1. Introduction
Orthoflavivirus omskense (formerly known as Omsk hemorrhagic fever virus, OHFV) is a tick-borne flavivirus that causes Omsk hemorrhagic fever (OHF), a severe zoonotic disease, which is endemic to Western Siberia of the Russian Federation [
1]. OHFV is most frequently transmitted to humans after the bite of ticks of the genera
Dermacentor and
Ixodes. Humans can also become infected through contact with infected muskrats (
Ondatra zibethicus) [
2,
3]. During the 20th century, several OHF outbreaks were reported in the former USSR; however, this infection was found only in Western Siberia [
4]. The most severe outbreak was reported from 1946 to 1958, during which more than 1000 cases of Omsk hemorrhagic fever infection were detected [
5]. In this century, no cases of OHF have been observed until 2022 [
4]. In 2022, OHF was detected in the Republic of Kazakhstan that was the first case of the infection registered outside the Russian Federation. OHFV was detected in the cerebrospinal fluid of a patient who died of encephalitis of unknown etiology in Almaty city, located 1000 km from the nearest region, where OHFV had previously been reported [
6].
The origin of OHFV is believed to be associated with a host-jumping event, when the virus transitioned from its primary vector, the
Ixodes persulcatus tick, to a new host, the muskrat [
3]. This host shift is thought to have occurred between 1931 and 1947, coinciding with the introduction of muskrats into Western Siberia [
5,
7]. The appearance of muskrats in this region created an ecological niche that allowed OHFV to adapt and establish itself as a distinct viral entity. Phylogenetic studies suggest that OHFV evolved from the tick-borne encephalitis virus (TBEV), with the virus undergoing adaptive amino acid substitutions in its E protein, which facilitated its transition to the muskrat host [
1,
3,
8]. This adaptation enabled OHFV to utilize the muskrat as both a reservoir and an amplifying host, leading to its establishment in Western Siberia [
7].
Currently, the primary natural reservoir of OHFV is the muskrat, which plays a crucial role in maintaining the virus in the environment, as they are highly susceptible to infection and can experience fatal epizootics [
3,
9]. The virus is maintained within muskrat populations through metaxenosis, a process where the virus is transmitted between different host species. In addition to muskrats, other small mammals and ticks may also contribute to the maintenance and circulation of OHFV in natural foci. Ticks, particularly
Dermacentor pictus and
Ixodes persulcatus, serve as both vectors and reservoirs of the virus. The virus can be transmitted vertically (from parent to offspring) and horizontally (between ticks and hosts) within tick populations [
2].
Omsk hemorrhagic fever is characterized by high fever, hemorrhagic manifestations, and vascular dysfunction, leading to significant morbidity [
1]. The incubation period of OHF lasts an average 3-7 days. In all cases, the disease is characterized by continuous fever with a temperature of 39°C-40°C with the following symptoms: cough, headache, muscle aches, diarrhea, abdominal pain, rehydration, bleeding from the nose, mouth and uterus, as well as skin hemorrhages. Fever may be accompanied by chills that last 8-15 days. With the course of the disease, primary hemorrhagic complications progress, and in very severe cases, there are gastrointestinal and pulmonary bleeding [
4]. Usually, the duration of the disease is 1-2 weeks, after which 50-70% of patients recover without any complications. In 30-50% of cases, the second phase of the disease occurs; it lasts 5-14 days and is characterized by high fever and symptoms of diffuse encephalitis, such as continuous headache and meningitis. Bruises appear on the skin at pressure or injection sites. In addition, the lungs and kidneys may be affected, and bronchitis and pneumonia can appear. Chronic forms of OHF in humans have not been reported. In children, meningitis has been registered in 41% of cases [
1].
The OHFV genome is a positive-sense single-stranded RNA of ~10,8 kb in length, with the open reading frame (ORF) is approximately 10,200 bases [
7]. OHFV ORF is flanked by the untranslated regions (UTRs). The 5’ untranslated region of OHFV contains a 5’ cap and has a sequence of about 30 nucleotides, which is not typical for other tick-borne flaviviruses [
10]. OHFV ORF encodes a polyprotein that undergoes post-translational processing by viral and cellular proteases and consists of three structural (C, prM, E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) [
11].
