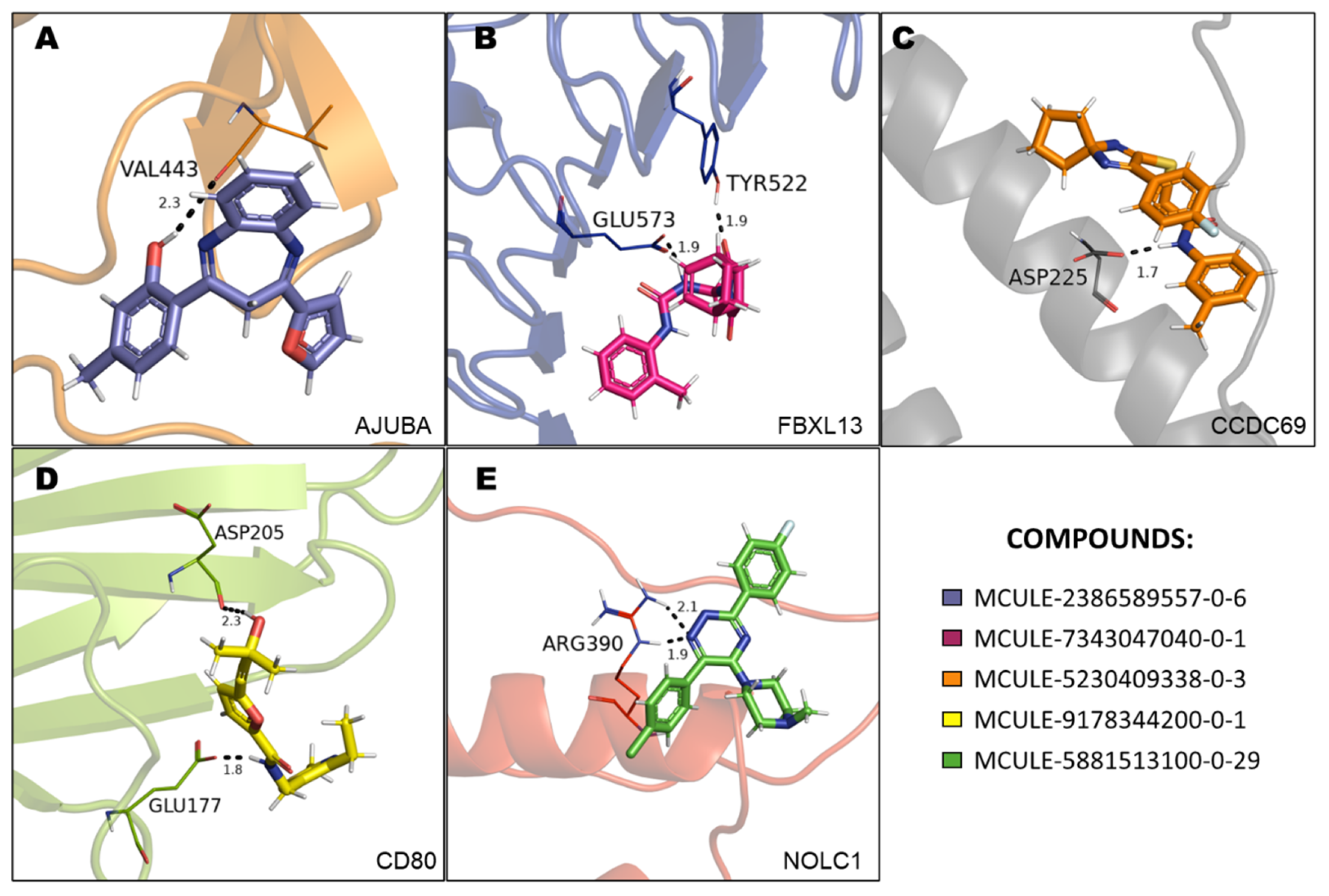
3.6. Molecular Docking (MD) Validation
| Protein-Ligand Complex | Docking Score | Predicted Toxicity | |||
|---|---|---|---|---|---|
| Ligand-ID | Target | DockThor | Mcule | LD50 | Class |
| MCULE-2386589557-0-6 | AJUBA | -8.4 | -7.3 | 2500mg/kg | V |
| MCULE-7343047040-0-1 | FBXL13 | -7.4 | -8.3 | 5400mg/kg | VI |
| MCULE-5230409338-0-3 | CCDC69 | -7.133 | -5.6 | 1000mg/kg | IV |
| MCULE-9178344200-0-1 | CD80 | -7.773 | -5.6 | 600mg/kg | IV |
| MCULE-5881513100-0-29 | NOLC1 | -7.260 | -7.5 | 640mg/kg | IV |
3.7. In Silico Pharmacokinetics Prediction
| ADME Properties | (1) | (2) | (3) | (4) | (5) |
|---|---|---|---|---|---|
| Molecular weight | 316.35 | 339.39 | 395.49 | 332.42 | 383.85 |
| TPSA | 58.09 | 78.51 | 79.12 | 103.60 | 45.15 |
| cLogP | 3.81 | 2.05 | 4.52 | 2.73 | 3.72 |
| LogS | -4.75 | -3.38 | -5.04 | -3.49 | -4.73 |
| GI absorption | High | High | High | High | High |
| BBB permeant | Yes | No | No | No | Yes |
| P-gp substrate | No | Yes | No | No | Yes |
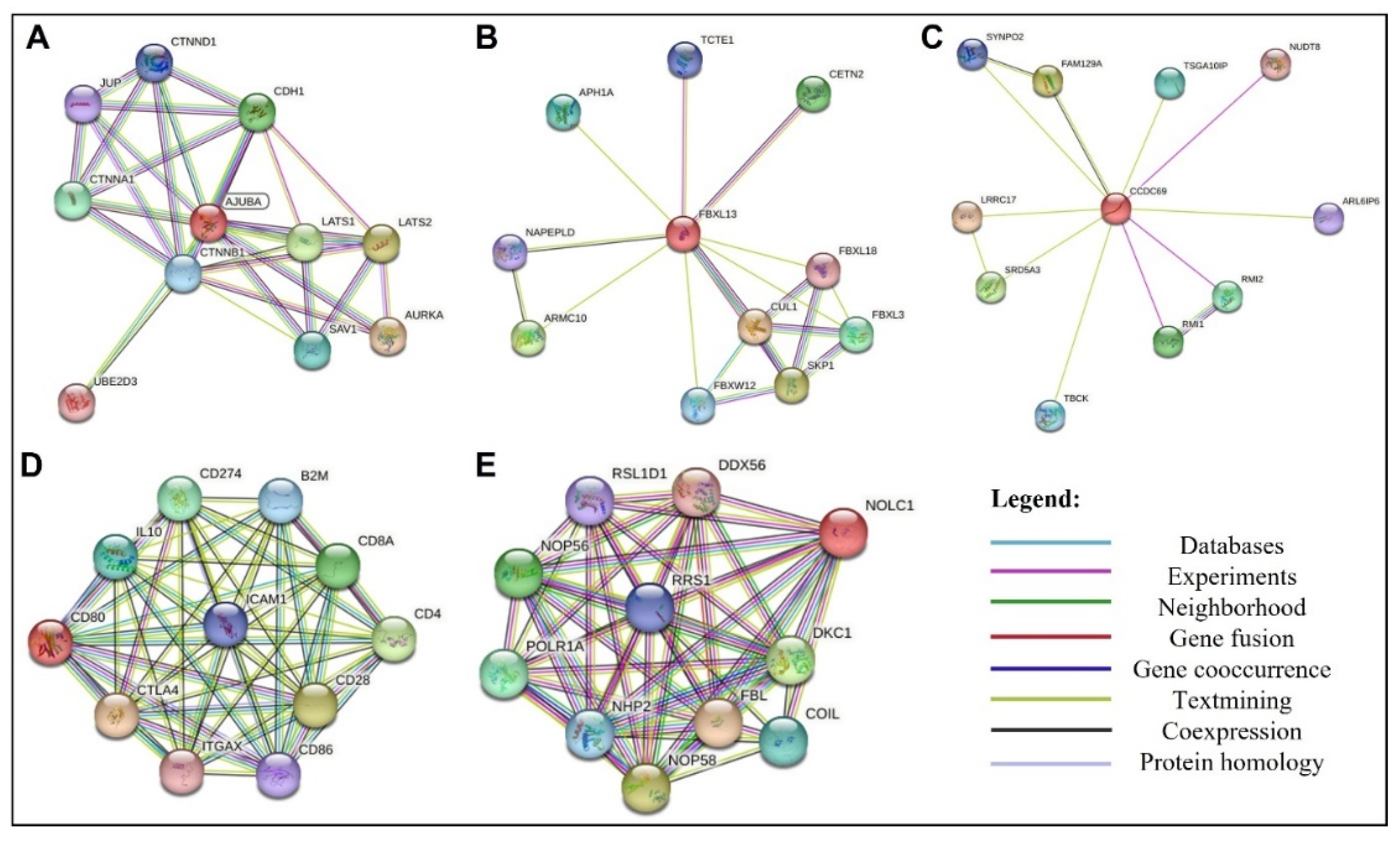
3.8. Protein-Protein Interaction Network
4. Discussion
5. Conclusions
Funding
Acknowledgments
Declaration of Competing Interests
References
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric Cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ao, X.; Wang, Y.; Li, X.; Wang, J. Long Non-Coding RNA in Gastric Cancer: Mechanisms and Clinical Implications for Drug Resistance. Front. Oncol. 2022, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- INCA. Estimativa 2023 : Incidência de Câncer No Brasil / Instituto Nacional de Câncer; 2022; ISBN 9786588517093. [Google Scholar]
- LAUREN, P. THE TWO HISTOLOGICAL MAIN TYPES OF GASTRIC CARCINOMA: DIFFUSE AND SO-CALLED INTESTINAL-TYPE CARCINOMA. AN ATTEMPT AT A HISTO-CLINICAL CLASSIFICATION. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Sasako, M.; Sakuramoto, S.; Katai, H.; Kinoshita, T.; Furukawa, H.; Yamaguchi, T.; Nashimoto, A.; Fujii, M.; Nakajima, T.; Ohashi, Y. Five-Year Outcomes of a Randomized Phase III Trial Comparing Adjuvant Chemotherapy with S-1 versus Surgery Alone in Stage II or III Gastric Cancer. J. Clin. Oncol. 2011, 29, 4387–4393. [Google Scholar] [CrossRef]
