Submitted:
24 June 2024
Posted:
26 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis
2.1.1. Gathering of Target Genes
2.1.1.1. QRC Target Genes
2.1.1.2. GC Target Genes
2.1.2. Common Genes Analysis
2.1.3. Gene Ontology and Functional Annotation Assay
2.1.4. Protein-to-Protein Analysis
2.1.5. Survival Curves and Immune System Infiltration
2.1.6. Molecular Docking Analysis
3. Results
3.1. Target Genes
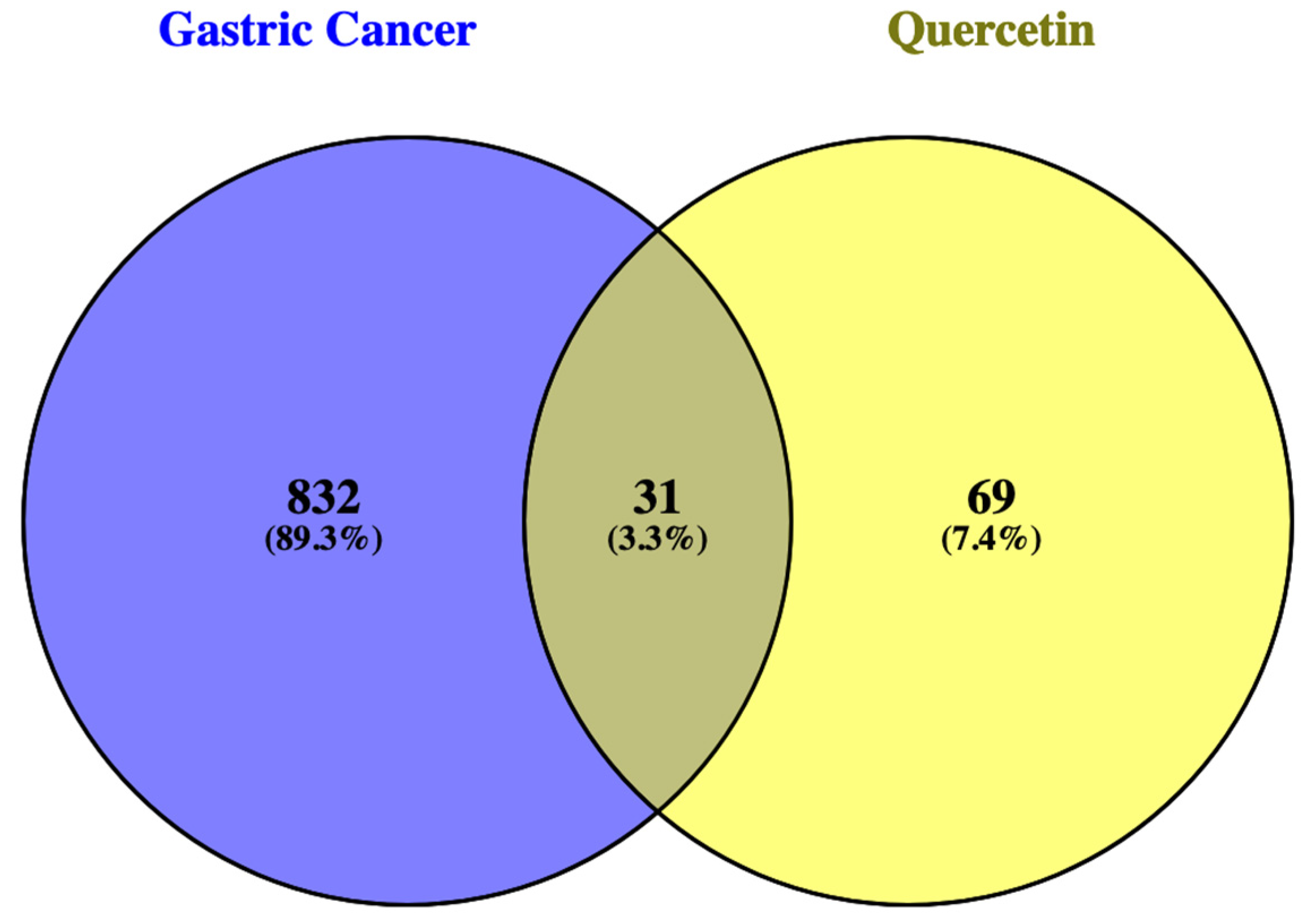
3.1.1. Common Genes between GC and QRC.
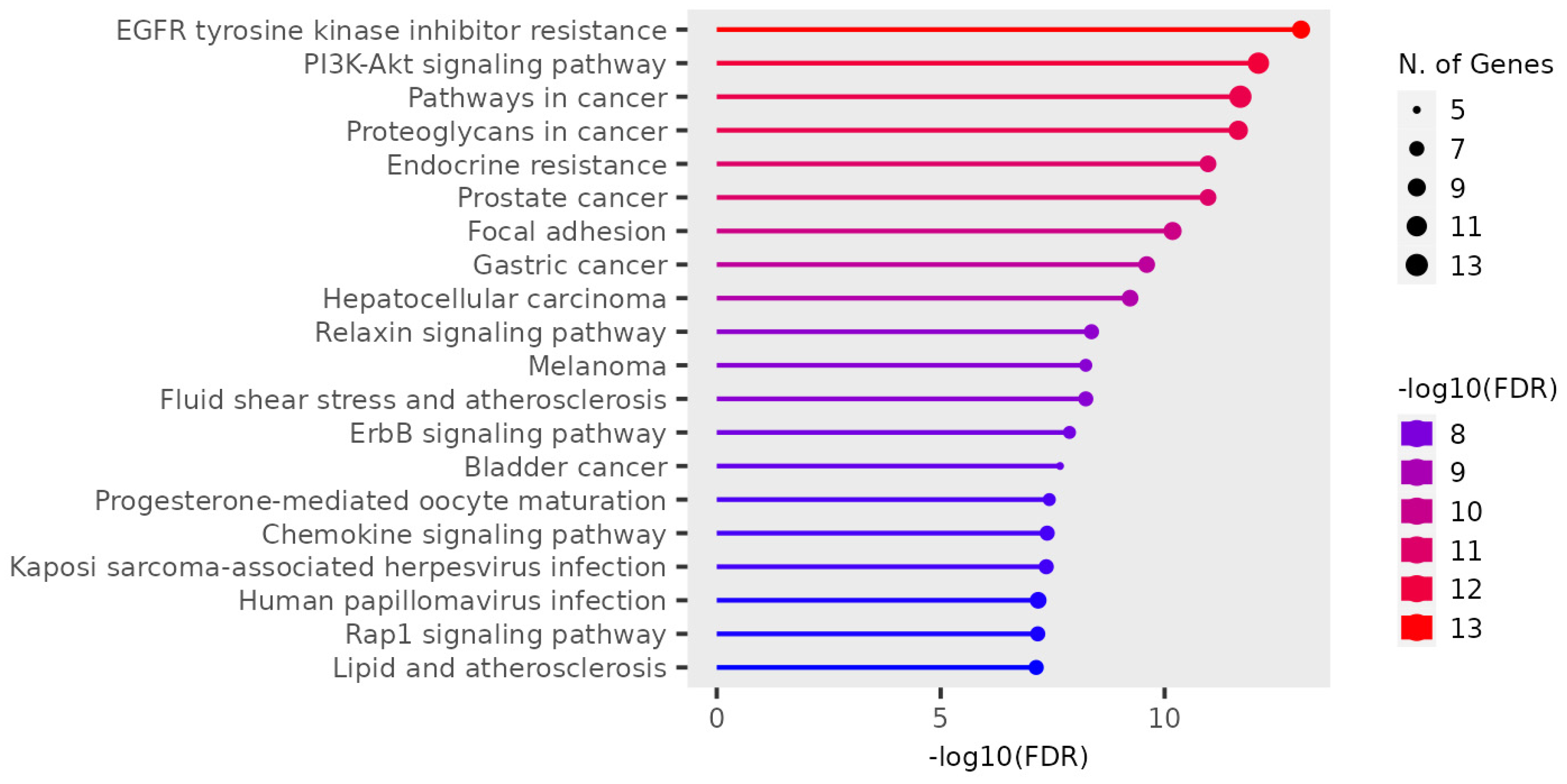
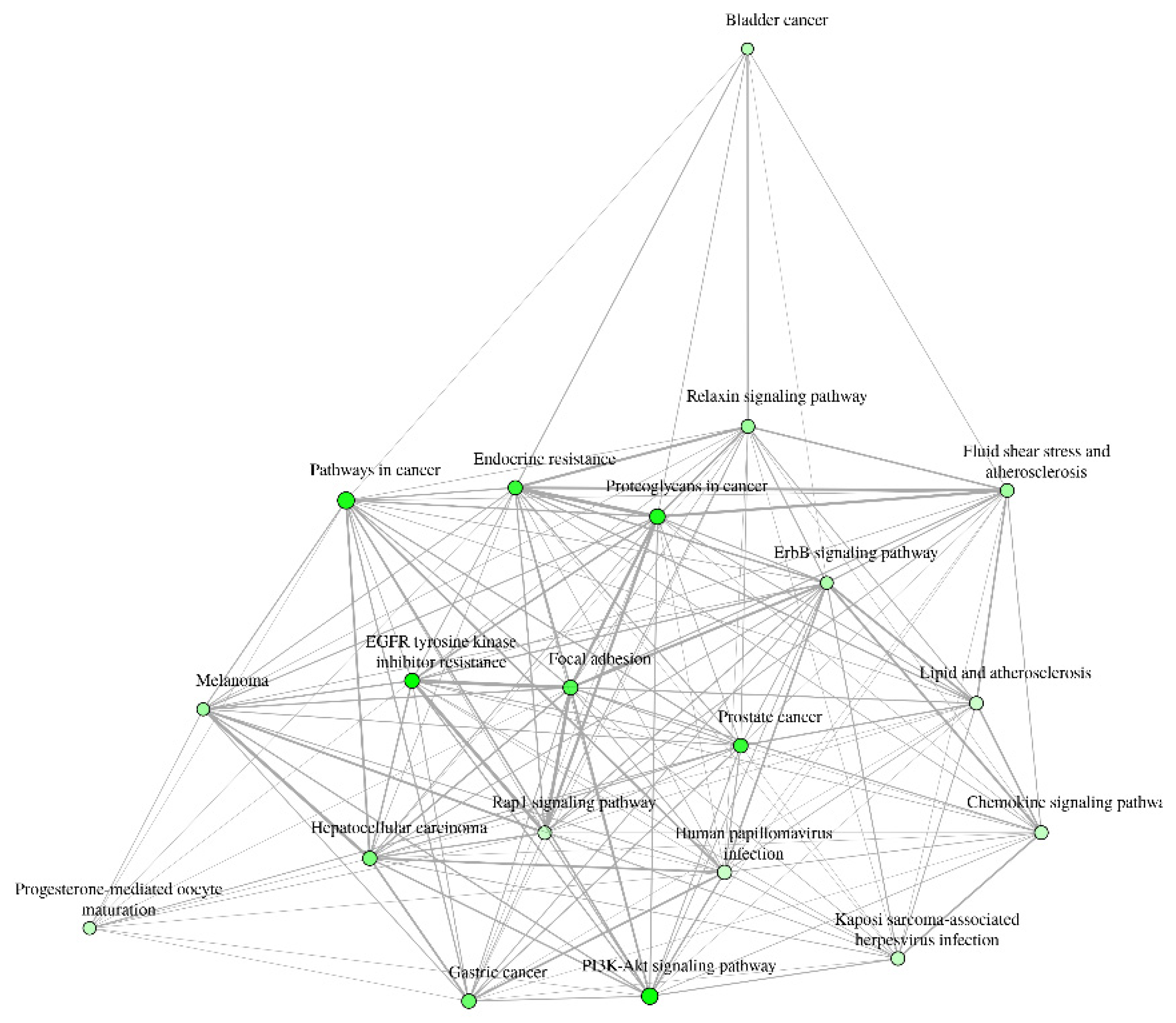
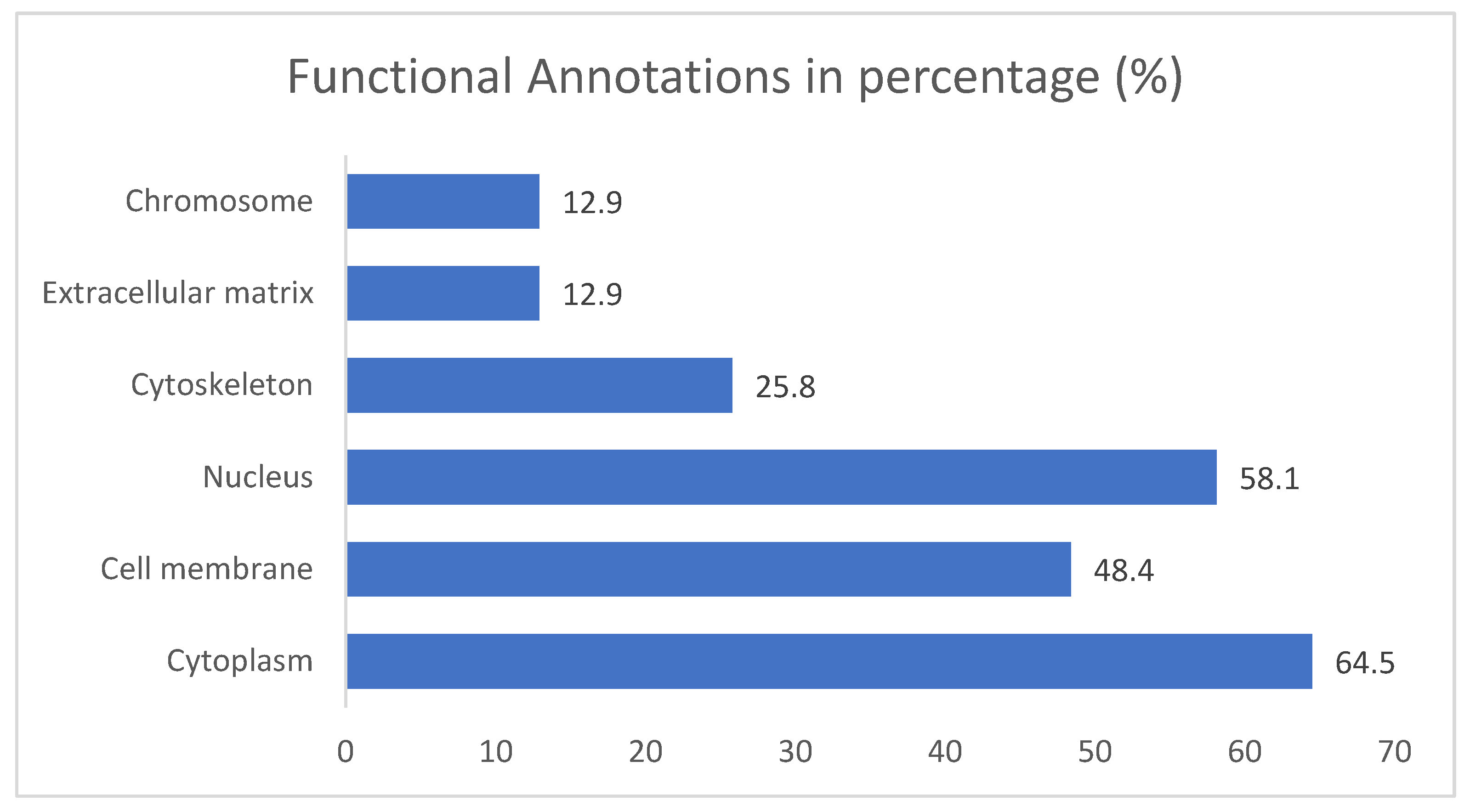
3.2. Gene Ontology Test (GO)
3.2.1.