Non-structural flavivirus protein 1 (NS1) is a conserved protein of ~352 amino acid residues in length with a molecular mass ranging from 46 kDa to 55 kDa depending on glycosylation status [
12,
13]. NS1 is the only flaviviral nonstructural protein secreted by infected cells [
14]. It is known that NS1 of mosquito-borne flaviviruses plays an important role in the viral pathogenesis of hemorrhagic fevers caused by the viruses, particularly West Nile and dengue viruses (WNV and DENV) [
15,
16]. The ability of this protein to affect the endothelial permeability formed by the endothelium of various tissues has been demonstrated [
15,
17]. NS1-mediated vascular leakage has been extensively studied in mosquito-borne flaviviruses, namely DENV, Zika virus, Japanese encephalitis virus (JEV), and WNV [
18,
19]. Recently, the ability of the NS1 protein of TBEV to affect the permeability of human lung microvascular endothelial cell (HLMVEC) has been reported [
20]. The pathogenesis of OHF and the role of the OHFV NS1 protein in it remains poorly understood.
Endothelial cells form a crucial barrier that regulates vascular integrity and permeability [
21]. Disruption of this barrier could contribute to hemorrhagic symptoms observed in OHF. In this study, the ability of the OHFV NS1 protein to affect the microvascular endothelial permeability in various endothelial types was investigated. The ability of this protein to increase the permeability of endothelium-derived human lung microvascular endothelial cells (HLMVEC) and human umbilical vein endothelial cells was demonstrated using the solute flux assay and TEER. Using RNAseq, we found increased mRNA level of genes associated with cellular stress responses, vascular signaling, and cell–cell junction regulation in OHFV NS1-treated HLMVEC cells.
2. Materials and Methods
2.1. Sera and Mammalian Cells
The plasmid pET32a-OHFV_NS1-sof, the plasmid pSB, Escherichia coli XL1-Blue cells and CHO-s cells, were obtained from the Collection of Extremophile Microorganisms and Type Cultures of ICBFM SB RAS. Endothelial cells HLMVEC was kindly provided by dr. Andrey Markov ICBFM SB RAS. Endothelial cells HUVEC was kindly provided by dr. Sargis Khachatryan State Novosibirsk Regional Clinical Hospital. TBEV positive sera were obtained from the Collection of Extremophile Microorganisms and Type Cultures of ICBFM SB RAS.
CHO-s cells were cultured using CD FortiCHO medium (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 2 mM GlutaMax I (Thermo Fisher Scientific, Waltham, MA, USA) and 1-x antibiotic antimycotic solution (Thermo Fisher Scientific, Waltham, MA, USA) in CO2-shaker at 150 rpm, 8% CO2 and 37° C.
HLMVEC and HUVEC were cultured using EndoGRO-MV Complete Culture Media (Sigma Aldrich, St. Louis, MO, USA) in 24 wells plate treated with Collagen, Type I from rat tail (Sigma Aldrich, St. Louis, MO, USA).
2.2. Phylogenetic Analysis Of NS1 Amino Acids Sequences
Amino acid sequences of the nonstructural protein 1 (NS1) from selected flaviviruses were obtained from the GenBank database. Multiple sequence alignment was carried out using the MUSCLE algorithm implemented in
MEGA 12. A phylogenetic tree was inferred using the Maximum Likelihood method in MEGA 12, applying the LG model with a discrete Gamma distribution (+G) to account for rate variation among sites and allowing for a proportion of invariant sites (+I). The reliability of the inferred tree topology was assessed using 1,000 bootstrap replicates. The final tree was visualized using the integrated tree viewer in MEGA and manually annotated for clarity. OHFV NS1 protein glycosylation sites were predicted using the NetNglyc server, which predicts N- linked glycosylation sites in human proteins [
22].
2.3. Construction of a Plasmid Encoding the NS1 OHFV Protein
The OHFV NS1 protein gene was amplified by PCR using primers Start_NS1_OHFV_56_pSB 5’- CCGTTGATATCGACGTTGGATGTGCTGTGGACACTGA -3’ and End_NS1_ OHFV_56_pSB 5’- CCGTTGGATCCGTGGTGATGGTGATGGTGAGCCACCACCATCGAGCGCAC-3’ and the pET32a-OHFV_NS1 as a matrix [10.3390/v16071032]. The expression plasmid pSB and PCR fragment were then cleaved by restriction endonucleases EcoRV and BamHI (Sibenzyme, Novosibirsk, Russia) and combined in a ligation reaction. E coli XL1-Blue cells (recA1, endA1, gyrA96, thi, hsdR17(rK-, mK+), supE44, relA1, lac, [F′, proAB+, laclqZΔM15, Tn10(Tetr)]) were transformed with the resulting ligation product and seeded on LB-agar with ampicillin at a dose of 50 μg/ml and cultured. Individual colonies of E. coli cells containing plasmid pSB-OHFV_NS1 were screened by PCR using the same primers. PCR amplification conditions were as follows: 5 min at 95 °C, followed by 30 cycles of 30 s at 95 °C, 20 s at 56 °C, 1.5 min at 72 °C, and a final elongation of 6 min at 72 °C. Obtained PCR products were assessed by electrophoresis in 1% agarose gel. The accuracy of the insertion of the gene encoding the NS1 OHFV protein was confirmed by Sanger sequencing using primers NS1_SEQ_55U 5’- ACCAGAGTGATCGAGGCTGGGG -3`and NS1_SEQ_55L 5’- CAGCGACGTAATCCCCCGTATG -3`. The resulting plasmid was pSB-OHFV_NS1, which encodes an OHFV NS1 protein with a His-tag at the C-terminus.
