1. Introduction
Seed quality traits play a fundamental role in crop production, influencing both agronomic performance and consumer preferences. These traits include seed size, oil content, protein composition, starch accumulation, germination rate, seed vigor, and longevity, all of which contribute to yield potential, nutritional value, and marketability. Improving seed quality is a major goal in crop breeding programs, as it directly impacts food security, industrial processing, and sustainable agriculture (Ronald, 2011; Wimalasekera, 2015; Qaim et al., 2020). However, seed quality traits are typically controlled by multiple genes and are influenced by environmental factors, making their genetic dissection highly complex.
1.1. Traditional QTL Mapping and Its Limitations
Over the past few decades, significant progress has been made in identifying genetic loci associated with seed quality traits using quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS). Traditional QTL mapping involves linkage analysis in biparental populations, allowing for the identification of genomic regions associated with phenotypic variation. While effective, this approach is often constrained by limited genetic diversity, low mapping resolution, and extensive time requirements for population development (Varshney et al., 2021). On the other hand, GWAS leverages natural genetic variation in diverse populations to detect marker-trait associations at a higher resolution (Zhu et al., 2008). However, GWAS is prone to false positives due to population structure and requires large sample sizes to achieve sufficient statistical power (Korte and Farlow, 2013).
Despite the utility of these methods, traditional QTL mapping approaches face challenges in accurately predicting seed quality traits due to the polygenic nature of these traits, gene-environment interactions, and the complexity of underlying biological networks. The emergence of high-throughput genotyping and phenotyping technologies has led to an explosion of genomic and phenomic data, necessitating more advanced computational approaches to effectively analyze and interpret these datasets.
1.2. The Role of AI and ML in QTL Mapping
Artificial intelligence (AI) and machine learning (ML) have emerged as transformative tools in plant genomics, offering novel computational frameworks for handling high-dimensional data and improving QTL identification. ML algorithms, including deep learning, support vector machines (SVMs), random forests (RFs), and Bayesian networks, can efficiently process complex genomic datasets, uncover hidden patterns, and improve trait prediction accuracy (Ma et al., 2018; Montesinos-Lopez et al., 2021). Unlike traditional statistical models, ML approaches can capture non-linear relationships between genetic markers and phenotypic traits, making them particularly well-suited for studying polygenic traits such as seed quality.
In recent years, AI and ML have been successfully applied in various aspects of crop breeding, including genomic selection, multi-omics data integration, and predictive modeling of agronomic traits (Desta and Ortiz, 2014; Crossa et al., 2017). These approaches enable the rapid identification of key genetic markers and provide insights into gene interactions and regulatory networks underlying seed quality traits. Additionally, AI-driven genomic selection models have demonstrated superior performance in predicting breeding values, allowing for more efficient selection of high-quality seed varieties in breeding programs.
Given the growing role of AI in plant genomics, this review aims to explore the application of AI and ML in identifying QTL associated with seed quality traits. First, it provides an overview of key seed quality traits and their genetic basis, emphasizing their importance in crop breeding and the challenges associated with their genetic dissection. Next, it discusses various AI and ML techniques used in QTL mapping and genomic prediction, highlighting how these approaches improve the accuracy and efficiency of trait identification compared to traditional methods. Furthermore, the review examines case studies that demonstrate AI-driven QTL discovery for seed quality traits, showcasing successful applications of ML models in different crop species. In addition, it addresses the challenges and limitations of AI-based QTL mapping, including issues related to data quality, model interpretability, computational complexity, and biological validation. Finally, it outlines future research directions and opportunities for integrating AI in crop breeding programs, focusing on emerging technologies and interdisciplinary collaborations that could further enhance the precision and applicability of AI in plant genomics. By synthesizing recent advancements in AI-driven QTL mapping, this review provides valuable insights into how AI can revolutionize the genetic improvement of seed quality traits, ultimately contributing to the development of high-yielding, high-quality crop varieties.
2. Seed Quality Traits and their Genetic Basis
Seed quality traits encompass a wide range of physical, biochemical, and physiological characteristics that influence crop productivity and post-harvest quality. These traits can be broadly categorized into:
Physical Traits: Seed size, shape, weight, and texture, which affect germination and processing quality (Lurstwut and Pornpanomchai, 2017).
Biochemical Traits: Oil, protein, sugars, isoflavones, fatty acids, fiber contents, etc. which influence nutritional quality and industrial applications (Kassem, 2021).
Physiological Traits: Germination rate, seed vigor, dormancy, and longevity, which are critical for seed storage and crop establishment (Reed et al., 2022).
The genetic regulation of these traits is highly complex, often governed by multiple QTLs and influenced by environmental factors (Kassem, 2021). High-throughput phenotyping techniques, such as near-infrared spectroscopy (NIRS) and hyperspectral imaging, have enabled the precise measurement of seed quality traits, providing large datasets for AI-driven analyses (Montesinos-Lopez et al., 2021, 2024). Advances in genomics, including next-generation sequencing (NGS) and genotyping-by-sequencing (GBS), have further facilitated the identification of genetic markers associated with seed quality traits. Integrating AI and ML in this domain offers a powerful approach to deciphering complex genotype-phenotype relationships and enhancing the efficiency of marker-assisted breeding.
