1. Introduction
Orthoflavivirus encephalitidis, formerly known as tick-borne encephalitis virus (TBEV), is a flavivirus found in at least 27 countries in Europe and Asia. Tick-borne encephalitis (TBE) is diagnosed in 10,000–15,000 patients each year [
1,
2]. The infection is most often transmitted through the bite of an infected tick and, in rare cases, through the consumption of infected unpasteurized dairy products [
3,
4]. The virus can cause infection of varying severity, from asymptomatic infection to serious disease of the central nervous system, resulting in a variety of neurological symptoms and potential long-term outcomes, including death. The clinical manifestations of TBE consist of the first phase characterized by headache and fever and the second phase, in which myelitis can cause changes in consciousness, tremor, ataxia, and paresis [
5]. Three recognized subtypes of the virus were described, namely the European (TBEV-Eu), Siberian (TBEV-Sib), and Far Eastern (TBEV-FE) subtypes, as well as two recent Baikal (TBEV-Blk) and Himalayan (TBEV-Him) subtypes, which are characterized by different disease outcomes [
2].
The TBEV genome is a positive-sense single-stranded RNA of approximately 11 kb in length [
6]. The open reading frame is >10,000 bases long and is flanked by untranslated regions (UTRs). It encodes a polyprotein that undergoes co- and post-transcriptional processing and consists of three structural and seven non-structural proteins [
7,
8].
Non-structural protein 1 (NS1) of flaviviruses is a highly conserved protein of approximately 352 amino acid residues s with a molecular mass of 46 to 55 kDa, depending on glycosylation [
9]. Among flaviviral non-structural proteins, NS1 is the only one secreted by infected cells [
10]. NS1 glycosylation is important for its effective secretion and interaction with complement [
11]and NS1 is a known inhibitor of complement and activator of toll-like receptors [
12,
13,
14]. If NS1 is deglycosylated, the efficiency and replication rate of flaviviruses decrease [
15]. NS1 can be found as a monomer (intracellular protein), dimer (membrane-bound protein), and hexamer (secreted protein) [
16]. Intracellular NS1 plays a central role in flaviviral replication, while secreted and membrane-bound NS1 is involved in the anti-flavivirus immune response [
17]. Flaviviral NS1 contains a glycosyl-phosphatidylinositol (GPI) anchor moiety, which facilitates the attachment of NS1 to the lipid rafts at the cell surface. GPI-anchored proteins are known to activate or inactivate intracellular signaling pathways when binding to specific antibodies or natural ligands [
18].
The presence of secreted form of NS1 (sNS1) was reported in circulation during primary and secondary infections that elicits high concentration of IgG. The anti-NS1 antibodies cross-react with a wide range of host proteins, namely human blood clotting factors, integrin/adhesion proteins, and components of extracellular matrix [
19]. It was shown that monoclonal and polyclonal anti-NS1 antibodies can interact with fibronectin, plasma fibronectin and peptides containing the RGD (arginine-glycine-aspartic acids) motif, which alters the normal functioning of the vascular system. This likely underlies vascular leakage in patients with flavivirus infections [
20]. It was found that deglycosylated NS1 mutant N207Q has a reduced ability to trigger endothelial layer glycocalyx (EGL) disruption and does not cause endothelial hyperpermeability in West Nile virus and Zika virus infections [
12,
18,
19].
It has previously been shown that NS1 proteins of mosquito-borne flaviviruses selectively bind to a variety of human endothelial cells isolated from brain, dermis, lung, liver, and ileal vein [
21]. Treatment with NS1 proteins alters endothelial permeability in vitro and induces tissue-specific vascular leakage in mouse models of flavivirus infections, reflecting the pathophysiology of each flavivirus. Moreover, exposure to NS1 of different mosquito-borne flaviviruses leads to differential disruption of endothelial glycocalyx components (EGL), resulting in endothelial hyperpermeability [
21]. However, the molecular determinants of NS1, which are necessary to trigger EGL disruption, and the cellular pathways involved in this process remain unknown [
22]. Only the impact of the NS1 proteins of mosquito-borne flaviviruses on vascular permeability was described [
12,
18,
19,
20,
21] and no data on the impact of the TBEV NS1 protein on vascular permeability were published.
In this study, the effect of TBEV NS1 protein on vascular permeability was examined using human umbilical vein endothelial cells (HUVEC) and human lung microvascular endothelial cells (HLMVEC) as in vitro models of the vascular endothelium. The obtained data indicated that TBEV NS1 induced endothelial hyperpermeability of HLMVECs but not HUVECs, and activation of TNF-α and other inflammatory signaling pathways mediate the effects of the TBEV NS1 protein.
2. Materials and Methods
2.1. Sera, Plasmids, Bacterial and Mammalian Cells
Sera from patients with confirmed TBE (54% males and 46% females) hospitalized at Novosibirsk Infectious Disease Clinical Hospital No. 1 from April to September 2017-2019, and sera from healthy volunteers [
42] were used in this study. Volunteers were healthy adults without chronic diseases (including autoimmune diseases), who had not been infected and/or hospitalized for at least six months. Informed consent was obtained from each patient and volunteer. The study was approved by the Ethical Committee of Novosibirsk Clinical Hospital of Infectious Diseases No. 1. Sera were stored at -70°C after collection. All serum samples were tested for the presence of TBEV RNA by RT-PCR and for anti-TBEV protein E antibodies by specific ELISA (both, Vector-Best, Novosibirsk, Russia).
Plasmids pET32a-TBEV_NS1-sof and pSB, E. coli XL1-Blue cells and HEK293 cell line, were obtained from the Collection of Extremophile Microorganisms and Type Cultures of ICBFM SB RAS. Endothelial cells HLMVEC was kindly provided by dr. Andrey Markov, ICBFM SB RAS. Endothelial cells HUVEC was kindly provided by dr. Sargis Khachatryan, State Novosibirsk Regional Clinical Hospital. TBEV positive sera were obtained from the Collection of Extremophile Microorganisms and Type Cultures of ICBFM SB RAS.
HEK293 cells were cultured using DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 2 mM GlutaMax I (Thermo Fisher Scientific, Waltham, MA, USA) and 1-x antibiotic antimycotic solution (Thermo Fisher Scientific, Waltham, MA, USA). HLMVEC and HUVEC were cultured with EndoGRO-MV Complete Culture Media (Sigma Aldrich, St. Louis, MO, USA) in 24-well plates treated with rat tail collagen, type I (Sigma Aldrich, St. Louis, MO, USA).
2.2. Construction of a Shuttle Plasmid Encoding the TBEV NS1 Protein
The gene encoding TBEV NS1 protein was amplified by PCR using primers Start_NS1_TBEV_56_pSB 5’-GATGTTGGCTGTGCTGTGGACACTG-3’ and End_NS1_TBEV_56_pSB 5’-GTGATGGTGATGGTGATGTGCCACCACCATTGAGCGGACA-3’ and the pET32a-TBEV_NS1-sof as matrix [
42]. The expression plasmid pSB and the PCR fragment were then digested with the restriction endonucleases EcoRV and BamHI (Sibenzyme, Novosibirsk, Russia) and combined in a ligation reaction.