2.4. OHFV NS1 Protein Production and Purification
The suspension CHO-s cells were grown in 125 ml Erlenmeyer Flask until reached cell density 2×106 cells/ml. Then cells were co-transfected with obtained plasmid pSB-OHFV_NS1 and pSB100x, encoded gene of sleeping beauty transposase using PeiPRO transfection reagent (Polyplus, Strasbourg, France). Efficacy of transfection were assessed using flowcytometry and confocal microscopy by evaluated signal level of GFP in transfected cells 24 hours after transfection. 48 hours after transfection GFP-positive cells were sorted using cell sorter SH800 (Sony Biotechnology inc., CA, USA) into 24-well plate, containing selective media (CD FortiCHO, 2 mM glutamine, 1x solution antibiotic-antimycotic, 10 µg/ml puromycin). Selective media were replaced each 3-4 days to fresh.
The OHFV NS1 protein was purified from the culture medium using metal-chelate chromatography on Ni-NTA agarose (Qiagen, Germany) according to the manufacturer’s instructions. Purified OHFV NS1 protein was dialyzed to phosphate buffer solution (PBS) and concentrated using Amicon centrifuge concentrators with a cutoff 30 kDA to a concentration of 1 mg/mL. Purified OHFV NS1 was sterilized using a 0.22 μm syringe filter and stored at +4 C.
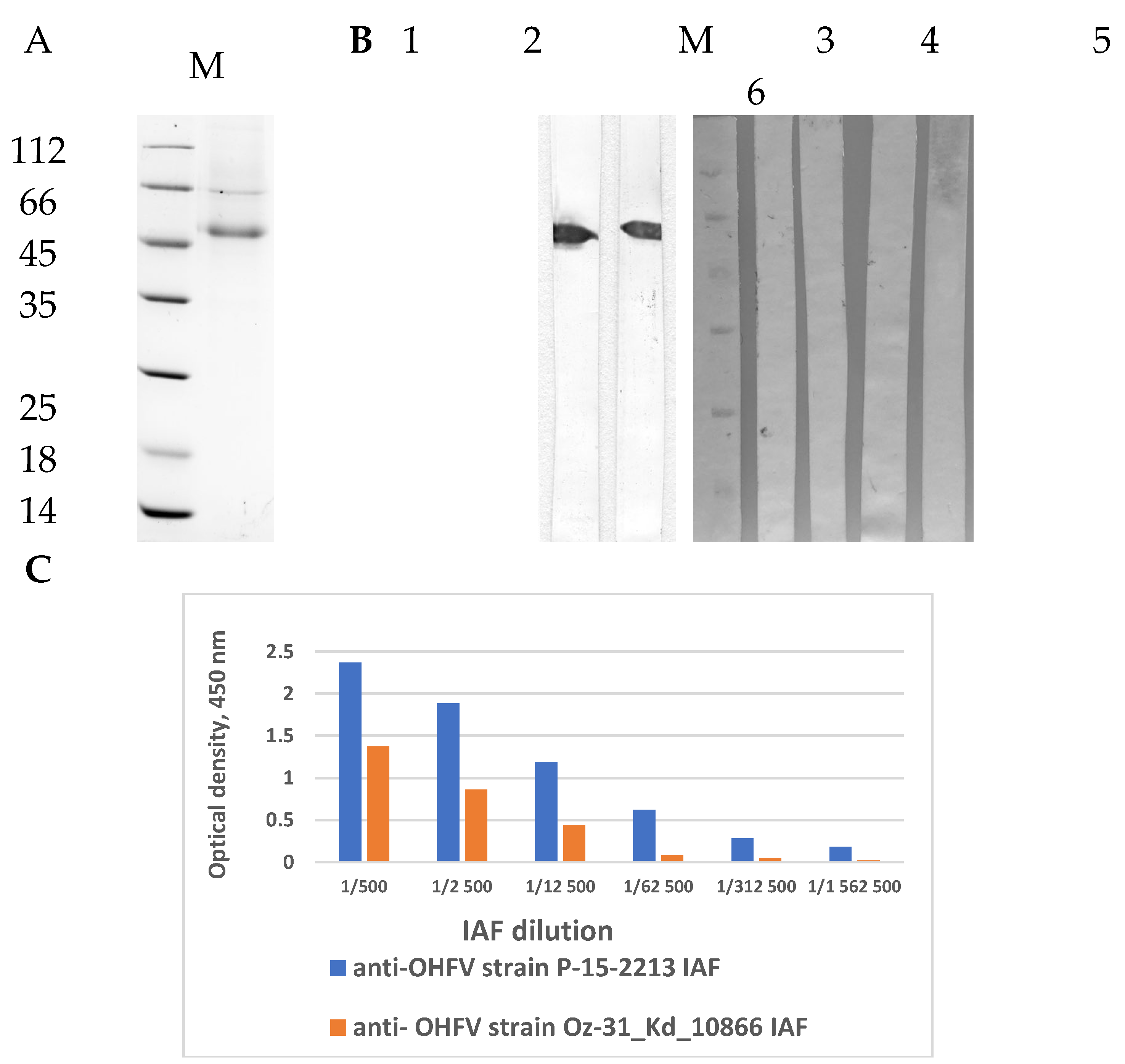
2.5. ELISA and Western Blot Analysis