3. AI and ML Techniques for QTL Mapping
3.1. Overview of AI and ML in Genomics
Artificial intelligence, particularly ML, has transformed the field of genomics by enabling high-throughput analysis of complex genetic datasets. ML models can efficiently handle large-scale omics data, uncover hidden patterns, and improve QTL prediction accuracy. Some of the most commonly used ML approaches in genomics include:
Support Vector Machines (SVM): Effective in high-dimensional genomic datasets for classification and regression tasks.
Random Forest (RF): An ensemble learning method that improves feature selection and predictive accuracy in genomic studies.
Deep Learning (DL): Neural networks, including convolutional neural networks (CNNs) and recurrent neural networks (RNNs), for analyzing multi-omics data (Ma et al., 2018; Nguyen and Wang, 2020; Montesinos-Lopez et al., 2021).
These AI techniques have been successfully applied in genomic selection, phenotypic prediction, and integrative omics analyses, making them valuable tools for QTL mapping in crop breeding (Crossa et al., 2017).
3.2. ML for QTL Mapping
Traditional QTL mapping approaches often rely on linear models, which may not effectively capture complex genetic interactions. ML models offer a non-parametric approach, allowing for the detection of epistatic interactions and gene-environment effects (Montesinos-Lopez et al., 2021, 2024).
Table 1 provides a comparative analysis of various ML models applied in QTL mapping, highlighting their strengths, limitations, and suitability for genomic prediction (
Table 1). Recent studies have demonstrated the application of ML in QTL discovery, including:
Feature Selection for Genetic Markers: RF and SVM are used to rank genetic markers based on their importance in explaining phenotypic variance.
Genomic Prediction Using DL Models: CNNs and deep neural networks (DNNs) improve the accuracy of genomic selection for seed quality traits (Montesinos-Lopez et al., 2021).
Integration of Multi-Omics Data: AI-driven multi-omics integration enhances the power of QTL detection and trait prediction (Nguyen and Wang, 2020).
By leveraging these AI techniques, researchers can accelerate the discovery of key genomic regions controlling seed quality traits and optimize breeding programs.
4. Integrating Multi-Omics Data Using AI/ML for QTL Discovery
Recent advances in genomics have highlighted the importance of integrating multi-omics data—genomics, transcriptomics, proteomics, metabolomics, and phenomics—to improve the accuracy of QTL mapping. Each of these omics layers provides valuable insights into gene regulation, metabolic pathways, and phenotypic expression. However, the integration of multi-omics data poses significant computational challenges due to the high dimensionality and heterogeneity of datasets. AI and ML techniques provide powerful solutions for handling and analyzing such complex datasets, allowing for more precise identification of QTL associated with seed quality traits.
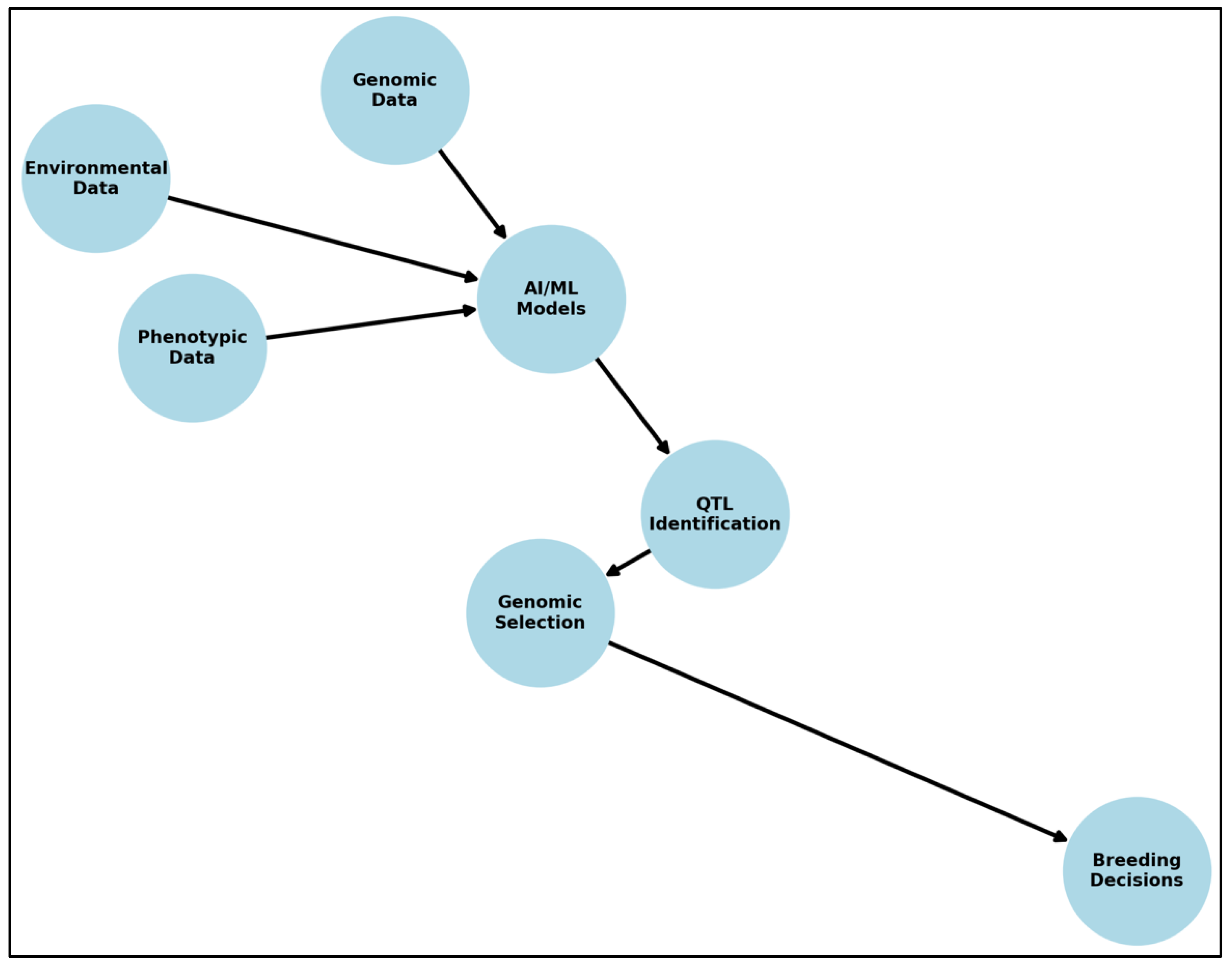
Figure 1 illustrates the AI/ML workflow for QTL mapping, demonstrating how genomic, phenotypic, and environmental data are processed through ML models to enhance QTL identification and genomic selection (
Figure 1).