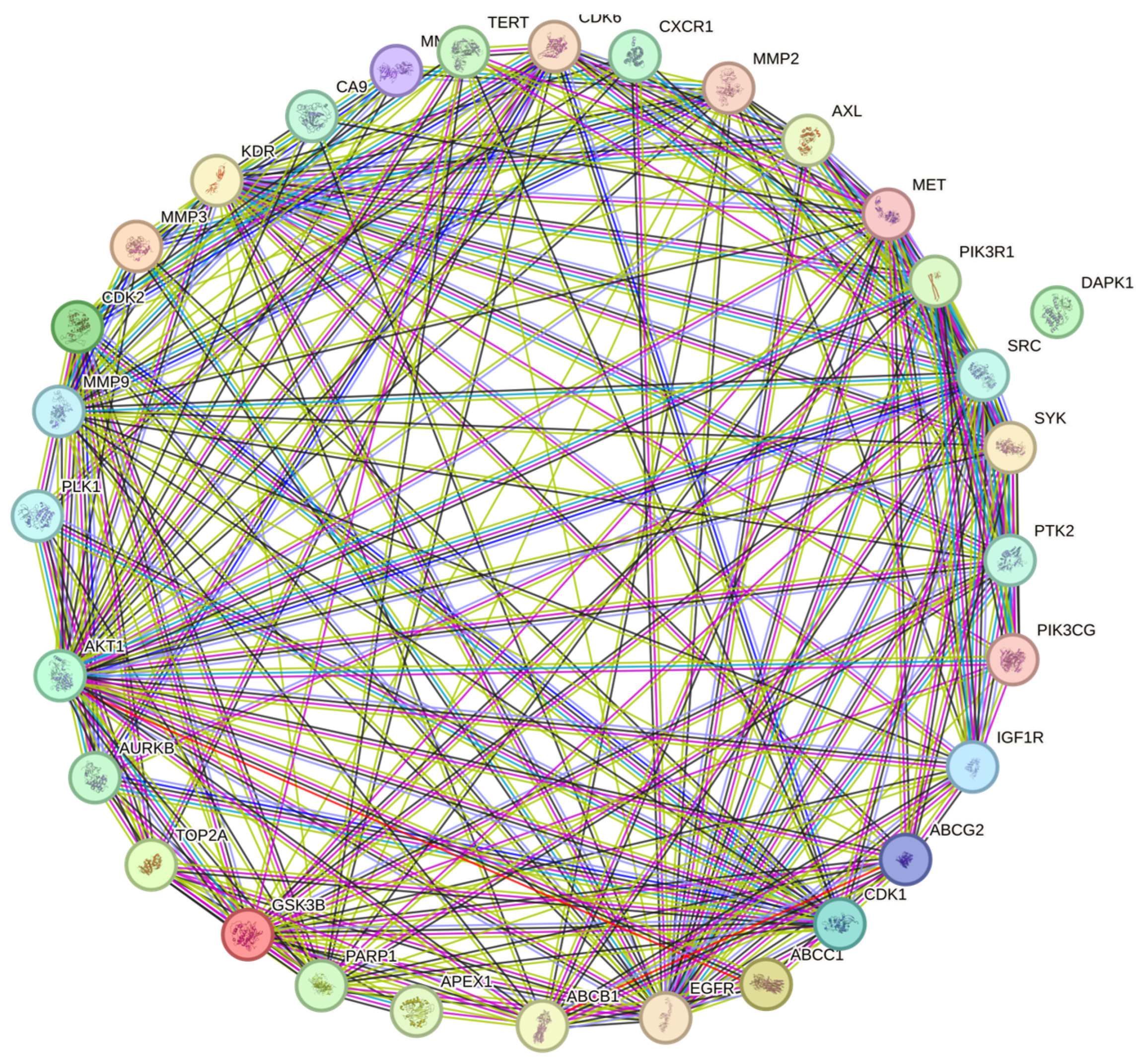
3.3. Protein-to-Protein Analysis
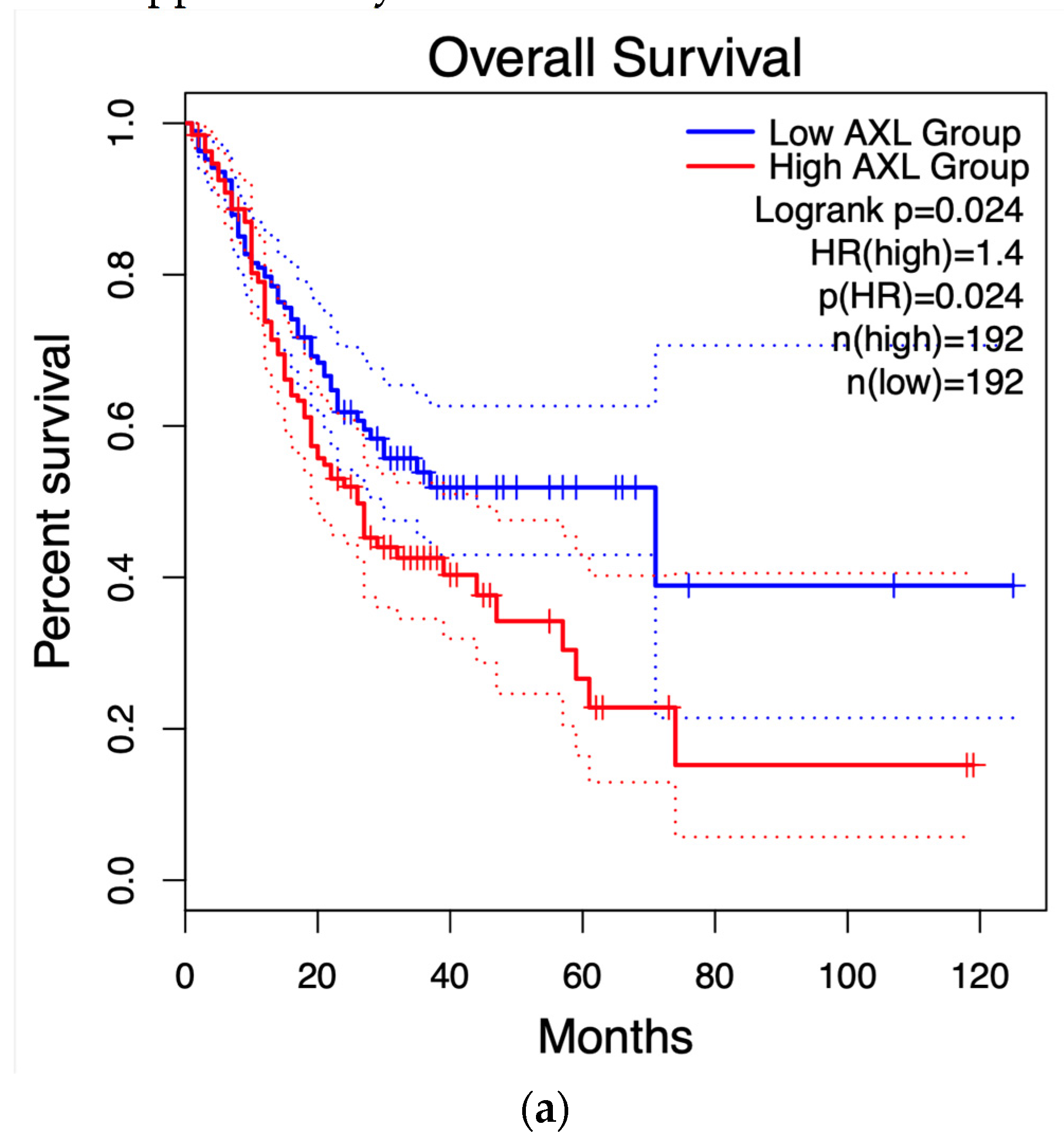
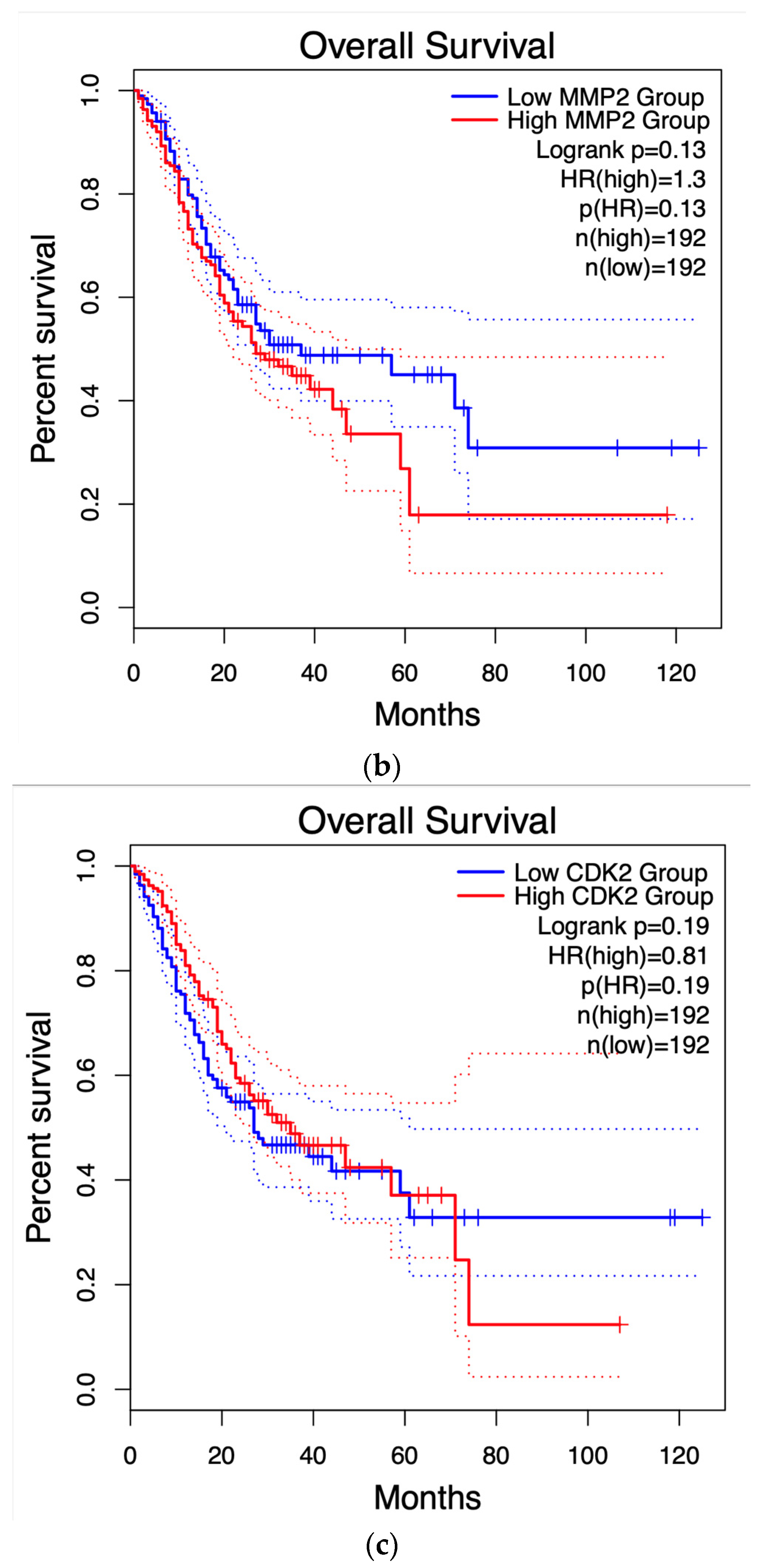
3.4. Survival Graphs
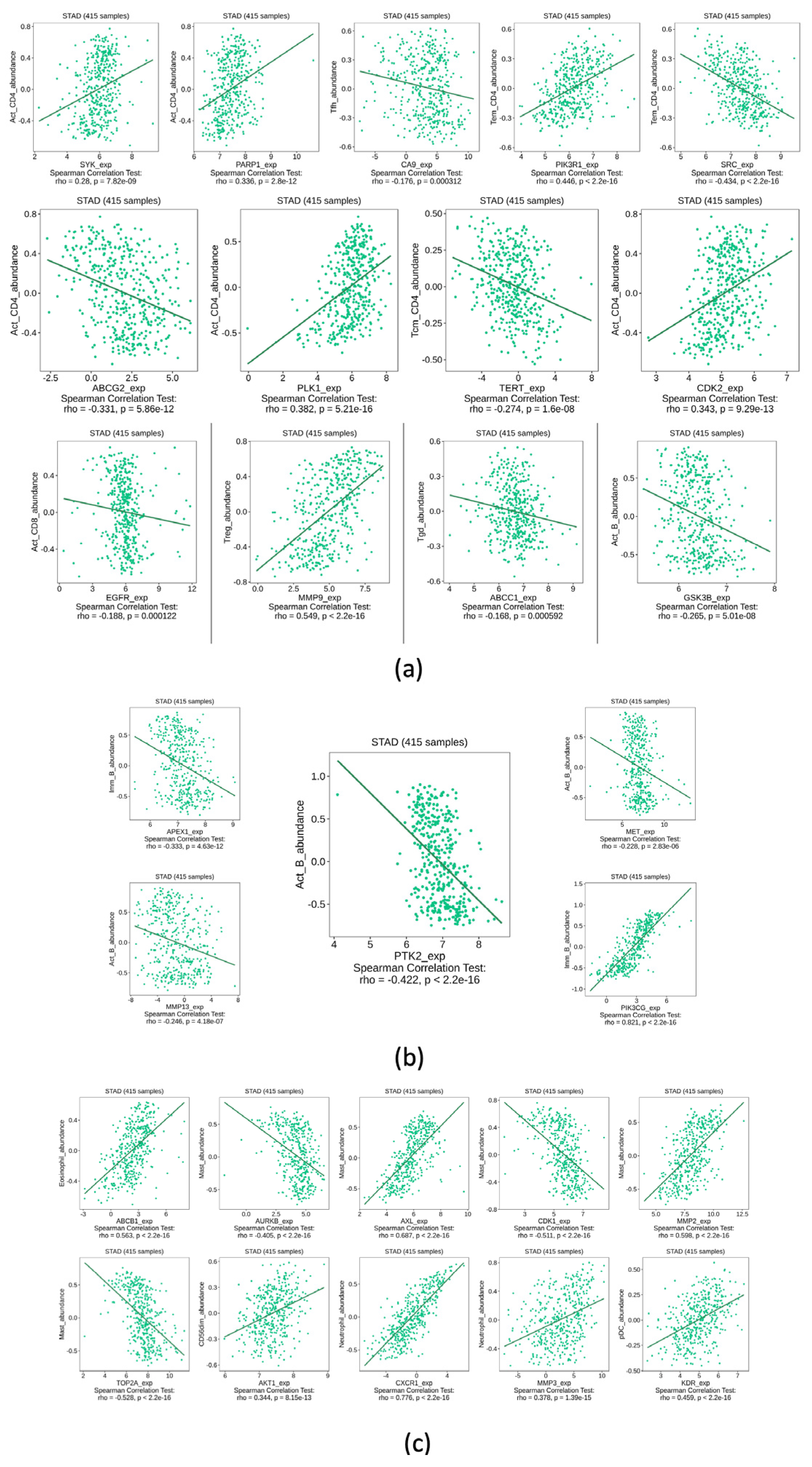
3.5. Immune System Infiltration
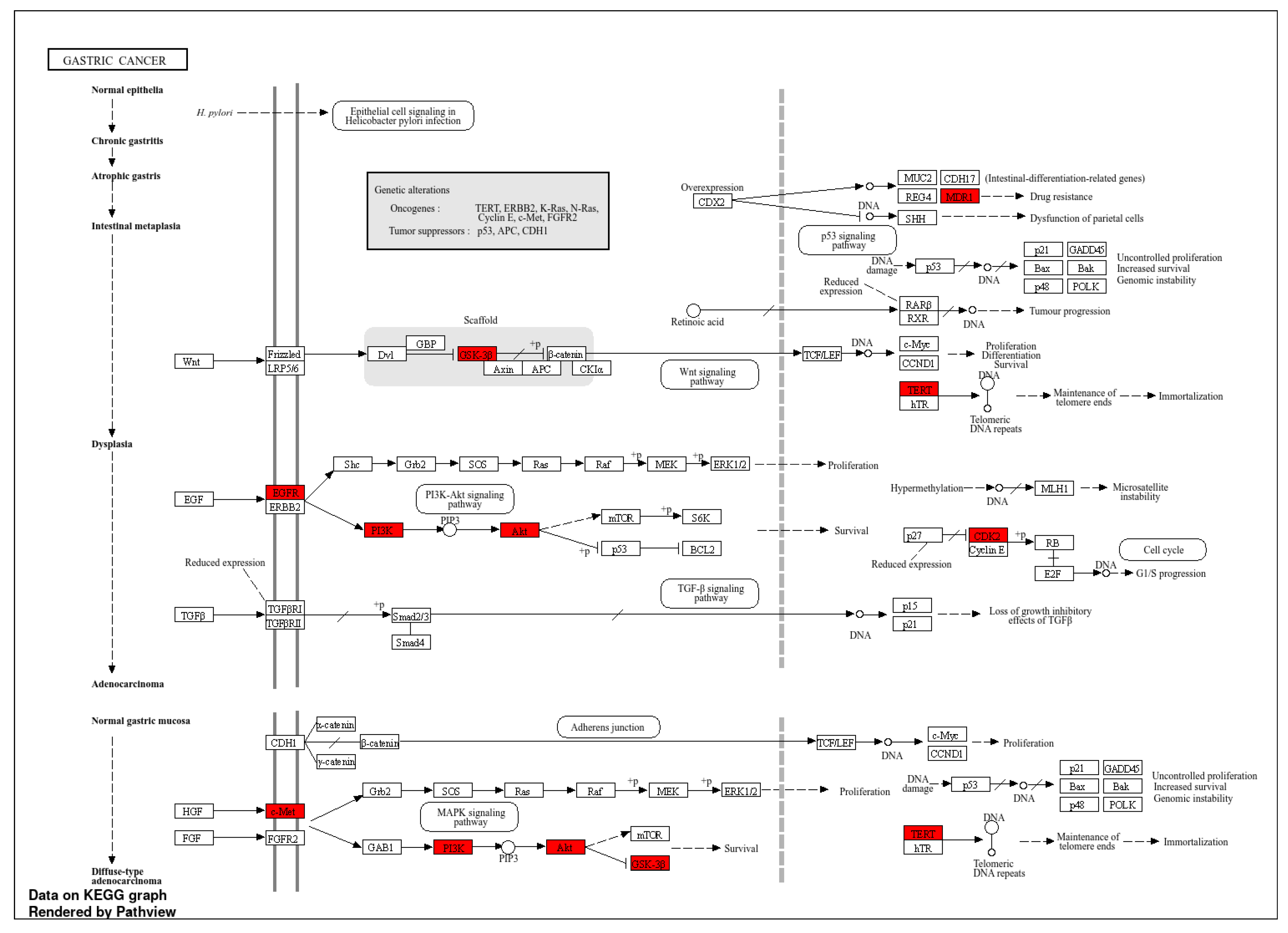
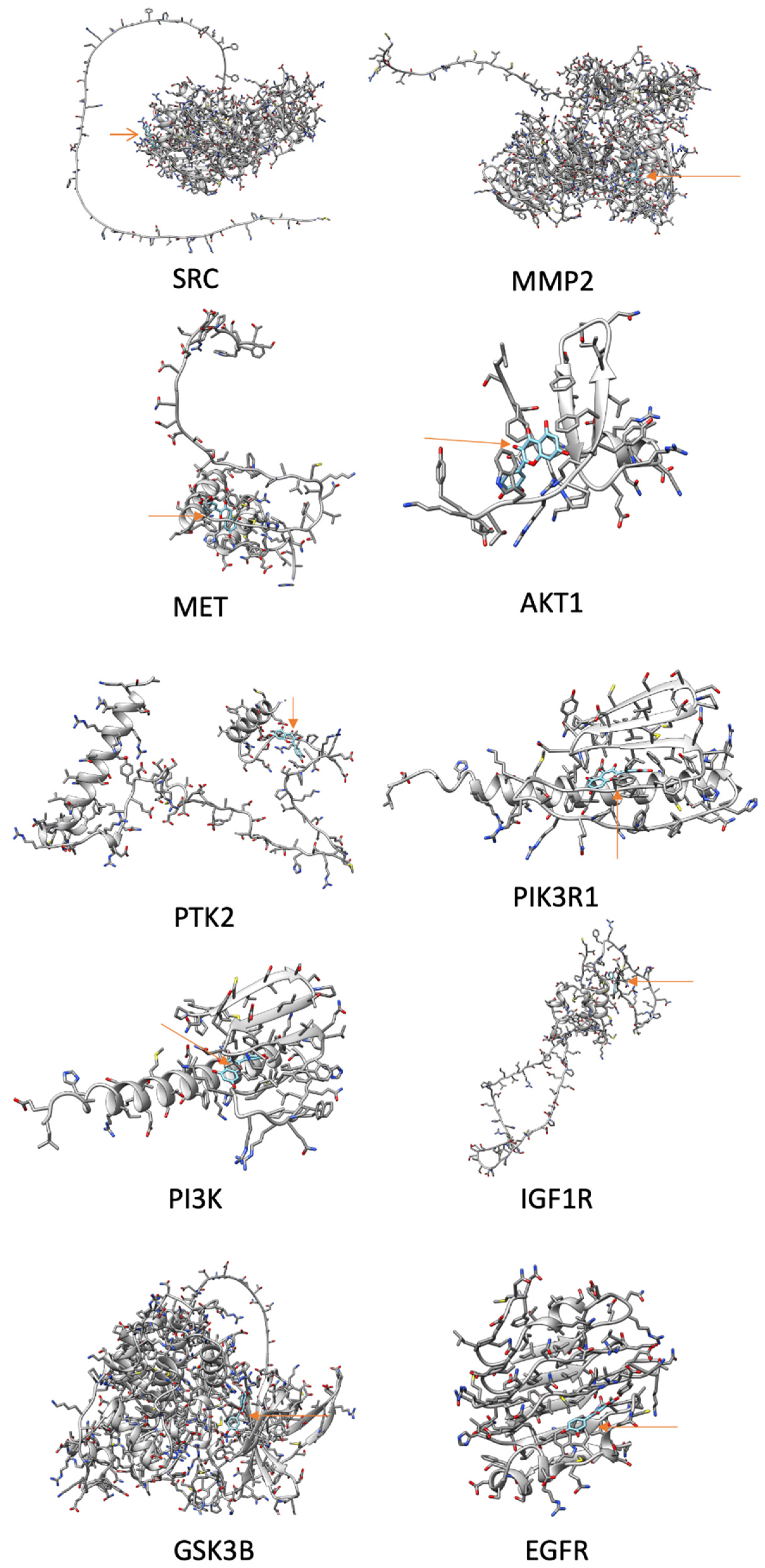
3.6. Docking Analysis
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sitarz R, Skierucha M, Mielko J, Offerhaus J, Maciejewski R, Polkowski W. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018 Feb;Volume 10:239–48.
- Mukkamalla SKR, Recio-Boiles A, Babiker HM. https://www.ncbi.nlm.nih.gov/books/NBK459142/. 2024. Gastric Cancer.