For indirect ELISA, 1 μg/ml of purified recombinant OHFV NS1 protein was adsorbed into the wells of 96-well polystyrene plates (Greiner, Kremsmünster, Austria), then the non-specific binding sites were blocked with 5% skim milk solution. Immune ascitic fluids obtained from OHFV-infected mice at a dilution of 1:5000 or monoclonal antibodies against NS1 TBEV protein (N=5) at a concentration of 10 μg/ml were added to the wells [
23,
24]. Then, wells were incubated with Anti-Mouse IgG (Fc specific) HRP conjugated antibody produced in rabbit (Biosan, Novosibirsk, Russia). Immune complexes were detected using 3,3’,5,5’-tetramethylbenzidine (TMB, Applichem, Solon, OH, Germany). Optical density was assessed at a wavelength of 450 nm using a microplate reader iMark (Bio-Rad, Hercules, CA, USA).
The purified recombinant OHFV NS1 protein were fractionated using 12.5% PAGE and then transfer to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). The nonspecific binding sites were blocked with 3% bovine serum albumin solution (BSA, Amresco, Solon, OH, USA). The membrane was incubated with immune ascitic fluids obtained from OHFV-infected mice at a dilution of 1:5000. The membrane was then incubated with Anti-Mouse IgG (Fc specific)–Peroxidase antibody produced in rabbit (Biosan, Novosibirsk, Russia). Immune complexes were detected using 4-chloro-1-naphthol (Applichem, Darmstadt, Germany). Monoclonal antibodies against NS1 TBEV protein were used as a negative control.