- Leichman, L.; Silberman, H.; Leichman, C.G.; Spears, C.P.; Ray, M.; Muggia, F.M.; Kiyabu, M.; Radin, R.; Laine, L.; Stain, S.; et al. Preoperative Systemic Chemotherapy Followed by Adjuvant Postoperative Intraperitoneal Therapy for Gastric Cancer: A University of Southern California Pilot Program. J. Clin. Oncol. 1992, 10, 1933–1942. [Google Scholar] [CrossRef]
- Ajani, J.A.; Mansfield, P.F.; Lynch, P.M.; Pisters, P.W.; Feig, B.; Dumas, P.; Evans, D.B.; Raijman, I.; Hargraves, K.; Curley, S.; et al. Enhanced Staging and All Chemotherapy Preoperatively in Patients with Potentially Resectable Gastric Carcinoma. J. Clin. Oncol. 1999, 17, 2403–2411. [Google Scholar] [CrossRef]
- Guan, W.L.; He, Y.; Xu, R.H. Gastric Cancer Treatment: Recent Progress and Future Perspectives. J. Hematol. Oncol. 2023 161 2023, 16, 1–28. [Google Scholar] [CrossRef]
- Joshi, S.S.; Badgwell, B.D. Current Treatment and Recent Progress in Gastric Cancer. CA. Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef]
- Miller, Z.; Kim, K.S.; Lee, D.M.; Kasam, V.; Baek, S.E.; Lee, K.H.; Zhang, Y.Y.; Ao, L.; Carmony, K.; Lee, N.R.; et al. Proteasome Inhibitors with Pyrazole Scaffolds from Structure-Based Virtual Screening. J. Med. Chem. 2015, 58, 2036–2041. [Google Scholar] [CrossRef]
- Rohr, M.; Beardsley, J.; Nakkina, S.P.; Zhu, X.; Aljabban, J.; Hadley, D.; Altomare, D. A Merged Microarray Meta-Dataset for Transcriptionally Profiling Colorectal Neoplasm Formation and Progression. Sci. data 2021, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Discovery and Ranking of the Most Robust Prognostic Biomarkers in Serous Ovarian Cancer. GeroScience 2023. [Google Scholar] [CrossRef] [PubMed]
- Maués, J.H. da S.; Ribeiro, H.F.; Pinto, G.R.; Lopes, L. de O.; Lamarao, L.M.; Pessoa, C.M.F.; Moreira-Nunes, C. de F.A.; Carvalho, R.M. de; Assumpção, P.P.; Rey, J.A. Gastric Cancer Cell Lines Have Different MYC-Regulated Expression Patterns but Share a Common Core of Altered Genes. Can. J. Gastroenterol. Hepatol. 2018, 2018. [Google Scholar]
- Riquelme, I.; Tapia, O.; Espinoza, J.A.; Leal, P.; Buchegger, K.; Sandoval, A.; Bizama, C.; Araya, J.C.; Peek, R.M.; Roa, J.C. The Gene Expression Status of the PI3K/AKT/MTOR Pathway in Gastric Cancer Tissues and Cell Lines. Pathol. Oncol. Res. 2016, 22, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, F.P.; Lucena da Silva, E.; Souza, P.F.N.; Lima, L.B.; Amaral, J.L.; Zuercher, W.; Albuquerque, L.M.; Rabenhorst, S.H.B.; Moreira-Nunes, C.A.; Amaral de Moraes, M.E.; et al. Kinase Inhibitor Screening Reveals Aurora-a Kinase Is a Potential Therapeutic and Prognostic Biomarker of Gastric Cancer. J. Cell. Biochem. 2021, 122, 1376–1388. [Google Scholar] [CrossRef]