4.1. AI-Based Approaches for Multi-Omics Integration
ML models can efficiently process and integrate multi-omics data to identify the key genomic regions associated with seed quality traits. Some of the most widely used AI-based approaches include:
Deep Neural Networks (DNNs): These models can learn hierarchical representations from multi-omics data, allowing for better QTL prediction.
Bayesian Networks: These probabilistic graphical models help in modeling gene-trait interactions across multiple omics layers.
Multi-View Learning: This technique integrates different types of omics data while maintaining their unique contributions to the phenotype (Nguyen and Wang., 2020).
Graph Neural Networks (GNNs): Used for modeling gene regulatory networks and detecting interactions between genes and metabolites (Hasibi et al., 2024).
4.2. Case Studies on AI-Driven Multi-Omics QTL Mapping
Several studies have successfully applied AI-driven multi-omics integration for QTL discovery in crops. For example:
Soybean Seed Quality: AI models integrating genomics and metabolomics have identified key QTLs controlling oil and protein content in soybean.
Rice Grain Quality: Deep learning-based multi-omics integration has improved genomic prediction for starch composition and amylose content in rice (Montesinos-Lopez et al., 2021, 2024).
Wheat Seed Germination: ML-driven transcriptomic analysis has led to the identification of genes regulating seed dormancy and vigor in wheat (Montesinos-Lopez et al., 2021, 2024).
By leveraging these AI techniques, researchers can gain deeper insights into the genetic mechanisms underlying seed quality traits, ultimately enhancing breeding efficiency.
5. Case Studies on AI-Driven QTL Discovery
The application of AI and ML in QTL mapping has gained momentum in recent years, enabling researchers to identify genetic loci associated with complex traits more efficiently. Several studies have demonstrated the power of ML in genomic prediction, SNP selection, and marker-trait association analysis, particularly for seed quality traits (Crossa et al., 2017; Ma et al., 2018; Kassem, 2025). This section highlights a case study applying ML to identify QTLs for seed mineral nutrients in soybean, illustrating the integration of AI techniques in genomic research.
5.1. ML-Based QTL Mapping for Seed Mineral Nutrients in Soybean
Seed mineral nutrients, such as Nickel (Ni), Molybdenum (Mo), Iron (Fe), Zinc (Zn), Boron (B), and Manganese (Mn), play essential roles in both plant metabolism and human nutrition. These micronutrients are key targets in crop improvement programs, as their accumulation in seeds is influenced by genetic factors and environmental interactions. Traditional QTL mapping approaches have provided valuable insights into the genetic control of these traits, but integrating ML methods offers a more data-driven approach to enhance prediction accuracy and marker selection.
5.1.1. Genetic Basis of Seed Mineral Nutrients
Recent studies utilized a Forrest × Williams 82 Recombinant Inbred Line (RIL) population grown in two different environments—Spring Lake, NC (2018), and Carbondale, IL (2020)—to identify QTLs influencing mineral nutrient accumulation (Bellaloui et al., 2023, 2024). Genetic analyses revealed varying levels of heritability among different minerals, with Ni exhibiting higher heritability (H² = 0.311) compared to Mo, which showed strong gene-by-environment interactions and lower heritability (Bellaloui et al., 2023). These findings indicate the complexity of mineral accumulation in soybean seeds, where both genetic and environmental factors contribute to trait variation.
5.1.2. QTL Identification and Candidate Genes
Using 2,075 single nucleotide polymorphisms (SNPs), QTL mapping identified significant loci associated with seed mineral content. SNPs on chromosomes 8, 9, and 14 were strongly associated with Ni and Mo accumulation (Bellaloui et al., 2023). Additionally, Fe and Zn accumulation was linked to candidate genes such as iron-ion binding oxidoreductase and zinc finger proteins (Bellaloui et al., 2024), while the accumulation of B and Mn was associated with genes encoding ATP-binding ABC transporters and metal detoxification proteins. These findings align with previous studies that have reported mineral-associated QTLs on multiple soybean chromosomes, reinforcing the polygenic nature of these traits.