E. coli XL1-Blue cells (recA1, endA1, gyrA96, thi, hsdR17(rK-, mK+), supE44, relA1, lac, [F′, proAB+, laclqZΔM15, Tn10(Tetr)]) were transformed with the resulting ligation product and seeded on LB agar with ampicillin at a dose of 50 μg/ml and cultured. Single colonies of
E. coli cells containing plasmid pSB-TBEV_NS1-sof were screened by PCR using the same primers. PCR amplification conditions were as follows: 5 min at 95 °C, followed by 30 cycles of 30 s at 95 °C, 20 s at 56 °C, 1.5 min at 72 °C, and a final extension of 6 min at 72 °C. The resulting PCR products were assessed by electrophoresis in 1% agarose gel. The accuracy of the insertion of the gene encoding the TBEV NS1 protein was confirmed by Sanger sequencing using primers E_SEQ_55U 5’- ACCAGAGTGATCGAGGCTGGGGG -3` and E_SEQ_55L 5’-CAGCGACGTAATCCCCCGTATG-3`. The resulting plasmid was pSB-TBEV_NS1-sof, which encodes a TBEV NS1 protein with a His-tag at the C-terminus.
2.3. TBEV NS1 Protein Production and Purification
The adherent HEK293 cells were grown in a 6-well plate until they reached 70-80% confluence. Then the cells were co-transfected with the obtained plasmid pSB-TBEV_NS1-sof and pSB100x, encoding the Sleeping Beauty transposase gene, using PeiPRO transfection reagent (Polyplus, Strasbourg, France). The efficacy of transfection was assessed by flow cytometry and confocal microscopy by evaluating the signal level of GFP in transfected cells 24 hours after transfection. Cells were dissociated by trypsinization and resuspended in phosphate buffer (PBS). The, 48 hours after transfection GFP-positive cells were sorted into 24-well plates containing selective media (IMDM, 10 FCS, 2 mM glutamate, 1× antibiotic-antimycotic solution, 5 µg/ml puromycin) using a cell sorter SH800 (Sony Biotechnology Inc., CA, USA). Selective medium was replaced with fresh medium every 3-4 days.
TBEV NS1 protein was purified from the culture medium by metal chelate chromatography on Ni-NTA agarose (Qiagen, Germany) according to the manufacturer’s instructions. Purified TBEV NS1 protein was dialyzed to PBS and concentrated to a concentration of 1 mg/mL using Amicon centrifugal concentrators with a cutoff of 30 kDA. Purified TBEV NS1 was sterilized using a 0.22 μm syringe filter and stored at +4 C.
2.4. ELISA and Western Blot Analysis
For indirect ELISA, 1 μg/mL of purified recombinant TBEV NS1 protein was added to each well of 96-well polystyrene plates (Greiner, Kremsmünster, Austria) and, after blocking the non-specific binding sites with 5% skim milk solution, sera from TBE-positive patients at a dilution of 1:500 (N=26) or monoclonal antibodies against TBEV NS1 protein (N=5) at a concentration of 10 μg/mL were added [
23,
42]. After washing, the wells were incubated with rabbit anti-mouse IgG (Fc specific) HRP conjugated antibody (Biosan, Novosibirsk, Russia) or anti-human IgG (Fc specific) HRP conjugated monoclonal antibody X-53 (Biosan, Novosibirsk, Russia) for one hour at 37ºC. Immune complexes were visualized using tetramethylbenzi-dine-3,3,5,5 (TMB, Applichem, Solon, OH, Germany). Absorbance was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA). The affinity constant of mAbs was determined by ELISA as previously described by Matveev et al. [
43].
To determine the cutoff level for ELISA with sera from TBE patients, sera obtained from conditionally healthy donors who had not previously had TBE were used as a control. The mean optical density level of the control sera plus one standard deviation was taken as cutoff. The ELISA signal levels of sera from TBE volunteers above the cutoff were considered positive.
The purified recombinant TBEV NS1 protein was separated using 12.5% PAGE and then transfer to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). After blocking the nonspecific binding sites with 3% bovine serum albumin solution (BSA, Amresco, Solon, OH, USA), the membrane was incubated with anti-TBEV NS1 protein monoclonal antibodies (N=4) [
23,
42]. The membrane was then incubated with Anti-Mouse IgG (Fc specific)–Peroxidase antibody produced in rabbit (Biosan, Novosibirsk, Russia). Immune complexes were revealed using 4-chloro-1-naphthol (Applichem, Darmstadt, Germany). Immune ascitic fluid obtained from TBEV-infected mice was used as a positive control.
2.5. Solute Flux Assay
To evaluate the influence of TBEV NS1 protein on the transit of macromolecules through the human epithelial cell monolayer, HLMVEC or HUVEC cells (60,000 or 80,000 cells per insert, respectively) were grown on collagen-coated PC Membrane Cell Culture Inserts for 24 well plates with a pore size of 0.4 μm and a diameter of 6.5 mm in a volume of 300 μL of EndoGRO-MV Complete Culture Media per insert. Each insert was transferred inside a well of a 24-well plate containing 1.2 ml of EndoGRO-MV Complete Culture Media. One day before adding TBEV NS1, 50% of the medium was replaced with fresh EndoGRO-MV Complete Culture Media. Recombinant TBEV NS1 protein at a concentration of 10 μg/mL was added to an insert containing a monolayer of cells. Five hours after the start of the experiment, streptavidin conjugated with horseradish peroxidase (Sigma) was added to the insert at a final concentration of 1 μg/ml and incubated for 20 minutes at 37°C. Then, inserts were removed and 100 μl of culture fluid was collected from each well (lower chamber) of a 24-well plate. Horseradish peroxidase activity was determined using tetramethyl benzidine and the concentration of biotin-conjugated horseradish peroxidase from the insert into a well of a 24-well plate was determined by plotting a standard horseradish peroxidase curve. The signal was measured using an iMark plate reader (Bio-Rad). TNF-α (100 ng/mL) was used as a positive control and untreated cell monolayers were used as a negative control.