- Bustin, S.A.; Beaulieu, J.-F.; Huggett, J.; Jaggi, R.; Kibenge, F.S.B.; Olsvik, P.A.; Penning, L.C.; Toegel, S. MIQE Precis: Practical Implementation of Minimum Standard Guidelines for Fluorescence-Based Quantitative Real-Time PCR Experiments. BMC Mol. Biol. 2010, 11, 1–5. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- DeLano, W.L.; Lam, J. PyMOL: A Communications Tool for Computational Models. 2005. [Google Scholar]
- Kiss, R.; Sandor, M.; Szalai, F.A. Http://Mcule.Com: A Public Web Service for Drug Discovery. J. Cheminform. 2012, 4, 2012. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; AlAjmi, M.F.; Aatif, M.; Farhan, M.; Shafi, S. Identification of a Potential Inhibitor (MCULE-8777613195-0-12) of New Delhi Metallo-β-Lactamase-1 (NDM-1) Using In Silico and In Vitro Approaches. Molecules 2022, 27. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like Properties and the Causes of Poor Solubility and Poor Permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- De Magalhães, C.S.; Almeida, D.M.; Barbosa, H.J.C.; Dardenne, L.E. A Dynamic Niching Genetic Algorithm Strategy for Docking Highly Flexible Ligands. Inf. Sci. (Ny). 2014, 289, 206–224. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New Machine Learning and Physics-Based Scoring Functions for Drug Discovery. Sci. Rep. 2021, 11, 1–19. [Google Scholar] [CrossRef]
- Inbar, Y.; Schneidman-Duhovny, D.; Halperin, I.; Oron, A.; Nussinov, R.; Wolfson, H.J. Approaching the CAPRI Challenge with an Efficient Geometry-Based Docking. In Proceedings of the Proteins: Structure, Function and Genetics; Proteins, August 1 2005; Vol. 60; pp. 217–223. [Google Scholar]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers (Basel). 