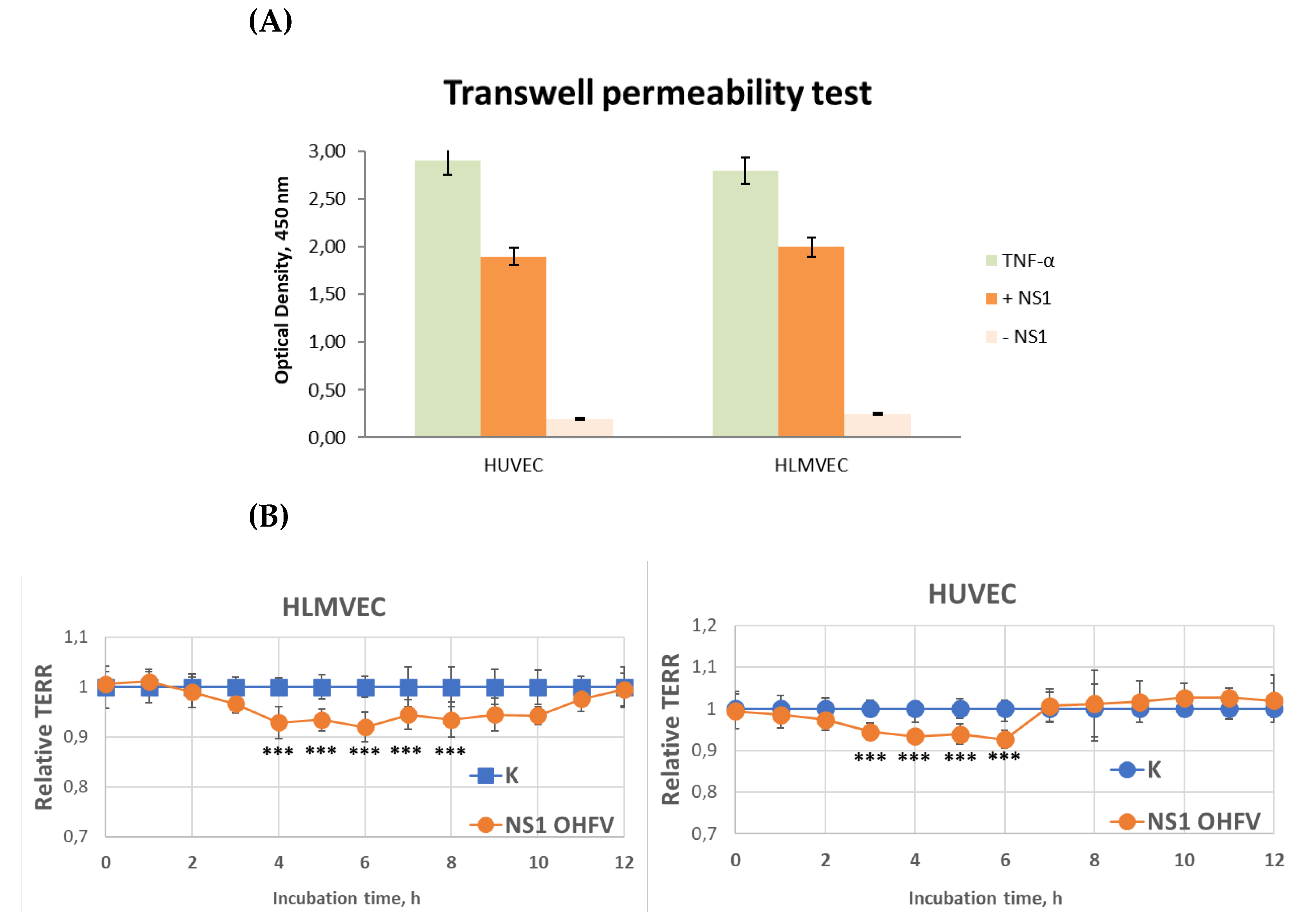
2.6. Solute Flux Assay
To assess the effect of OHFV NS1 protein on the transit of macromolecules through the human epithelial cell monolayer, HLMVEC or HUVEC cells (60,000 or 80,000 cells/insert, respectively) were grown on collagen-coated PC Membrane Cell Culture Inserts for 24 well plates with pore size 0.4 μm and diameter 6.5 mm in the volume of 300 μL of EndoGRO-MV Complete Culture Media per insert. Each insert was transferred inside a well of 24-well plate containing 1.2 ml of EndoGRO-MV Complete Culture Media. One day before the experiment, 50% of the medium was replaced with fresh EndoGRO-MV Complete Culture Media. Recombinant OHFV NS1 protein at a concentration of 10 μg/mL was then added to an insert containing a monolayer of cells. Five hours after the start of the experiment, streptavidin conjugated with horseradish peroxidase (Sigma) was added to the insert at a final concentration of 100 ng/ml and incubated for 20 minutes at 37°C. The inserts were removed, and 100 μl of culture fluid was collected from each well (lower chamber) of a 24-well plate. Horseradish peroxidase activity was determined using tetramethyl benzidine, and the concentration of biotin-conjugated horseradish peroxidase from the insert into a well of a 24-well plate was determined by plotting a standard horseradish peroxidase curve. The signal was measured using an iMark plate reader (Bio-Rad). TNF-α (100 ng/mL) was used as a positive control and untreated cell monolayers were used as a negative control.