2.6. Endothelial Permeability Test by Measure Trans-Endothelial Electrical Resistance (TEER)
The permeability of endothelial cells treated with recombinant TBEV NS1 protein was assessed by measuring TEER of these cells. A total of 50,000 HUVEC or HLMVEC cells were seeded in the PC Membrane Cell Culture Inserts for 24 wells plates with a pore size of 0.4 μm and a diameter of 6.5 mm (Wuxi NEST Biotechnology Co.,Ltd, China), in the volume of 300 μL of EndoGRO-MV Complete Culture Media per insert. Each insert was transferred inside a well of a 24-well plate containing 1.2 ml of EndoGRO-MV Complete Culture Media. 24-well plates with inserts were incubated at 37°C and 5% CO2 until TEER ranges of 150-180 ohm (Ω) were reached. Recombinant TBEV NS1 protein at a final concentration of 10 μg/mL was then added to an insert containing a monolayer of cells. TEER values were measured at consecutive 1-hour time points after addition of TBEV NS1 using an EVOM3 Epithelial Volt/Ohm (TEER) Meter (World Precision Instruments, Sarasota, FL. USA) with “wand” electrodes (World Precision Instruments, Sarasota, FL. USA). Endothelial permeability was calculated as relative TEER, using the following formula: (Ω treated endothelial cells - Ω medium)/(Ω untreated endothelial cells - Ω medium).
2.7. RNA-seq
A total of 100,000 cells of HLMVEC were seeded into the wells of a 12-well plate in a volume of 1 mL of EndoGRO-MV Complete Culture Media. The 12-well plate was incubated at 37°C and 5% CO2 until a monolayer of HLMVEC cells was formed. The cells were treated with 50 μL per well of recombinant TBEV NS1 protein at a concentration of 200 μg/mL in PBS or 50 μL of PBS alone and incubated at 37°C and 5% CO2 for 3 hours. The culture medium was then removed and 1 ml of TRIzol™ reagent (Thermo Fisher Scientific, Waltham, MA, USA) was added to each well to lyse the cells. Cell lysates were then snap frozen and stored at -70°C. RNA isolation and RNA seq were performed in the laboratory of Genomed LLC (Moscow, Russia).
2.8. Statistics
Statistical analysis of ELISA with TBEV-positive sera from patients with TBE and healthy volunteers was carried out using one-way ANOVA. Statistically significant differences between TBEV NS1 treated group and TBEV NS1 non-treated group were evaluated by two-way ANOVA analysis using Dunnett’s test for multiple comparisons. The Statistica 10 software package (StatSoft Inc., Tulsa, OK, USA) was used to perform statistical analysis.
3. Results
3.1. Production and Purification of TBEV NS1 Protein
To produce recombinant TBEV NS1 protein, the plasmid pSB-TBEV_NS1-sof was used. In this expression plasmid, the NS1 gene is located directly downstream the leader sequence of the human albumin gene in the same open reading frame. The 6His-tag coding sequence required for Ni-NTA purification is located immediately after the NS1 gene. The plasmid pSB-TBEV_NS1-sof contains an ITR for the Sleeping Beauty transposase, the puromycin N-acetyl transferase gene and the GFP gene (
Figure S1).
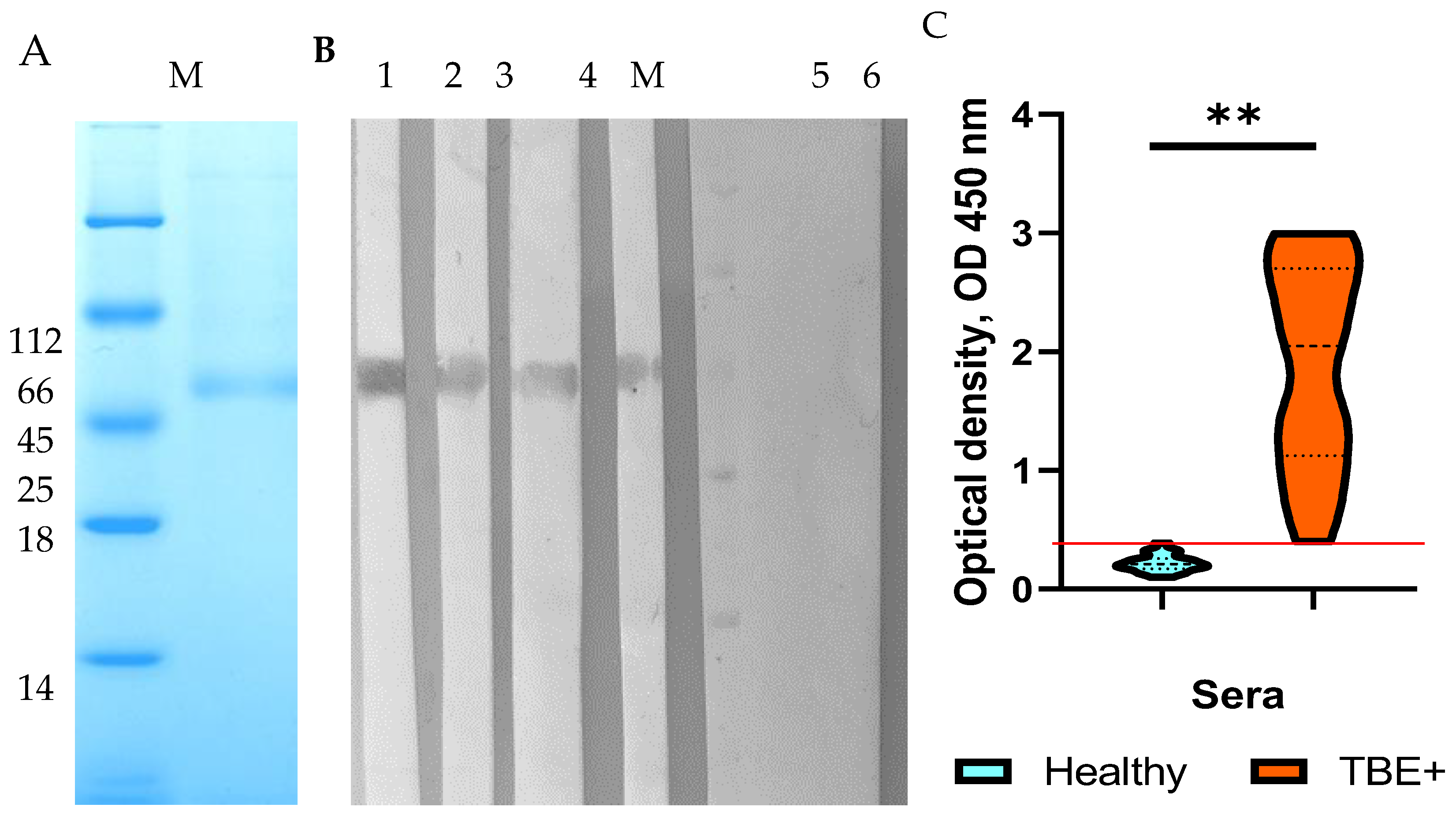
HEK293 cells were transfected with the constructed plasmid pSB-TBEV_NS1-sof. After selection with puromycin, a stable strain producing recombinant TBEV NS1 protein was obtained. The recombinant TBEV NS1 protein was purified from the culture medium by immobilized metal chelate affinity chromatography on Ni-NTA resin. Electrophoretic analysis of the purified recombinant TBEV NS1 protein indicated its homogeneity (
Figure 1a). The electrophoretic mobility of the purified NS1 protein was consistent with its theoretically predicted molecular mass of 52 kDa. The purity of the purified TBEV NS1 protein, as estimated by PAGE, was approximately 80%. A total of 3.5 mg of purified NS1 protein was obtained from 1 L of culture medium. The purified TBEV NS1 protein was concentrated to a concentration of 1 mg/mL in PBS, filtered through a 0.22 syringe filter and stored at 4 °C.