- World Health Organization. https://gco.iarc.fr/en. 2022. Global Cancer Observatory.
- Wipperman J, NT, & WT. Cervical Cancer: Evaluation and Management.. Am Fam Physician. 2018;97(7):4449–54.
- Herranz-López, Olivares-Vicente, Rodríguez Gallego E, Antonio Encinar, Pérez-Sánchez A, Ruiz-Torres et al. Quercetin metabolites from Hibiscus sabdariffa contribute to alleviate glucolipotoxicity-induced metabolic stress in vitro. Food Chem Toxicol. 2020;(144):111606.
- Herranz-López M, Olivares-Vicente M, Rodríguez Gallego E, Encinar JA, Pérez-Sánchez A, Ruiz-Torres V, et al. Quercetin metabolites from Hibiscus sabdariffa contribute to alleviate glucolipotoxicity-induced metabolic stress in vitro. Food and Chemical Toxicology. 2020 Oct 1;144.
- Azizi E, Fouladdel S, Komeili Movahhed T, Modaresi F, Barzegar E, Ghahremani MH, et al. Quercetin Effects on Cell Cycle Arrest and Apoptosis and Doxorubicin Activity in T47D Cancer Stem Cells. Asian Pacific Journal of Cancer Prevention. 2022 Dec 1;23(12):4145–54.
- Chen K, Rekep M, Wei W, Wu Q, Xue Q, Li S, et al. Quercetin Prevents In Vivo and In Vitro Myocardial Hypertrophy Through the Proteasome-GSK-3 Pathway. Cardiovasc Drugs Ther. 2018 Feb 1;32(1):5–21.
- Dhanya R, Arun KB, Syama HP, Nisha P, Sundaresan A, Santhosh Kumar TR, et al. Rutin and quercetin enhance glucose