5.1.3. ML Applications for SNP Identification
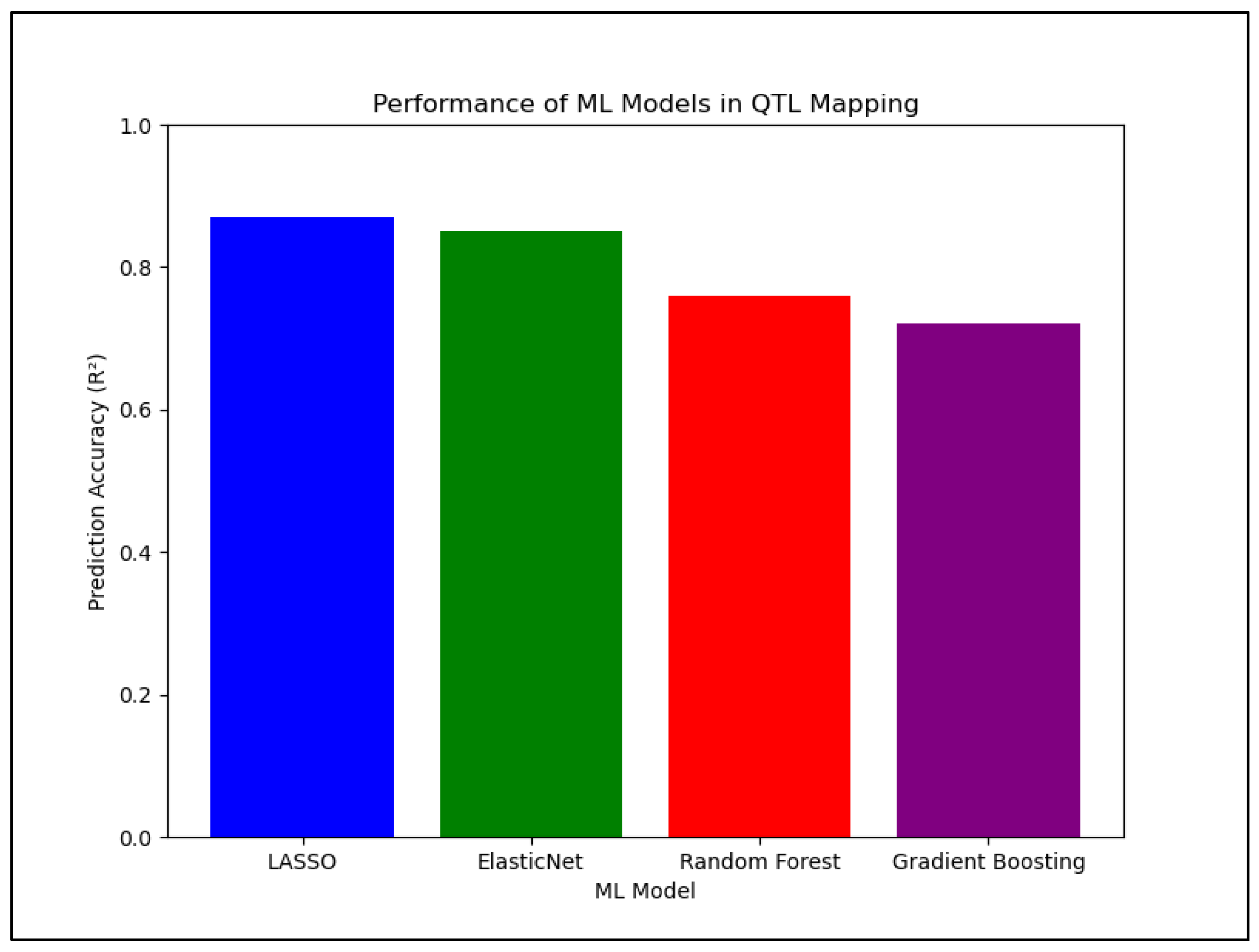
To improve SNP selection and QTL identification, Kassem (2025) applied multiple machine learning (ML) models—including LASSO Regression, Random Forest, Gradient Boosting, and ElasticNet—to the Carbondale, IL (2020) dataset to predict significant SNPs associated with mineral accumulation. As shown in
Figure 2, LASSO Regression and ElasticNet outperformed tree-based models, consistently yielding the lowest Root Mean Square Errors (RMSE) and the highest prediction accuracy (R² scores) across most mineral traits. In contrast, Random Forest and Gradient Boosting exhibited higher error rates and negative R² scores, indicating challenges in predictive accuracy and the need for improved feature selection (
Figure 2).
These results underscore the importance of choosing appropriate ML algorithms and fine-tuning hyperparameters to enhance SNP prediction. While ML-based feature selection provided valuable insights, traditional QTL mapping methods—such as Interval Mapping (IM) and Composite Interval Mapping (CIM)—were still recommended for validating SNP-trait associations and ensuring biological relevance (Kassem, 2025).
5.1.5. Future Directions and Integration with Breeding Programs
The integration of machine learning (ML) with traditional QTL mapping provides a powerful framework for identifying candidate genes and genomic regions associated with seed mineral content. By leveraging ML techniques, researchers can improve the precision of QTL detection, enhance genomic prediction accuracy, and ultimately facilitate marker-assisted selection (MAS) in crop breeding. However, to fully realize the potential of AI-driven QTL mapping, several key research directions must be pursued.