3.2. Charachterization and Immunological Properties of the Recombinat TBEV NS1 Protein
The immunological properties and antigenic profile of the recombinant NS1 protein were evaluated by ELISA and Western blot analysis using monoclonal antibodies NS1-1.3, NS1-1.6, NS1-2.299, NS1-2.290, and NS1-2.44. These monoclonal antibodies were previously obtained by hybridoma technology by fusing splenocytes from mice infected with a sublethal dose of TBEV strain Sofjin with the murine myeloma cell line SP 2/0. Importantly, the monoclonal antibodies were selected using purified native TBEV NS1 protein [
23]. Western blot analysis indicated that monoclonal antibodies NS1-1.6, NS1-2.299, NS1-2.290, and NS1-2.44 revealed a band corresponding to the recombinant NS1 protein (
Figure 1b). In ELISA, the above monoclonal antibodies bound to the HEK293-derived NS1 protein with the dissociation constants (Kd) similar to those of the native TBEV NS1 protein, whereas binding to the
Escherichia coli derived TBEV Trx-NS1 protein [
23] was 2-10 times lower than that of the native protein (
Table 1,
Figure S2).
In addition, sera from patients with TBE and healthy donors [
24] were used to test their binding to the HEK293-derived TBEV NS1 protein. The results indicated that the recombinant protein was detected by more than 95% of sera containing antibodies to the TBEV E protein, while control sera from healthy donors did not react with the HEK293-derived NS1 protein NS1 (
Figure 1c). Therefore, the obtained data confirmed that the antigenic profile of the engineered recombinant TBEV NS1 protein corresponds to that of the native TBEV NS1 protein.
3.3. Effect of Recombinant TBEV NS1 Protein on Human Endothelial Permeability In Vitro
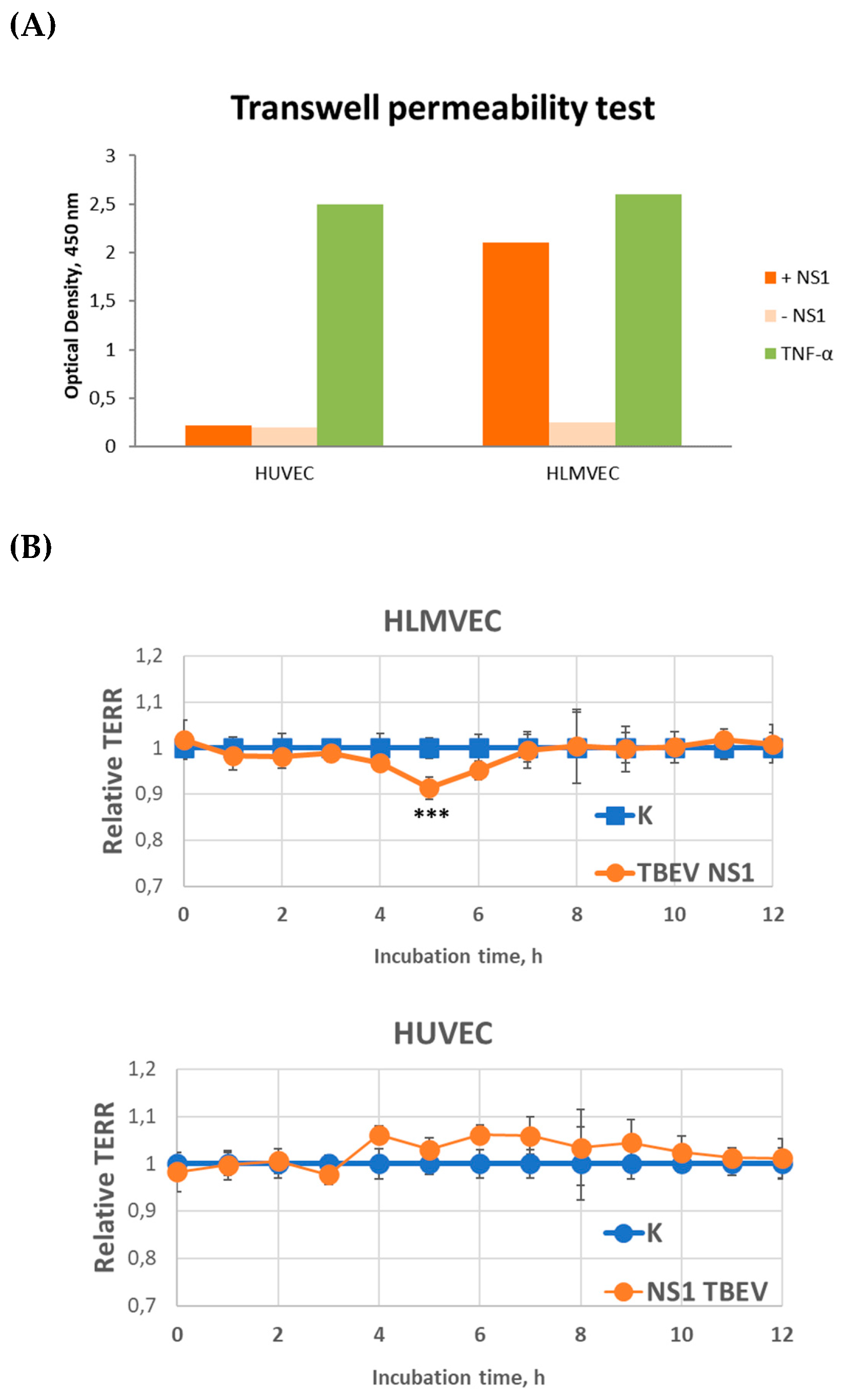
Two endothelial cell lines, HLMVEC and HUVEC, were used to study the effect of TBEV NS1 protein on endothelial permeability. The results showed that the TBEV NS1 protein had no effect on the permeability of the HUVEC endothelial monolayer in the solute flux assay (
Figure 2a). In contrast, a 4-fold increase in endothelial monolayer permeability was observed for HLMVEC cells compared to the negative control during the first 5 minutes of the experiment.