uptake in L6 myotubes under oxidative stress induced by tertiary butyl hydrogen peroxide. Food Chem. 2014 Sep;158:546–54.
- Chen WJ, Tsai JH, Hsu LS, Lin CL, Hong HM, Pan MH. Quercetin blocks the aggressive phenotype of triple-negative breast cancer by inhibiting igf1/ igf1r-mediated emt program. J Food Drug Anal. 2021;29(1):98–112.
- Henning SM, Wang P, Lee RP, Trang A, Husari G, Yang J, et al. Prospective randomized trial evaluating blood and prostate tissue concentrations of green tea polyphenols and quercetin in men with prostate cancer. Food Funct. 2020 May 1;11(5):4114–22.
- Costa LG, Garrick JM, Roquè PJ, Pellacani C. Mechanisms of Neuroprotection by Quercetin: Counteracting Oxidative Stress and More. Oxid Med Cell Longev [Internet]. 2016 [cited 2022 May 19];2016. Available from: https://pubmed.ncbi.nlm.nih.gov/26904161/.
- Zúñiga-Hernández SR, García-Iglesias T, Macías-Carballo M, Pérez-Larios A, Gutiérrez-Mercado YK, Camargo-Hernández G, et al. Targets and Effects of Common Biocompounds of Hibiscus sabdariffa (Delphinidin-3-Sambubiosid, Quercetin, and Hibiscus Acid) in Different Pathways of Human Cells According to a Bioinformatic Assay. Nutrients. 2024 Feb 19;16(4):566.
- Daina A, Michielin O, Zoete V. Swiss Target Prediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019 Jul 2;47(W1):W357–64.
- Rappaport N, Nativ N, Stelzer G, Twik M, Guan-Golan Y, Iny Stein T, et al. MalaCards: an integrated compendium for diseases and their annotation. Database. 2013 Jan 1;2013.
- Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020 Apr 15;36(8):2628–9.
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003 Aug 14;4(9):R60.
- Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005 Jul 1;21(13):3017–24.
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015 Jan 28;43(D1):D447–52.
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012 Nov 29;41(D1):D808–15.
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007 Jan 3;35(Database):D358–62.