2.7. Endothelial Permeability Test by Measure Trans-Endothelial Electrical Resistance (TEER)
The permeability of endothelial cells treated with recombinant OHFV NS1 proteins was assessed by measuring TEER of these cells. A total of 50,000 cells for HUVEC and HLMVEC were seeded in the PC Membrane Cell Culture Inserts for 24 wells plates with pore size 0.4 μm and diameter 6.5 mm (Wuxi NEST Biotechnology Co.,Ltd, China), in the volume of 300 μL of EndoGRO-MV Complete Culture Media per insert. Each insert was transferred inside a well of 24-well plate containing 1.2 ml of EndoGRO-MV Complete Culture Media. 24-well plates with inserts were incubated at 37°C and 5% CO2 until TEER ranges of 150-180 ohm (Ω) were reached. Recombinant OHFV NS1 protein at a final concentration of 10 μg/mL was then added to an insert containing a monolayer of cells. TEER values were measured at consecutive 1-hour time points after the treatment of recombinant protein using an EVOM3 Epithelial Volt/Ohm (TEER) Meter (World Precision Instruments, Sarasota, FL. USA). TEER values were measured at consecutive 1-hour time points after addition of test proteins using an epithelial volt-ohmmeter (EVOM) with “wand” electrodes (World Precision Instruments). Endothelial permeability was calculated as relative TEER, using the following formula: (Ω treated endothelial cells - Ω medium)/(Ω untreated endothelial cells - Ω medium).
2.8. RNA-seq
A total of 100,000 cells of HLMVEC were seeded in the wells of 12 well plate in the volume of 1 mL of EndoGRO-MV Complete Culture Media. 12-well plate was incubated at 37°C and 5% CO2 until monolayer of HLMVEC cells was formed. The cells was treated with 50 μl per well recombinant OHFV NS1 protein in the concentration of 200 μg/ml in PBS or only 50 μl PBS and incubated for 3 hours at 37 °C and 5% CO2. After culture media was removed and 1 ml of TRIzol™ Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was added to each well to lyse cells. Then, cell lysates were frozen and storage at -70 °C. RNA isolation and RNA seq was carried out in the laboratory of Genomed LLC (Moscow, Russia).
2.9. Statistics
Statistically significant differences between OHFV NS1 treated endothelium cells group and OHFV NS1 non-treated endothelium cells group were evaluated by two-way ANOVA analysis using Dunnett’s test for multiple comparisons. The Statistica 10 software package (StatSoft Inc., Tulsa, OK, USA) was used to perform statistical analysis.
4. Discussion
Endothelial cells form a crucial barrier that regulates vascular integrity and permeability [
21,
26]. Disruption of this barrier could contribute to hemorrhagic symptoms observed in OHF. Emerging evidence suggests that flavivirus NS1 proteins can interact with endothelial cells in a tissue-specific manner, leading to increased permeability and vascular dysfunction [
17,
27].
The close phylogenetic relationship between OHFV and TBEV supports the hypothesis that these viruses share a recent common ancestor and may have diverged following ecological or geographic isolation events. The phylogenetic positioning of OHFV suggests that it evolved within the tick-borne flavivirus lineage, likely originating from a common ancestral virus circulating in Eurasia. Its close association with TBEV, particularly the Siberian subtype, is consistent with their overlapping geographic ranges and shared vector species (e.g., Dermacentor reticulatus, Ixodes persulcatus). This phylogenetic evidence supports earlier hypotheses proposing that OHFV may have diverged from a TBEV-like ancestor in Western Siberia, possibly due to host adaptation, ecological niche specialization, or historical dispersal patterns.
Other tick-borne flaviviruses, including KFDV AHFV, and POWV, occupy distinct clades, indicating a deeper evolutionary divergence from the OHFV/TBEV lineage. Similarly, mosquito-borne flaviviruses such as DENV, JEV, and WNV form separate and evolutionarily distant lineages, clearly demarcated from the tick-borne cluster. Although OHFV is clinically classified as a hemorrhagic fever, phylogenetic analyses based on NS1 protein sequences consistently place it in close evolutionary proximity to members of the TBEV complex, such as TBEV, Louping ill virus (LIV), and Turkish sheep encephalitis virus (TSEV). In contrast, other hemorrhagic tick-borne flaviviruses, namely KFDV and AHFV, cluster separately within the tick-borne group. This phylogenetic distinction suggests that, despite overlapping clinical features, the hemorrhagic phenotype in OHFV likely emerged independently from that of KFDV and AHFV. Taken together, these facts support a model of convergent evolution, in which hemorrhagic manifestations evolved in parallel within distinct tick-borne flavivirus lineages through separate ecological and host-driven adaptations, rather than being inherited from a common hemorrhagic ancestor.