The influence of TBEV NS1 protein on endothelial cell permeability was further verified by TEER measurements and the obtained results confirmed the data from the solute flux assay. For HLMVEC cells, an 8.7% decrease in the monolayer resistance was observed compared to the control, which peaked after 5 hours, indicating increased permeability of the human pulmonary capillary endothelial layer induced by the TBEV NS1 protein (
Figure 2b). Monolayer resistance returned to its initial level 24 hours after the start of the experiment. No effect of TBEV NS1 protein on the permeability of HUVEC line was observed (
Figure 2а). The obtained results are consistent with previously reported data on the NS1 proteins of mosquito-borne flaviviruses. To determine the potential molecular mechanisms responsible for this process, RNA sequencing (RNA-seq) was performed.
3.4. Transcription Profile of Human Lung Capillary Endothelial Cells Threated with TBEV NS1 Recombinant Protein
Given the specific effect of NS1 protein on permeability of HLMVECs, but not HUVECs, a transcriptional analysis of the former was performed using RNA-seq. Differential gene expression analysis was performed comparing HLMVEC cells treated with TBEV NS1 protein and untreated control cells. The quality of the sequenced raw FASTQ files was assessed using FastQC software (version 0.11.9), and the data were mapped to the Ensembl human genome reference version hg38 using STAR, resulting in read counts per gene and sample. Genes with a maximum read count of less than 10 in all samples were excluded from further analysis. Expression levels were compared by values obtained using Qualimap software. Normalization was performed by dividing the read count of each gene by the average read count for the respective sample and scaling by the highest average read count observed between the two samples. Differences in gene expression were calculated as the log2 ratio of read counts for each gene in the NS1-treated sample relative to the control (C) sample.
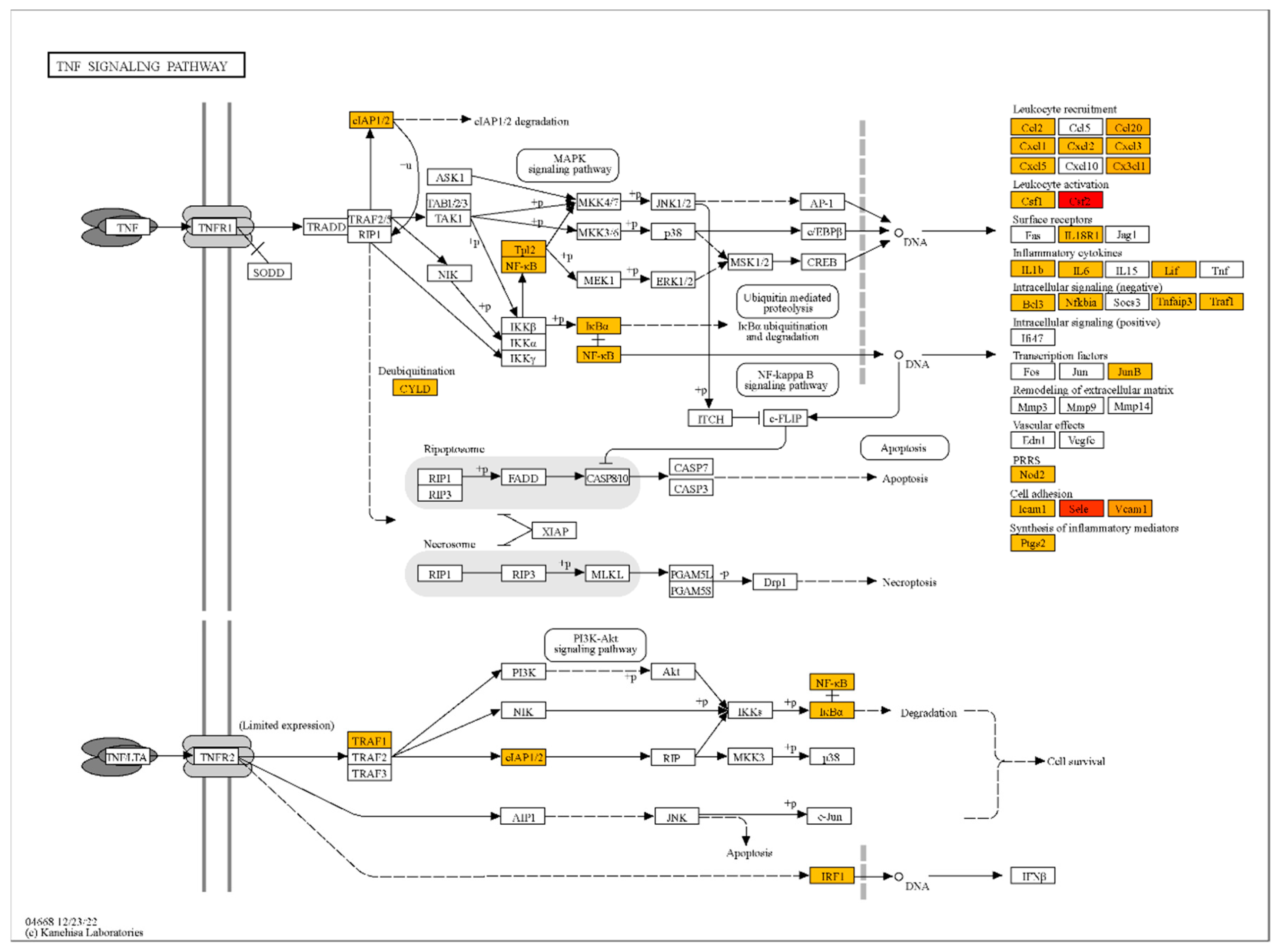
The analysis revealed that NS1 treatment significantly upregulated the mRNA levels of genes associated with endothelial activation, inflammation, and immune response in HLMVECs. Key genes with the highest fold changes included CSF2 (logFC = 9.2), SELE (logFC = 8.7), and VCAM1 (logFC = 6.9), which are involved in cytokine regulation and cell adhesion (
Table S1, S2, S3). Gene ontology term enrichment analysis was performed using ShinyGO for functional enrichment analysis to identify significantly overrepresented biological pathways. It was demonstrated, that genes associated with TNF-α signaling (TRAF1, TNFAIP2, NFKBIZ) and oxidative stress regulation (SOD2) were significantly upregulated, suggesting that NS1 activates pathways critical for endothelial dysfunction and vascular inflammation (
Figure 3). Chemokines such as CXCL1, CXCL8 and CCL2 were strongly upregulated, indicating a robust inflammatory response (
Table S1, S2). These findings highlight the role of NS1 in promoting vascular inflammation through upregulation of cytokines and adhesion molecules.