One crucial area for improvement is the refinement of ML models. Enhancing hyperparameter tuning, feature selection strategies, and model interpretability will be essential for increasing the accuracy and reliability of predictions. Advanced ML techniques, such as ensemble learning and deep neural networks, could be explored to further optimize the identification of significant genetic markers.
Expanding the genetic datasets used in ML training is another priority. Larger and more genetically diverse populations will strengthen QTL validation, improving the generalizability of AI-driven genomic predictions across different soybean varieties and environmental conditions. The inclusion of multi-omics data, such as transcriptomics and metabolomics, could further enhance the predictive power of ML models.
Additionally, functional validation of candidate genes is necessary to confirm the biological relevance of AI-identified genomic regions. Cutting-edge gene-editing tools like CRISPR-Cas9 can be used to experimentally verify the role of key genes in mineral nutrient accumulation, ensuring that computationally predicted loci translate into practical breeding applications.
Finally, AI-assisted breeding pipelines should be developed to integrate ML-driven QTL mapping into soybean breeding programs. These pipelines can streamline the selection of nutrient-rich cultivars, accelerating the breeding cycle and enhancing the nutritional quality of crops. By incorporating AI-driven decision-making into traditional breeding workflows, researchers can make more data-informed selections, ultimately contributing to improved agricultural sustainability and food security.
5.2. ML Applications for Other Seed Quality Traits
Beyond seed mineral nutrient accumulation, machine learning (ML) and deep learning (DL) approaches have been increasingly utilized to predict and map QTLs for seed quality traits, including seed morphology, oil and protein content, seed yield, image-based trait assessment, and biochemical composition (
Table 2). The following studies demonstrate the growing impact of AI and ML in genomic selection and plant phenotyping (
Table 2).
5.2.1. ML for Seed Morphology and Phenotyping
The ability to analyze seed morphology is essential for understanding genetic variation, domestication, and breeding selection. Seki and Toda (2022) applied instance segmentation neural networks to analyze seed shape in lettuce (Lactuca spp.) and identified 11 QTLs related to seed area, width, length, circularity, and eccentricity. Notably, three QTLs—qLWR-3.1, qECC-3.1, and qCIR-3.1—were located in a genomic region previously associated with lettuce domestication traits, highlighting the genetic basis of seed shape selection (Seki and Toda, 2022).
High-throughput phenotyping techniques have also been enhanced using AI-driven tools. AIseed, a novel automated seed image analysis software, enables large-scale assessment of seed shape, color, and texture, reducing the need for manual annotation (Tu et al., 2023). Additionally, QTG-Finder2, a ML algorithm, was designed to accelerate the discovery of causal genes linked to quantitative traits in plants, facilitating genetic analysis across multiple crop species (Lin et al., 2020).
A study by Miranda et al. (2023) applied convolutional neural networks (CNNs) to analyze RGB seed images, achieving 98% segmentation accuracy and predicting hundred-seed weight (HSW) with high precision (Miranda et al., 2023). Similarly, Duc et al. (2023) used Random Forest (RF) and Multiple Linear Regression (MLR) to predict HSW in soybean, achieving R² values of 0.98 and 0.94, respectively (Duc et al., 2023).
Deep learning approaches are also being optimized using synthetic datasets. A study on barley seed morphology demonstrated that neural networks can be effectively trained with synthetic data, eliminating the need for extensive manual annotation, making high-throughput seed phenotyping more scalable (Toda et al., 2020).
5.2.2. ML for Seed Oil, Protein, and Biochemical Composition
Oil and protein content are key determinants of seed quality. Yoosefzadeh-Najafabadi et al. (2023) applied support vector regression (SVR)-mediated GWAS to map QTLs linked to soybean seed protein and oil content, identifying a higher number of relevant QTLs compared to conventional FarmCPU-based GWAS (Yoosefzadeh-Najafabadi et al., 2023). Similarly, Parsaeian et al. (2020) demonstrated how ANN models integrated with image processing can be used for non-invasive estimation of oil and protein content in sesame (Parsaeian et al., 2020).
Machine learning has also improved biochemical trait assessment in seeds. X-ray imaging combined with CNNs has been used to assess tomato seed quality, revealing that low-opacity seeds with minimal damage had the highest germination rates. The Mask R-CNN deep learning model effectively categorized seed quality into four classes, demonstrating the potential for non-destructive seed viability assessment (Pessoa et al., 2023).
5.2.3. ML for Seed Yield and Genomic Selection
ML models have been successfully used to predict seed yield and genomic selection traits in crops. A study on rapeseed (Brassica napus L.) found that Nu-SVR (support vector regression) with a quadratic polynomial kernel function achieved the highest performance in predicting seed yield (SY), with an R² value of 0.86 (Shahsavari et al., 2023). The study also demonstrated that combining multilayer perceptron neural networks (MLPNNs) with feature selection techniques allowed accurate yield predictions using only three agronomic traits (Shahsavari et al., 2023).
Similarly, XGBoost-based gene regulatory network analysis has been applied to seed cotton yield prediction. This study combined expression quantitative trait loci (eQTL) mapping with ML-driven gene prioritization, identifying NF-YB3, FLA2, and GRDP1 as key regulators of seed size and yield in cotton (Zhao et al., 2023).
5.2.4. ML and Deep Learning in Multi-Omics and High-Throughput Plant Phenotyping
Multi-omics strategies are increasingly being used to enhance seed quality and nutritional traits. A study on pigeonpea (Cajanus cajan) integrated genomics, transcriptomics, proteomics, and metabolomics to identify genes regulating seed size, protein content, and disease resistance, providing insights into breeding nutrient-dense legume varieties (Singh et al., 2020).