Phylogenetic analysis of the amino acid sequence of the OHFV NS1 protein showed that it is clustered distantly from the NS1 proteins of flaviviruses causing hemorrhagic fevers and carried by mosquitoes (DENV, YFV) with an identity level of less than 50% and ticks (KDFV and AHFV) with an identity level of less than 75%. OHFV NS1 is phylogenetically closest to TBEV NS1 of the Siberian subtype (identity level more than 85%). According to one version, OHFV originated from TBEV [
3]. Notably, a single outbreak of TBEV of the Far Eastern subtype with hemorrhagic form has been detected in 1999 in Novosibirsk region [
28] and no new cases of tick-borne encephalitis with hemorrhagic form were reported. Probably, this outbreak was caused by a combined infection with TBEV and a pathogen, which was undetected that time.
Bioinformatics analysis showed that OHFV NS1 protein has a theoretical N-linked glycosylation pattern similar to TBEV NS1. OHFV NS1 has three putative N-linked glycosylation sites at residues N85, N207 and N223. TBEV and LIV are known to have three putative N-linked glycosylation sites at residues 85, 207 and 223 [
29]. Most members of the genus Flavivirus have two N-linked glycosylation sites N130 and N207, including JEV, ZIKV, and all four serotypes of DENV [
30]. Some representatives of mosquito-borne flaviviruses such as WNV, SLEV and MVEV have a third glycosylation site located at residue N175 [
31]. Probably n-linked glycosylation of NS1 protein of flaviviruses does not affect the appearance of hemorrhagic symptoms.
The predicted structure of OHFV NS1 was compared with the predicted structure of TBEV NS1, showing their structural similarities [
25]. Nevertheless, differences in the antigenic profiles of OHFV NS1 and TBEV NS1 have been proved previously [
25] as monoclonal antibodies against TBEV NS1 and sera from volunteers with confirmed TBE did not bind OHFV NS1.
The ability of TBEV NS1 to influence endothelial permeability was previously investigated. This study showed that TBEV NS1 demonstrates tissue specificity to endothelial cells. TBEV NS1 increased endothelial permeability formed by HLMVEC cells but not HUVECs. RNAseq indicated that treatment of HLMVEC cells with TBEV NS1 activated the TNF-signaling pathway [
20].
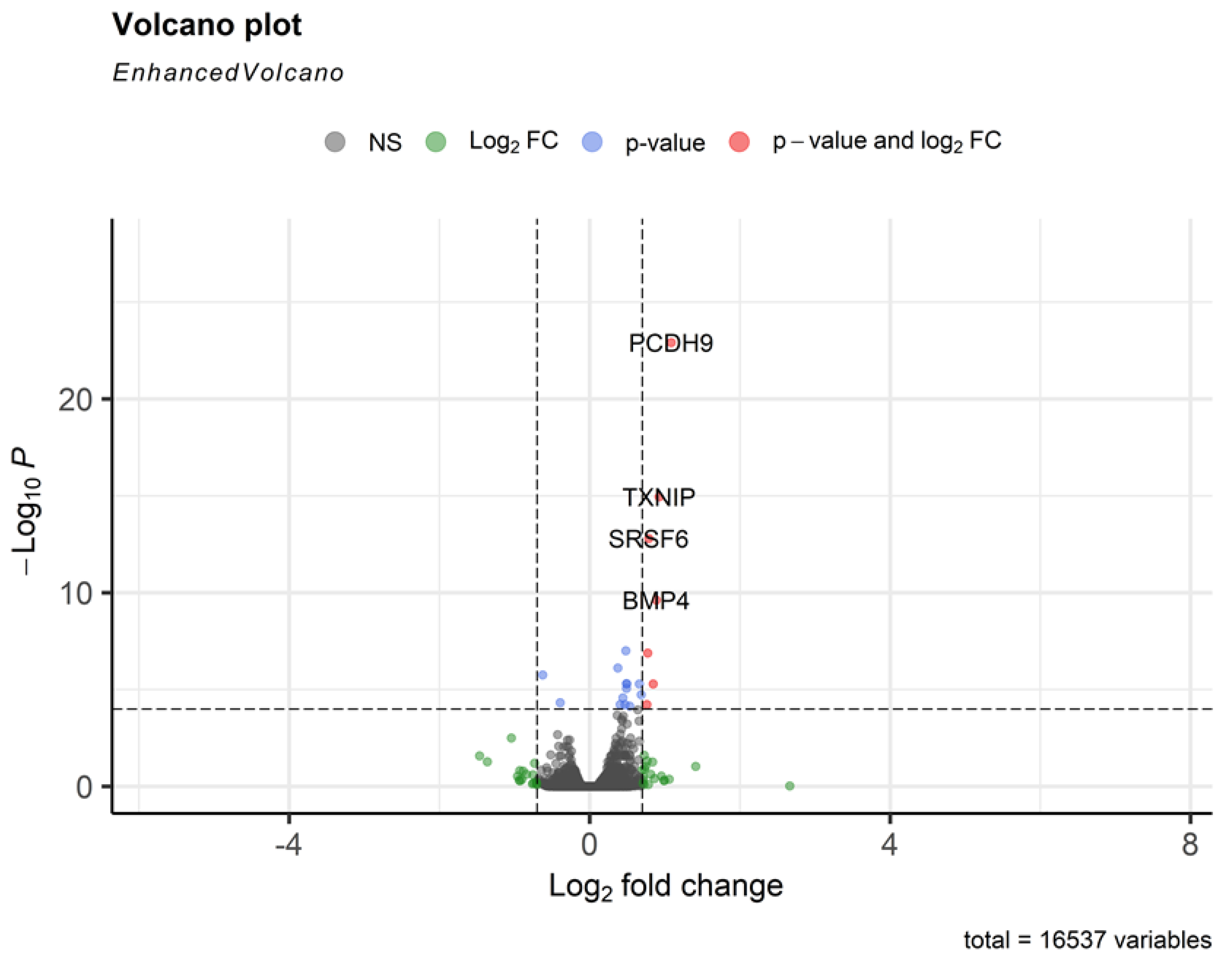