4. Discussion
In most cases, TBEV infection occurs after a tick bite. The viral particles penetrate the body with saliva during tick feeding. The virus then multiplies locally in subcutaneous tissues, including Langerhans cells (macrophage of the skin) and neutrophils of the skin. Migrating monocytes/macrophages produce TBEV [
25] and these cells can serve as vectors for transporting viral particles to lymph nodes. Replication of the virus in the lymph nodes leads to its dissemination into the bloodstream and the onset of viraemia. TBE is usually characterized by a biphasic course [
2]. During primary viraemia, the virus affects various peripheral organs and tissues; however, infection often ceases at this stage and seroconversion occurs without any obvious clinical signs [
26]. The biphasic nature of TBE reflects the initial spread of the virus to peripheral tissues, which triggers a cytokine response, followed in some cases by penetration of the virus into the central nervous system (CNS), and a second neurological phase of the disease is recorded. It is known that the blood-brain barrier (BBB), consists of endothelial cells, astrocytes, pericytes and adjacent neurons and plays a crucial role in brain homeostasis as well as in the formation of its microenvironment [
27]. Virus penetration through the BBB are prerequisites for CNS infection. This can lead to BBB dysfunction characterized by increased permeability, hypercellularity, and encephalopathy.
There are a number of various routes, by which virus particles can cross the BBB: (i) induction of BBB permeability directly [
28]; (ii) infection of microvascular endothelial cells from the BBB front line [
29,
30,
31]; (iii) direct axonal retrograde transport from infected peripheral neurons spreading across neuromuscular junctions from muscles to somatic motor neurons in the spinal cord; (iv) infection of olfactory neurons and spread to the olfactory bulb [
32]. A so-called ‘Trojan horse’ mechanism has also been described for Zika and West Nile viruses, in which the virus is transported by infected immune cells that enter the CNS from the peripheral blood [
33,
34]. It has been shown that TBEV, Langat virus, West Nile Virus, and Japanese encephalitis virus can enter CNS without disrupting BBB, and BBB permeability increases as a result of cytokine release in response to flavivirus replication in the brain [
35,
36,
37,
38]. Langat virus can probably also utilize an infection pathway via the olfactory nerve. After peripheral infection of mice, this virus was found first in the olfactory bulb and then spread to other brain regions [
39]. It is still not entirely clear whether TBEV enters the brain via olfactory neurons or otherwise. It should be noted that most studies were performed in vitro or in animal models; however, such models may not adequately reflect the sequence of events occurring in the human body when TBEV penetrates the BBB.
Once TBEV enters the human brain, it multiplies in the large neurons of the anterior horns of the spinal cord, medulla oblongata, pontine, dentate nucleus, Purkinje cells and corpus striatum [
2]. The main histological inflammatory responses observed after TBEV infection of the brain are lymphocytic-meningeal and perivascular infiltrates, as well as microglia proliferation with glial nodule formation and neuronophagia [
2].
How TBEV crosses the BBB is not clear, and various mechanisms can be probably involved in this process. One hypothesis is that TBEV neuroinvasion occurs by direct infection of microvascular endothelial cells. In an in vitro BBB model, it was shown that only 5% of microvascular endothelial cells were infected after the addition of TBEV; however, the infection persisted and spread through the BBB [
40]. Notably, a primary brain vascular endothelial cell line was used in the study [
40], which probably lead to the presence of a subpopulation of neuronal cells that could be infected with TBEV.
Our data indicated that the NS1 protein of TBEV variously affects the permeability of vascular endothelial cells. Exposure of the endothelial cells of the small capillaries of the lung with this protein leads to an increase in their permeability, while it has no effect on the endothelial cells of the umbilical capillaries. The obtained data are consistent with studies of the effects of NS1 proteins of mosquito-borne flaviviruses, for which these proteins have been shown to differentially affect the permeability of different types of endothelium [
21]. A disadvantage of this work is the lack of experiments with brain vascular endothelial cells. It was previously indicated that these cells are the most sensitive to treatment with different the mosquito-borne flavivirus NS1 proteins, with the exception of NS1 YFV, significantly reduced the permeability of the brain vascular endothelial monolayer [
21].
Analysis of the enrichment of signaling and metabolic pathways with genes in the samples of NS1-treated cells compared to untreated samples showed the highest enrichment for the TNF signaling pathway (Table 4). Other signaling pathways detected were also associated with infectious and inflammatory processes. Thus, it is likely that treatment of endothelial cells with the TBEV NS1 protein triggers inflammatory response in the endothelium, similar to the reaction induced by TNFa, which leads to disruption of the endothelial integrity through gap junctions between cells and thereby can contribute to the overcoming of the BBB by TBEV.
Previously performed RNA-seq of dendritic cells treated with TBEV NS1[
41] showed preferential downregulation of the genes encoding cytokines (TNF, Il1a, Il1b, Il6, Il12a, Il12b, Il15, Il27 and Il33), chemokines (CCL5, CXCL5, CVCL9, CXCL10, CXCL11 and CXCL16), and IFN-stimulated genes (ISGs, ISG15 and ISG20), co-stimulatory molecules (CD40, CD80, CD83 and CD86) in treated cells compared to untreated cells. Our data indicated that endothelial cells treated with TBEV NS1 increased the expression of genes involved in cytokine regulation and cell adhesion (SELE, VCAM1), genes encoding chemokines (CXCL1, CXCL8, and CCL2) and linked to TNF-α signaling (TRAF1, TNFAIP2, NFKBIZ), as well as oxidative stress regulation (SOD2). This difference in results can be explained by the fact that when the NS1 protein appears in the bloodstream, TBEV can affect different types of cells in different ways. On the one hand, TBEV NS1 is able to suppress proliferation and activation of T cells and inhibit the response to cytokine-mediated signaling in immune cells, and on the other hand, it can activate inflammatory signaling and change cell adhesion when interacting with endothelial cells. Given the multidirectional effect of TBEV NS1 on different cells, the role of this protein in the pathogenesis of TBE is underestimated and the mechanisms of protein interaction with various tissues require further investigation.