AI-driven high-throughput phenotyping tools have also been developed for dynamic panicle trait analysis in rice. Panicle-iAnalyzer, a deep learning-based pipeline, enables the measurement of panicle traits (i-traits) and plant growth metrics, supporting rice breeding programs (Geng et al., 2024). Furthermore, a comprehensive review of deep learning-based segmentation techniques has explored how CNNs and instance segmentation models can enhance plant phenotyping by overcoming challenges like background noise and lighting variability (Tang et al., 2024).
These studies demonstrate the growing impact of ML in seed quality research, showcasing its ability to enhance trait prediction, accelerate breeding cycles, and facilitate high-throughput phenotyping in diverse crop species.
6. Challenges and Limitations of AI/ML in QTL Mapping
Despite the remarkable advancements in AI and ML for QTL mapping, several challenges and limitations remain. These challenges need to be addressed to ensure the reliability and applicability of AI-driven approaches in crop breeding.
6.1. Data Quality and Availability
One of the primary challenges in applying AI to QTL mapping is the need for large, high-quality datasets. ML algorithms require extensive training data to generate accurate predictions. However, plant breeding programs often have limited datasets due to the cost and time required for phenotyping and genotyping (Montesinos-Lopez et al., 2021, 2024). Furthermore, missing data and batch effects in multi-omics datasets can introduce biases in AI models.
6.2. Model Interpretability and Biological Validation
AI models, particularly deep learning algorithms, often function as “black boxes,” making it difficult to interpret the biological significance of the predictions (Montesinos-Lopez et al., 2021, 2024). While these models can accurately predict trait variations, they may not provide clear mechanistic insights into gene function and regulation. Interpretability techniques, such as SHAP (Shapley Additive Explanations) and attention mechanisms, are being developed to enhance transparency in AI-driven QTL analysis. However, biological validation of AI-predicted QTLs remains a major challenge, requiring experimental confirmation through gene knockout or overexpression studies.
6.3. Computational Complexity and Scalability
Processing high-dimensional genomic and phenomic datasets requires significant computational resources. Training deep learning models on large-scale multi-omics data can be time-consuming and computationally expensive. Cloud-based platforms and high-performance computing (HPC) infrastructure are increasingly being used to address these scalability issues (Montesinos-Lopez et al., 2021, 2024; Hasibi et al., 2024). However, accessibility to such resources remains a limitation for many research institutions.
6.4. Ethical and Regulatory Concerns
The use of AI in plant genomics raises ethical and regulatory concerns, particularly regarding data privacy and intellectual property rights. Large-scale genomic datasets are often shared across institutions and countries, necessitating robust data governance frameworks to ensure ethical use and equitable benefits. Additionally, AI-driven genomic prediction models may favor elite breeding programs, potentially widening the gap between resource-rich and resource-poor breeding initiatives (Nguyen and Wang., 2020).
Addressing these challenges will be crucial for the widespread adoption of AI in QTL mapping and plant breeding.
7. Future Directions and Opportunities
AI and ML have immense potential to revolutionize QTL mapping and accelerate crop improvement. Future research should focus on enhancing the accuracy, interpretability, and applicability of AI-driven models.
7.1. Integration of AI with Emerging Technologies
The next frontier in AI-driven QTL mapping will involve integrating AI with emerging technologies such as:
Quantum Computing: To accelerate the processing of large-scale genomic datasets.
Edge Computing: For real-time genomic prediction and phenotyping.
Synthetic Biology: To design AI-guided gene editing strategies for improving seed quality traits.
7.2. AI-Assisted Breeding Pipelines
Developing AI-assisted breeding platforms that combine genomics, phenomics, and environmental data will enable precision breeding. AI-driven decision support systems can help breeders prioritize candidate lines with optimal seed quality traits.
7.3. Enhanced Model Interpretability
Developing explainable AI models will be essential for bridging the gap between computational predictions and biological validation. Incorporating functional genomics and network-based approaches can improve model interpretability.
7.4. Open-Source AI Platforms for Genomics
Collaborative efforts to develop open-source AI tools for QTL mapping can democratize access to cutting-edge AI technologies in plant breeding. Initiatives such as the AI for Agriculture Innovation Consortium aim to create publicly available AI-driven genomic prediction models (Montesinos-Lopez et al., 2021, 2024).
By addressing these future directions, AI-driven QTL mapping can significantly contribute to sustainable agriculture and global food security.
8. Conclusion
AI and ML are transforming the field of QTL mapping by providing innovative solutions for analyzing complex genomic datasets and predicting seed quality traits. By leveraging advanced computational models, researchers can enhance the accuracy and efficiency of QTL identification, leading to improved breeding strategies. However, challenges related to data quality, model interpretability, and computational scalability must be addressed to fully realize the potential of AI in plant genomics. Future research should focus on integrating AI with emerging technologies, improving model transparency, and fostering collaborative AI-driven breeding initiatives. By embracing these advancements, AI-powered QTL mapping can play a pivotal role in developing high-quality, climate-resilient crop varieties, ensuring global food security.