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011 Jan 1;39(Database):D561–8.
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017 Jan 4;45(D1):D362–8.
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009 Jan 1;37(Database):D412–6.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019 Jan 8;47(D1):D607–13.
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021 Jan 8;49(D1):D605–12.
- Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023 Jan 6;51(D1):D638–46.
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019 Jul 2;47(W1):W556–60.
- Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor–immune system interactions. Bioinformatics. 2019 Oct 15;35(20):4200–2.
- Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011 Jul 1;39(suppl):W270–7.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003 Nov;13(11):2498–504.
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chao H, Chen L, et al. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023 Jan 6;51(D1):D488–508.
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug 26;596(7873):583–9.
- Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022 Jan 7;50(D1):D20–6.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605–12.
- Huang AY, Xiong Z, Liu K, Chang Y, Shu L, Gao G, et al. Identification of kaempferol as an OSX upregulator by network pharmacology-based analysis of qianggu Capsule for osteoporosis. Front Pharmacol. 2022 Sep 23;13.
- Kolobkov DS, Sviridova DA, Abilev SK, Kuzovlev AN, Salnikova LE. Genes and Diseases: Insights from Transcriptomics Studies. Genes (Basel). 2022 Jun 28;13(7):1168.
- Yan L, Zhang Z, Liu Y, Ren S, Zhu Z, Wei L, et al. Anticancer Activity of Erianin: Cancer-Specific Target Prediction Based on Network Pharmacology. Front Mol Biosci. 2022 Mar 17;9.
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov;414(6859):105–11.
- Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol. 2020 Apr;61:167–79.
- Levantini E, Maroni G, Del Re M, Tenen DG. EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol. 2022 Oct;85:253–75.
- Lin X, Zhuang S, Chen X, Du J, Zhong L, Ding J, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Molecular Therapy. 2022 Feb;30(2):688–702.
- Song X, Traub B, Shi J, Kornmann M. Possible Roles of Interleukin-4 and -13 and Their Receptors in Gastric and Colon Cancer. Int J Mol Sci. 2021 Jan 13;22(2):727.
- Zhang SC, Hu ZQ, Long JH, Zhu GM, Wang Y, Jia Y, et al. Clinical Implications of Tumor-Infiltrating Immune Cells in Breast Cancer. J Cancer. 2019;10(24):6175–84.
- Lei ZN, Teng QX, Tian Q, Chen W, Xie Y, Wu K, et al. Signaling pathways and therapeutic interventions in gastric cancer. Signal Transduct Target Ther. 2022 Oct 8;7(1):358.
- Chou CC, Yang JS, Lu HF, Ip SW, Lo C, Wu CC, et al. Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells. Arch Pharm Res. 2010 Aug;33(8):1181–91.
- Kim M, Park J, Bouhaddou M, Kim K, Rojc A, Modak M, et al. A protein interaction landscape of breast cancer. Science (1979). 2021 Oct;374(6563).
- Kim J, Yoo JM, Park JS, Kim J, Kim SG, Seok YJ, et al. Anti-angiogenic effect of mountain ginseng in vitro and in vivo: Comparison with farm-cultivated ginseng. Mol Med Rep. 2021 Jun 29;24(2):615.
- Sigstedt SC, Hooten CJ, Callewaert MC, Jenkins AR, Romero AE, Pullin MJ, et al. Evaluation of aqueous extracts of Taraxacum officinale on growth and invasion of breast and prostate cancer cells. Int J Oncol. 2008 May;32(5):1085–90.
- Villegas-Comonfort S, Serna-Marquez N, Galindo-Hernandez O, Navarro-Tito N, Salazar EP. Arachidonic acid induces an increase of β-1,4-galactosyltransferase I expression in MDA-MB-231 breast cancer cells. J Cell Biochem. 2012 Nov 18;113(11):3330–41.
- Wei C, Wang B, Peng D, Zhang X, Li Z, Luo L, et al. Pan-Cancer Analysis Shows That ALKBH5 Is a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancer Types Including Gliomas. Front Immunol. 2022 Apr 4;13.
- Zhu C, Wei Y, Wei X. AXL receptor tyrosine kinase as a promising anti-cancer approach: functions, molecular mechanisms and clinical applications. Mol Cancer. 2019 Dec 4;18(1):153.
- Han S, Wang Y, Ge C, Gao M, Wang X, Wang F, et al. Pharmaceutical inhibition of AXL suppresses tumor growth and invasion of esophageal squamous cell carcinoma. Exp Ther Med. 2020 Sep 2;20(5):1–1.
- Tai KY, Shieh YS, Lee CS, Shiah SG, Wu CW. Axl promotes cell invasion by inducing MMP-9 activity through activation of NF-κB and Brg-1. Oncogene. 2008 Jul 3;27(29):4044–55.
- Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011 Mar;144(5):646–74.
- Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015 Dec 8;15(1):577.
- MacLeod MKL, Clambey ET, Kappler JW, Marrack P. CD4 memory T cells: What are they and what can they do? Semin Immunol. 2009 Apr;21(2):53–61.
- Duan Z, Li Y, Qiu Y, Shen Y, Wang Y, Zhang Y, et al. CD39 expression defines exhausted CD4 + T cells associated with poor survival and immune evasion in human gastric cancer. Clin Transl Immunology. 2024 Jan 18;13(3).
- Dolina JS, Van Braeckel-Budimir N, Thomas GD, Salek-Ardakani S. CD8+ T Cell Exhaustion in Cancer. Front Immunol. 2021 Jul 20;12.
- Reina-Campos M, Scharping NE, Goldrath AW. CD8+ T cell metabolism in infection and cancer. Nat Rev Immunol. 2021 Nov 12;21(11):718–38.
- Rao S, Schieber AMP, O’Connor CP, Leblanc M, Michel D, Ayres JS. Pathogen-Mediated Inhibition of Anorexia Promotes Host Survival and Transmission. Cell. 2017 Jan;168(3):503-516.e12.
- Komi DEA, Redegeld FA. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin Rev Allergy Immunol. 2020 Jun 29;58(3):313–25.
- Davis BP, Rothenberg ME. Eosinophils and Cancer. Cancer Immunol Res. 2014 Jan 1;2(1):1–8.
- Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011 Jul 30;32(10):2149–59.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