5. Conclusions
In conclusion, the aim of this study was to comprehensively characterize the effects of TBEV NS1 on endothelial barrier function and the intracellular signaling cascades it triggers, in order to better understand its contribution to TBEV pathogenesis and identify potential therapeutic targets.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, Y.A.K. and A.L.M.; methodology, B.I.K., Y.A.K. and A.L.M.; software, B.I.K., Y.A.K., A.A.K. and A.L.M.; validation, Y.A.K. and B.I.K; formal analysis, B.I.K., Y.A.K., L.A.E., A.A.K. and A.L.M.; investigation, B.I.K., Y.A.K., A.A.K., A.O.S., L.A.E., S.M.K. and A.L.M.; resources, Y.A.K., N.V.T.; data curation, Y.A.K. and B.I.K.; writing—original draft preparation, B.I.K., Y.A.K., N.V.T. and A.L.M.; writing—review and editing, A.L.M. and N.V.T.; visualization, B.I.K. and A.L.M.; supervision, A.L.M.; project administration, Y.A.K. and N.V.T.; funding acquisition, Y.A.K. and N.V.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 22-74-10103,
https://rscf.ru/project/22-74-10103/. Plasmid pET32a-NS1_TBEV-sof and CHO-S cells was obtained from the Collection of Extremophile Microorganisms and Type Cultures of ICBFM SB RAS, which is supported by the Ministry of Education and Science, Project No. 125012300671-8.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the ethical committee of the Novosibirsk Infectious Diseases Clinical Hospital No. 1 (protocol code №3, date of approval 15.07.2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
authors are very grateful to Dr. Andrey Markov from ICBFM SB RAS for donating HLMVEC cells.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TBEV |
Tick-borne encephalitis virus |
| TBE |
Tick-borne encephalitis |
| NS1 |
Non-structural protein 1 of flaviviruses |
| EGL |
Endothelial glycocalyx components |
| HUVEC |
Human umbilical vein endothelial cells |
| HLMVEC |
Human lung microvascular endothelial cells |
| sNS1 |
secreted form of NS1 |
| GPI |
Glycosyl-phosphatidylinositol |
References
- Chiffi, G.; Grandgirard, D.; Leib, S.L.; Chrdle, A.; Růžek, D. Tick-Borne Encephalitis: A Comprehensive Review of the Epidemiology, Virology, and Clinical Picture. Rev Med Virol 2023, 33, e2470. [Google Scholar] [CrossRef] [PubMed]
- Ruzek, D.; Avšič Županc, T.; Borde, J.; Chrdle, A.; Eyer, L.; Karganova, G.; Kholodilov, I.; Knap, N.; Kozlovskaya, L.; Matveev, A.; et al. Tick-Borne Encephalitis in Europe and Russia: Review of Pathogenesis, Clinical Features, Therapy, and Vaccines. Antiviral Res 2019, 164, 23–51. [Google Scholar] [CrossRef]
- Ličková, M.; Fumačová Havlíková, S.; Sláviková, M.; Klempa, B. Alimentary Infections by Tick-Borne Encephalitis Virus. Viruses 2022, 14, 56. [Google Scholar] [CrossRef]
- Michelitsch, A.; Wernike, K.; Klaus, C.; Dobler, G.; Beer, M. Exploring the Reservoir Hosts of Tick-Borne Encephalitis Virus. Viruses 2019, 11, 669. [Google Scholar] [CrossRef]
- Mittova, V.; Tsetskhladze, Z.R.; Motsonelidze, C.; Palumbo, R.; Vicidomini, C.; Roviello, G.N. Tick-Borne Encephalitis Virus (TBEV): Epidemiology, Diagnosis, Therapeutic Approaches and Some Molecular Aspects—An Updated Review. Microbiology Research 2024, 15, 2619–2649. [Google Scholar] [CrossRef]
- Formanová, P.; Černý, J.; Bolfíková, B.Č.; Valdés, J.J.; Kozlova, I.; Dzhioev, Y.; Růžek, D. Full Genome Sequences and Molecular Characterization of Tick-Borne Encephalitis Virus Strains Isolated from Human Patients. Ticks and Tick-borne Diseases 2015, 6, 38–46. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Strongin, A.Y. Structural and Functional Parameters of the Flaviviral Protease: A Promising Antiviral Drug Target. Future Virol 2010, 5, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Assenberg, R.; Mastrangelo, E.; Walter, T.S.; Verma, A.; Milani, M.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Mancini, E.J. Crystal Structure of a Novel Conformational State of the Flavivirus NS3 Protein: Implications for Polyprotein Processing and Viral Replication. J Virol 2009, 83, 12895–12906. [Google Scholar] [CrossRef]
- Perera, D.R.; Ranadeva, N.D.; Sirisena, K.; Wijesinghe, K.J. Roles of NS1 Protein in Flavivirus Pathogenesis. ACS Infect Dis 2024, 10, 20–56. [Google Scholar] [CrossRef]
- Fisher, R.; Lustig, Y.; Sklan, E.H.; Schwartz, E. The Role of NS1 Protein in the Diagnosis of Flavivirus Infections. Viruses 2023, 15, 572. [Google Scholar] [CrossRef]
- Somnuke, P.; Hauhart, R.E.; Atkinson, J.P.; Diamond, M.S.; Avirutnan, P. N-Linked Glycosylation of Dengue Virus NS1 Protein Modulates Secretion, Cell-Surface Expression, Hexamer Stability, and Interactions with Human Complement. Virology 2011, 413, 253–264. [Google Scholar] [CrossRef]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue Virus NS1 Protein Activates Cells via Toll-like Receptor 4 and Disrupts Endothelial Cell Monolayer Integrity. Sci Transl Med 2015, 7, 304ra142. [Google Scholar] [CrossRef] [PubMed]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Antagonism of the Complement Component C4 by Flavivirus Nonstructural Protein NS1. J Exp Med 2010, 207, 793–806. [Google Scholar] [CrossRef]
- Conde, J.N.; Silva, E.M.; Barbosa, A.S.; Mohana-Borges, R. The Complement System in Flavivirus Infections. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Fan, J.; Liu, Y.; Yuan, Z. Critical Role of Dengue Virus NS1 Protein in Viral Replication. Virol Sin 2014, 29, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A Multifaceted Enigmatic Viral Protein. Virol J 2016, 13, 131. [Google Scholar] [CrossRef]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 Crystal Structures Reveal a Surface for Membrane Association and Regions of Interaction with the Immune System. Science 2014, 343, 881–885. [Google Scholar] [CrossRef]
- Noisakran, S.; Dechtawewat, T.; Avirutnan, P.; Kinoshita, T.; Siripanyaphinyo, U.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Sittisombut, N. Association of Dengue Virus NS1 Protein with Lipid Rafts. J Gen Virol 2008, 89, 2492–2500. [Google Scholar] [CrossRef]
- Falconar, A.K. The Dengue Virus Nonstructural-1 Protein (NS1) Generates Antibodies to Common Epitopes on Human Blood Clotting, Integrin/Adhesin Proteins and Binds to Human Endothelial Cells: Potential Implications in Haemorrhagic Fever Pathogenesis. Arch Virol 1997, 142, 897–916. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-H.; Shyu, H.-F.; Wang, Y.-M.; Sun, D.-S.; Shyu, R.-H.; Tang, S.-S.; Huang, Y.-S. Facilitation of Cell Adhesion by Immobilized Dengue Viral Nonstructural Protein 1 (NS1): Arginine-Glycine-Aspartic Acid Structural Mimicry within the Dengue Viral NS1 Antigen. J Infect Dis 2002, 186, 743–751. [Google Scholar] [CrossRef]
- Puerta-Guardo, H.; Glasner, D.R.; Espinosa, D.A.; Biering, S.B.; Patana, M.; Ratnasiri, K.; Wang, C.; Beatty, P.R.; Harris, E. Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell Rep 2019, 26, 1598–1613.e8. [Google Scholar] [CrossRef]
- Wang, C.; Puerta-Guardo, H.; Biering, S.B.; Glasner, D.R.; Tran, E.B.; Patana, M.; Gomberg, T.A.; Malvar, C.; Lo, N.T.N.; Espinosa, D.A.; et al. Endocytosis of Flavivirus NS1 Is Required for NS1-Mediated Endothelial Hyperpermeability and Is Abolished by a Single N-Glycosylation Site Mutation. PLoS Pathog 2019, 15, e1007938. [Google Scholar] [CrossRef] [PubMed]
- Andrey, M.; Yana, K.; Olga, G.; Bogdana, K.; Sergey, T.; Lyudmila, E.; Nina, T. Tick-Borne Encephalitis Nonstructural Protein NS1 Expressed in E. Coli Retains Immunological Properties of the Native Protein. Protein Expr Purif 2022, 191, 106031. [Google Scholar] [CrossRef] [PubMed]
- Kravchuk, B.I.; Khlusevich, Y.A.; Chicherina, G.S.; Yakimenko, V.V.; Krasnova, E.I.; Tikunova, N.N.; Matveev, A.L. Cross-Reactive Antibodies to the NS1 Protein of Omsk Hemorrhagic Fever Virus Are Absent in the Sera of Patients with Tick-Borne Encephalitis. Viruses 2024, 16, 1032. [Google Scholar] [CrossRef] [PubMed]
- Labuda, M.; Austyn, J.M.; Zuffova, E.; Kozuch, O.; Fuchsberger, N.; Lysy, J.; Nuttall, P.A. Importance of Localized Skin Infection in Tick-Borne Encephalitis Virus Transmission. Virology 1996, 219, 357–366. [Google Scholar] [CrossRef]
- Prevalence of Antibodies against Tick-Borne Encephalitis among Residents of North-Eastern Poland - PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/7784806/ (accessed on 10 March 2025).