References
- Abdipour, M.; Ramazani, S.H.R.; Younessi-Hmazekhanlu, M.; Niazian, M. Modeling Oil Content of Sesame (Sesamum indicum L.) Using Artificial Neural Network and Multiple Linear Regression Approaches. J. Am. Oil Chem. Soc. 2018, 95, 283–297.
- Bellaloui N, D Knizia, J Yuan, , TD Vuong, M Usovsky, Q Song, F Betts, T Register, E Williams, N Lakhssassi, H Mazouz , HT Nguyen, K Meksem, A Mengistu, and MA Kassem. Genetic Mapping for QTL Associated with Seed Nickel and Molybdenum Accumulation in the Soybean ‘Forrest’ by ‘Williams 82’ RIL Population. Plants 2023, 12(21), 3709. [CrossRef]
- Bellaloui N, D Knizia, J Yuan, Q Song, F Betts, T Register, E Williams, N Lakhssassi, H Mazouz, HT Nguyen, K Meksem, A Mengistu, and MA Kassem. Genomic regions and candidate genes for seed iron and seed zinc accumulation identified in the soybean ‘Forrest’ by ‘Williams 82’ RIL population. Int. J. Plant Biol. 2024, 15(2): 452-467. [CrossRef]
- Crossa, J., Perez-Rodriguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., delosCampos, G., Burgueño, J., Camacho-González, J. M., Pérez-Elizalde, S., Beyene, Y., Dreisigacker, S., Singh, R., Zhang, X., Gowda, M., Roorkiwal, M., Rutkoski, J., and Varshney, R. K. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Heredity, 2017, 22(11), 961–975. [CrossRef]
- Desta, Z.A., Ortiz, R. Genomic selection: genome-wide prediction in plant improvement. Trends in Plant Science 2014, 19(9), 592-601. [CrossRef]
- Emamgholizadeh, S.; Parsaeian, M.; Baradaran, M. Seed yield prediction of sesame using artificial neural network. Eur. J. Agron. 2015, 68, 89–96.
- Geng, Z., Lu, Y., Duan, L., Chen, H., Wang, Z., Zhang, J., Liu, Z., Wang, X., Zhai, R., Ouyang, Y., Yang, W. High-throughput phenotyping and deep learning to analyze dynamic panicle growth and dissect the genetic architecture of yield formation. Crop and Environment 2024, 3(1), 1-11. [CrossRef]
- Haider, S.A.; Naqvi, S.R.; Akram, T.; Umar, G.A.; Shahzad, A.; Sial, M.R.; Khaliq, S.; Kamran, M. LSTM Neural Network Based Forecasting Model for Wheat Production in Pakistan. Agronomy 2019, 9, 72.
- Hasibi, R., Michoel, T., Oyarzún, D.A. Integration of graph neural networks and genome-scale metabolic models for predicting gene essentiality. npj Syst Biol Appl 2024, 10, 24. [CrossRef]
- Kassem MA. QTL and Candidate Genes for Seed Mineral Nutrients and Application of Machine Learning Models to Predict Seed Composition Traits in Soybean. The International Plant and Animal Genome Conference (PAG 32), January 10-15, 2025, San Diego, CA. Oral Presentation.
- Kassem, M.A. Soybean Seed Composition: Protein, Oil, Fatty Acids, Amino Acids, Sugars, Mineral Nutrients, Tocopherols, and Isoflavones. Springer 2021. [CrossRef]
- Korte, A., Farlow, A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9, 29 (2013). [CrossRef]
- Lee, S.; Jeong, Y.; Son, S.; Lee, B. A Self-Predictable Crop Yield Platform (SCYP) Based on Crop Diseases Using Deep Learning. Sustainability 2019, 11, 3637.
- Lin, F., Lazarus, E.Z., Rhee, S,Y. A Generalized Machine-Learning Algorithm for Prioritizing QTL Causal Genes in Plants, G3 Genes|Genomes|Genetics 2020, 10(7): 2411–2421. [CrossRef]
- Lurstwut, B., Pornpanomchai, C. Image analysis based on color, shape and texture for rice seed (Oryza sativa L.) germination evaluation. Agriculture and Natural Resources 2017, 51(5), 383-389. [CrossRef]
- Ma, W., Qiu, Z., Song, J., Li, J., Cheng, Q., Zhai, J., and Ma, C. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 2018, 248, (5):1307–1318. [CrossRef]
- Montesinos-Lopez, O.A., Chavira-Flores, M., Kismiantini, Crespo-Herrera, L., Saint-Piere, C., Li, H., Fritsche-Neto, R., Al-Nowibet, K., Montesinos-Lopez, A., and Crossa, J. A review of multimodal deep learning methods for genomic-enabled prediction in plant breeding. Genetics, 2024, 228(4), iyae161. [CrossRef]
- Montesinos-Lopez, O.A., Montesinos-Lopez, A., Pérez-Rodríguez, P., Barrón-López, J.A., Martini, J.W.R., Fajardo-Flores, S,B., Gaytan-Lugo, L.S., Santana-Mancilla, P.C., and Crossa, J. A review of deep learning applications for genomic selection. BMC Genomics 2021, 22:19. [CrossRef]
- Nguyen, N.D., Wang, D. Multiview learning for understanding functional multi-omics. PLOS Comp. Biology 2020, 16(4): e1007677. [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Abdipour, M. Artificial neural network and multiple regression analysis models to predict essential oil content of ajowan (Carum copticum L.). J. Appl. Res. Med. Aromat. Plants 2018, 9, 124–131.