In contrary to TBEV NS1 protein, OHFV NS1 protein did not demonstrate tissue-specificity to endothelial cells. TBEV NS1 increased the permeability of the endothelium formed by both HLMVEC and HUVEC. RNAseq results showed that treatment of HLMVEC cells with TBEV NS1 activated the expression of TXNIP (a redox-responsive gene), SRSF6 (involved in RNA splicing), BMP4 (linked to vascular development), and PCDH9 (a protocadherin involved in cell adhesion). These transcriptional shifts suggest a coordinated activation of pathways related to cellular stress responses, vascular signaling, and cell–cell junction regulation.
NS1 proteins from various mosquito-borne flaviviruses have been shown to increase endothelial cell permeability in tissues associated with each flavivirus disease’s viral tropism. Specifically, NS1 from DENV, which causes systemic disease, causes hyperpermeability in endothelial cells of the lung, skin, umbilical vein, brain, and liver [
32]. NS1 from ZIKV, which affects the placenta and developing brain, causes hyperpermeability only in endothelial cells of the umbilical vein and brain [
19]. NS1 from YFV, which is systemic but causes predominantly hepatic lesions, had the strongest effect in hepatic endothelial cells permeability, with a slight increase in permeability in pulmonary endothelial cells [
17].
Although OHFV and TBEV are closely related species, the clinical manifestations of infection with these viruses differ significantly. Like dengue fever, OHF is a systemic disease associated with nasal, oral, uterine, and cutaneous hemorrhages. Probably, both OHFV NS1 and DENV NS1 can cause endothelial permeability disorders in a wide range of tissues and organs, promoting viral penetration into these organs and being one of the factors in the development of hemorrhagic complications. However, TBE manifests as meningitis, encephalitis or meningoencephalitis without hemorrhagic complications. One of the probable reasons for such differences in pathogenesis is the tissue specificity of TBEV NS1 to the endothelium.
Author Contributions
Conceptualization, Y.A.K. and A.L.M.; methodology, B.I.K., Y.A.K. and A.L.M.; software, B.I.K., Y.A.K., A.A.K. and A.L.M.; validation, Y.A.K. and B.I.K; formal analysis, B.I.K., Y.A.K., L.A.E., A.A.K. and A.L.M.; investigation, B.I.K., Y.A.K., A.A.K., A.O.S., L.A.E., S.M.K. and A.L.M.; resources, Y.A.K., N.V.T.; data curation, Y.A.K. and B.I.K.; writing—original draft preparation, B.I.K., Y.A.K., N.V.T. and A.L.M.; writing—review and editing, A.L.M. and N.V.T.; visualization, B.I.K. and A.L.M.; supervision, A.L.M.; project administration, Y.A.K. and N.V.T.; funding acquisition, Y.A.K. and N.V.T. All authors have read and agreed to the published version of the manuscript.