- Hawkins, B.T.; Davis, T.P. The Blood-Brain Barrier/Neurovascular Unit in Health and Disease. Pharmacol Rev 2005, 57, 173–185. [Google Scholar] [CrossRef]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.; Musso, D.; Ghawche, F. Zika Virus Infection Complicated by Guillain-Barre Syndrome--Case Report, French Polynesia, December 2013. Euro Surveill 2014, 19, 20720. [Google Scholar] [CrossRef]
- Ayala-Nunez, N.V.; Gaudin, R. A Viral Journey to the Brain: Current Considerations and Future Developments. PLOS Pathogens 2020, 16, e1008434. [Google Scholar] [CrossRef]
- Hsieh, J.T.; St John, A.L. Japanese Encephalitis Virus and Its Mechanisms of Neuroinvasion. PLoS Pathog 2020, 16, e1008260. [Google Scholar] [CrossRef]
- Chen, Z.; Li, G. Immune Response and Blood-Brain Barrier Dysfunction during Viral Neuroinvasion. Innate Immun 2021, 27, 109–117. [Google Scholar] [CrossRef]
- Watrin, L.; Ghawché, F.; Larre, P.; Neau, J.-P.; Mathis, S.; Fournier, E. Guillain-Barré Syndrome (42 Cases) Occurring During a Zika Virus Outbreak in French Polynesia. Medicine (Baltimore) 2016, 95, e3257. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Diamond, M.S. Animal Models of Zika Virus Infection, Pathogenesis, and Immunity. J Virol 2017, 91, e00009-17. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.W. Flaviviruses Are Neurotropic, but How Do They Invade the CNS? J Infect 2014, 69, 203–215. [Google Scholar] [CrossRef]
- Roe, K.; Kumar, M.; Lum, S.; Orillo, B.; Nerurkar, V.R.; Verma, S. West Nile Virus-Induced Disruption of the Blood–Brain Barrier in Mice Is Characterized by the Degradation of the Junctional Complex Proteins and Increase in Multiple Matrix Metalloproteinases. Journal of General Virology 2012, 93, 1193–1203. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Yu, L.; Cao, S.; Wang, K.; Yuan, J.; Wang, C.; Wang, K.; Cui, M.; Fu, Z.F. Viral Infection of the Central Nervous System and Neuroinflammation Precede Blood-Brain Barrier Disruption during Japanese Encephalitis Virus Infection. J Virol 2015, 89, 5602–5614. [Google Scholar] [CrossRef] [PubMed]
- Růžek, D.; Salát, J.; Singh, S.K.; Kopecký, J. Breakdown of the Blood-Brain Barrier during Tick-Borne Encephalitis in Mice Is Not Dependent on CD8+ T-Cells. PLoS One 2011, 6, e20472. [Google Scholar] [CrossRef]
- Weber, E.; Finsterbusch, K.; Lindquist, R.; Nair, S.; Lienenklaus, S.; Gekara, N.O.; Janik, D.; Weiss, S.; Kalinke, U.; Överby, A.K.; et al. Type I Interferon Protects Mice from Fatal Neurotropic Infection with Langat Virus by Systemic and Local Antiviral Responses. J Virol 2014, 88, 12202–12212. [Google Scholar] [CrossRef]
- Kurhade, C.; Zegenhagen, L.; Weber, E.; Nair, S.; Michaelsen-Preusse, K.; Spanier, J.; Gekara, N.O.; Kröger, A.; Överby, A.K. Type I Interferon Response in Olfactory Bulb, the Site of Tick-Borne Flavivirus Accumulation, Is Primarily Regulated by IPS-1. J Neuroinflammation 2016, 13, 22. [Google Scholar] [CrossRef]
- Palus, M.; Vancova, M.; Sirmarova, J.; Elsterova, J.; Perner, J.; Ruzek, D. Tick-Borne Encephalitis Virus Infects Human Brain Microvascular Endothelial Cells without Compromising Blood-Brain Barrier Integrity. Virology 2017, 507, 110–122. [Google Scholar] [CrossRef]
- Camarão, A.A.R.; Gern, O.L.; Stegmann, F.; Mulenge, F.; Costa, B.; Saremi, B.; Jung, K.; Lepenies, B.; Kalinke, U.; Steffen, I. Secreted NS1 Proteins of Tick-Borne Encephalitis Virus and West Nile Virus Block Dendritic Cell Activation and Effector Functions. Microbiology Spectrum 2023, 11. [Google Scholar] [CrossRef]
- Igolkina, Y.; Rar, V.; Krasnova, E.; Filimonova, E.; Tikunov, A.; Epikhina, T.; Tikunova, N. Occurrence and Clinical Manifestations of Tick-Borne Rickettsioses in Western Siberia: First Russian Cases of Rickettsia Aeschlimannii and Rickettsia Slovaca Infections. Ticks Tick Borne Dis 2022, 13, 101927. [Google Scholar] [CrossRef] [PubMed]
- Matveev, A.; Pyankov, O.; Khlusevich, Y.; Tyazhelkova, O.; Emelyanova, L.; Timofeeva, A.; Shipovalov, A.; Chechushkov, A.; Zaitseva, N.; Kudrov, G.; et al. Antibodies Capable of Enhancing SARS-CoV-2 Infection Can Circulate in Patients with Severe COVID-19. Int J Mol Sci 2023, 24, 10799. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).