- Niedbala, G. Application of artificial neural networks for multi-criteria yield prediction of winter rapeseed. Sustainability 2019, 11, 533.
- Niedbala, G.; Kozłowski, R.J. Application of Artificial Neural Networks for Multi-Criteria Yield Prediction of Winter Wheat. J. Agric. Sci. Technol. 2019, 21, 51–61.
- Niedbala, G.; Kurasiak-Popowska, D.; Stuper-Szablewska, K.; Nawracała, J. Application of Artificial Neural Networks to Analyze the Concentration of Ferulic Acid, Deoxynivalenol, and Nivalenol in Winter Wheat Grain. Agriculture 2020, 10, 127.
- Parsaeian, M.; Shahabi, M.; Hassanpour, H. Estimating Oil and Protein Content of Sesame Seeds Using Image Processing and Artificial Neural Network. J. Am. Oil Chem. Soc. 2020, 97, 691–702.
- Pessoa, H.P., Copati, M.G.F., Azevedo, A.M., Dariva, F.D., de Almeida, G.Q., Gomes, C.N. Combining deep learning and X-ray imaging technology to assess tomato seed quality. Scientia Agricola 2023, 80. [CrossRef]
- Qaim M. Role of New Plant Breeding Technologies for Food Security and Sustainable Agricultural Development. Applied Economic Perspectives and Policy 2020, 42(2), 129-150. [CrossRef]
- Ray, A.; Halder, T.; Jena, S.; Sahoo, A.; Ghosh, B.; Mohanty, S.; Mahapatra, N.; Nayak, S. Application of artificial neural network (ANN) model for prediction and optimization of coronarin D content in Hedychium coronarium. Ind. Crops Prod. 2020, 146, 112186.
- Reed, R.C., Bradford, K.J., Khanday, I. Seed germination and vigor: ensuring crop sustainability in a changing climate. Heredity 2022, 128, 450–459. [CrossRef]
- Ronald, P. Plant Genetics, Sustainable Agriculture and Global Food Security, Genetics 2011, 188(1), 11–20. [CrossRef]
- Sadeghi-Tehran, P.; Virlet, N.; Ampe, E.M.; Reyns, P.; Hawkesford, M.J. DeepCount: In-Field Automatic Quantification of Wheat Spikes Using Simple Linear Iterative Clustering and Deep Convolutional Neural Networks. Front. Plant Sci. 2019, 10.
- Seki, K., Toda, Y. QTL mapping for seed morphology using the instance segmentation neural network in Lactuca spp. QTL mapping for seed morphology using the instance segmentation neural network in Lactuca spp. Front. Plant Sci. 2022, 13:949470. [CrossRef]
- Shahsavari, M., Mohammadi, V., Alizadeh, B., Alizadeh, H. Application of machine learning algorithms and feature selection in rapeseed (Brassica napus L.) breeding for seed yield. Plant Methods 2023, 19, 57. [CrossRef]
- Singh, N., Rai, V., Singh, N.K. Multi-omics strategies and prospects to enhance seed quality and nutritional traits in pigeonpea. Nucleus 2020, 63, 249–256. [CrossRef]
- Tang, H., Kong, W., Nabukalu, P., Lomas, J.S., Moser, M., Zhang, J., Jiang, M., Zhang, X., Paterson, A.H., Yim, W.C. GRABSEEDS: extraction of plant organ traits through image analysis. Plant Methods 2024, 20, 140. [CrossRef]
- Toda, Y., Okura, F., Ito, J., Okada, S., Kinoshita, T., Tsuji, H., Saisho, D. Training instance segmentation neural network with synthetic datasets for crop seed phenotyping. Communications Biology 2020, 3, 173. [CrossRef]
- Tu, K., Wu, W., Cheng, Y., Zhang, H., Xu, Y., Dong, X., Wang, M., Sun, Q. AIseed: An automated image analysis software for high-throughput phenotyping and quality non-destructive testing of individual plant seeds. Computers and Electronics in Agriculture 2023, 207, 107740. [CrossRef]
- Varshney, R.K., Bohra, A., Yu, J., Garner, A., Zhang, Q., and Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends in Plant Science 2021, 26(6), 631-649. [CrossRef]
- Wimalasekera, R. (2015). Role of Seed Quality in Improving Crop Yields. In: Hakeem, K. (eds) Crop Production and Global Environmental Issues. Springer, Cham. [CrossRef]
- Yoosefzadeh-Najafabadi, M., Torabi, S., Tulpan, D., Rajcan, I., Eskandari, M. Application of SVR-Mediated GWAS for Identification of Durable Genetic Regions Associated with Soybean Seed Quality Traits. Plants. 2023; 12(14):2659. [CrossRef]
- Zhao, T., Wu, H., Wang, X., Zhao, Y., Wang, L., Pan, J., Mei, H., Han, J., Wang, S., Lu, K., Li, M., Gao, M., Cao, Z., Zhang, H., Wan, K., Li, J., Fang, L., Zhang, T., Guan, X. Integration of eQTL and machine learning to dissect causal genes with pleiotropic effects in genetic regulation networks of seed cotton yield. Cell Reports 2023, 42(9), 113111. [CrossRef]
- Zhu, C., Gore, M., Buckler, E.S., and Yu, J. Status and prospects of association mapping in Plants. The Plant Genome 2008, 1(1), 5-20. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).