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Borriello, A.; Caldarelli, I.; Basile, M.A.; Bencivenga, D.; Tramontano, A.; Perrotta, S.; della Ragione, F.; Oliva, A. The Tyrosine Kinase Inhibitor Dasatinib Induces a Marked Adipogenic Differentiation of Human Multipotent Mesenchymal Stromal Cells. PLoS One 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Xu, T.; Tan, Y.; Lv, W.; Zhao, C.; Wu, M.; Wu, Y.; Zhang, Q. CCDC69 Is a Prognostic Marker of Breast Cancer and Correlates with Tumor Immune Cell Infiltration. Front. Surg. 2022, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Martínez, F.D.; López-López, E.; Eurídice Juárez-Mercado, K.; Medina-Franco, J.L. Computational Drug Design Methods—Current and Future Perspectives. Silico Drug Des. Repurposing Tech. Methodol. 2019, 19–44. [Google Scholar] [CrossRef]
- Anthony, L.F.W.; Kanding, B.; Selvan, R. Carbontracker: Tracking and Predicting the Carbon Footprint of Training Deep Learning Models. arXiv Prepr. arXiv2007.03051 2020. [Google Scholar]
- Song, Y.; Ye, L.; Tan, Y.; Tong, H.; Lv, Z.; Wan, X.; Li, Y. Therapeutic Exosomes Loaded with SERPINA5 Attenuated Endometrial Cancer Cell Migration via the Integrin Β1/FAK Signaling Pathway. Cell. Oncol. 2022, 45, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Cui, M.; Li, C.; Li, H.; Dai, Y.; Cui, K.; Li, Z. Kaempferol Reverses Aerobic Glycolysis via MiR-339-5p-Mediated PKM Alternative Splicing in Colon Cancer Cells. J. Agric. Food Chem. 2021, 69, 3060–3068. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Song, L.; Cong, Q.; Wang, J.; Xu, H.; Chu, Y.; Li, Q.; Zhang, Y.; Zou, X.; Zhang, C. The LIM Protein AJUBA Promotes Colorectal Cancer Cell Survival through Suppression of JAK1/STAT1/IFIT2 Network. Oncogene 2017, 36, 2655–2666. [Google Scholar] [CrossRef] [PubMed]
- Dommann, N.; Sánchez-Taltavull, D.; Eggs, L.; Birrer, F.; Brodie, T.; Salm, L.; Baier, F.A.; Medova, M.; Humbert, M.; Tschan, M.P. The LIM Protein Ajuba Augments Tumor Metastasis in Colon Cancer. Cancers (Basel). 2020, 12, 1913. [Google Scholar] [CrossRef]
- Fung, E.; Richter, C.; Yang, H.; Schäffer, I.; Fischer, R.; Kessler, B.M.; Bassermann, F.; D’Angiolella, V. FBXL 13 Directs the Proteolysis of CEP 192 to Regulate Centrosome Homeostasis and Cell Migration. EMBO Rep. 2018, 19, e44799. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G. V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Schnerch, D.; Nigg, E.A. Structural Centrosome Aberrations Favor Proliferation by Abrogating Microtubule-Dependent Tissue Integrity of Breast Epithelial Mammospheres. Oncogene 2016, 35, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Wu, D.; Haruta, A.; Matsumura, F.; Wei, Q. Role of a Novel Coiled-Coil Domain-Containing Protein CCDC69 in Regulating Central Spindle Assembly. Cell cycle 2010, 9, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Xu, T.; Tan, Y.; Lv, W.; Zhao, C.; Wu, M.; Wu, Y.; Zhang, Q. CCDC69 Is a Prognostic Marker of Breast Cancer and Correlates with Tumor Immune Cell Infiltration. Front. Surg. 2022, 9, 879921. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Y.; Inoue, H.; Mori, M.; Akiyoshi, T. The Expression of Costimulatory Molecules CD80 and CD86 in Human Carcinoma Cell Lines: Its Regulation by Interferon Gamma and Interleukin-10. Cancer Immunol. Immunother. 1996, 43, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.A.; Long, T.M.; Atkinson, R.; Clements, V.; Ostrand-Rosenberg, S. Soluble CD80 Protein Delays Tumor Growth and Promotes Tumor-Infiltrating Lymphocytes. Cancer Immunol. Res. 2018, 6, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.Y.; Lu, L.; Wang, K.F.; Zhu, B.Y.; Wen, X.Z.; Peng, R.Q.; Ding, Y.; Li, D.D.; Li, J.J.; Li, Y.; et al. Low Expression of CD80 Predicts for Poor Prognosis in Patients with Gastric Adenocarcinoma. Future Oncol. 2019, 15, 473–483. [Google Scholar] [CrossRef]
- Peggs, K.S.; Quezada, S.A. Ipilimumab: Attenuation of an Inhibitory Immune Checkpoint Improves Survival in Metastatic Melanoma. Expert Rev. Anticancer Ther. 2010, 10, 1697–1701. [Google Scholar] [CrossRef]
- Zhai, F.; Wang, J.; Luo, X.; Ye, M.; Jin, X. Roles of NOLC1 in Cancers and Viral Infection. J. Cancer Res. Clin. Oncol. 2023. [Google Scholar] [CrossRef]
- Kong, F.; Shang, Y.; Diao, X.; Huang, J.; Liu, H. Knockdown of NOLC1 Inhibits PI3K-AKT Pathway to Improve the Poor Prognosis of Esophageal Carcinoma. J. Oncol. 2021, 2021. [Google Scholar] [CrossRef]
- L. Garner, A.; D. Janda, K. Protein-Protein Interactions and Cancer: Targeting the Central Dogma. Curr. Top. Med. Chem. 2010, 11, 258–280. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2020, 5. [Google Scholar] [CrossRef]
- Bahmanyar, S.; Kaplan, D.D.; DeLuca, J.G.; Giddings, T.H.; O’Toole, E.T.; Winey, M.; Salmon, E.D.; Casey, P.J.; Nelson, W.J.; Barth, A.I.M. β-Catenin Is a Nek2 Substrate Involved in Centrosome Separation. Genes Dev. 2008, 22, 91–105. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





