1. Introduction
Breast cancer remains the leading cause of cancer-related mortality in women worldwide, with its high incidence and mortality rates posing a critical public health burden. In 2022, there were approximately 2.3 million new cases and 666,000 deaths globally (Bray et al., 2024), reflecting its profound societal impact. The disease spans all post-pubertal age groups, with incidence rising progressively with age (Giaquinto et al., 2024). Clinically and molecularly heterogeneous, breast cancer is classified into distinct molecular subtypes—Luminal A, Luminal B, HER2+, and triple-negative breast cancer (TNBC)—each defined by unique biological features and prognostic profiles that dictate therapeutic efficacy (Xiong et al., 2025).
Current management strategies, including surgery, chemotherapy, radiotherapy, endocrine therapy, targeted therapies, and immunotherapy, remain foundational to breast cancer care. However, their effectiveness varies substantially across subtypes. TNBC, characterized by aggressive behavior, limited therapeutic options, and poor outcomes, exemplifies the urgent need for precision-driven innovations (Hu et al., 2024).
Precision medicine has underscored the importance of molecular biomarkers in guiding therapeutic decisions. Within this framework, metabolic reprogramming—particularly dysregulated fatty acid metabolism—has emerged as a hallmark of breast cancer pathogenesis (Liu et al., 2024). The lipid-rich mammary microenvironment facilitates dynamic crosstalk between tumor cells and adipocytes, driving carcinogenesis, proliferation, and therapy resistance (Yang et al., 2024). Despite its therapeutic promise, mechanistic insights into how altered fatty acid metabolism influences tumor progression and treatment responses remain incomplete, and targeted strategies for this pathway are underdeveloped. Consequently, identifying robust fatty acid metabolism-associated biomarkers is critical for advancing subtype-agnostic prognostic frameworks and therapeutic interventions.
Here, we developed a fatty acid metabolic signature for prognosis (FAMOUS) using transcriptomic data. FAMOUS-based risk stratification demonstrated robust prognostic utility across all molecular subtypes and therapeutic modalities, offering a universal tool to refine clinical decision-making.
2. Materials and Methods
2.1. Patients
Breast cancer patients were selected from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. Inclusion criteria: availability of transcriptomic data for FAMOUS construction; documented molecular subtype classification; pathologically confirmed breast cancer diagnosis; no prior exposure to radiotherapy, chemotherapy, targeted therapy, immunotherapy, or other anticancer treatments. Sequencing data and clinical annotations were retrieved from TCGA and GEO portals.
2.2. Gene Selection
Genes were curated from the KEGG_FATTY_ACID_METABOLISM, HALLMARK_FATTY_ACID_METABOLISM, and REACTOME_FATTY_ACID_ METABOLISM pathways. After deduplication, 156 genes were retained for analysis (
Table S1).
2.3. FAMOUS Construction
The TCGA breast cancer cohort was stratified into training (70%) and validation (30%) sets via stratified randomization. In the training set, prognostic genes linked to overall survival (OS) were identified using univariate Cox regression. Optimal expression thresholds for high/low gene expression subgroups were determined via the Survminer R package to maximize OS discrimination. Gene expression levels were binarized (0/1) relative to these thresholds (Ying et al., 2021). A Cox regression model, regularized by the least absolute shrinkage and selection operator (LASSO), refined gene selection through penalized likelihood optimization. The FAMOUS score was calculated as: FAMOUS score = ∑ (Gene expression level × Cox regression coefficient). Prognostic performance was validated in the TCGA validation set and external cohort GSE72245 (Jeschke et al., 2017).
2.4. Gene Set Enrichment Analysis (GSEA)
Differentially expressed genes (DEGs) between low- and high-FAMOUS-score subgroups were identified (adjusted p<0.05, |log2FC|>1). Pathway enrichment was analyzed via GSEA using REACTOME, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, implemented in NetworkAnalyst 3.0 (
https://www.networkanalyst.ca/).
2.5. Immune Infiltration Estimation
Immune cell abundance in the tumor microenvironment (TME) was estimated using the CIBERSORT algorithm with the LM22 gene signature (
https://cibersortx.stanford.edu/), based on transcriptomic data.
2.6. Statistical Analysis
Group differences were assessed using χ², Fisher’s exact, t-tests, or Mann-Whitney U tests, as appropriate. Survival outcomes were compared via Kaplan-Meier curves and log-rank tests. Multivariate Cox regression evaluated the independent prognostic value of FAMOUS, with hazard ratios (HRs) and 95% confidence intervals (CIs) reported. A two-sided p<0.05 defined statistical significance. Analyses were performed in R (v4.3.2), Bioconductor packages, and SPSS (v19.0).
3. Results
3.1. Patient Characteristics
The TCGA breast cancer cohort included 1,036 eligible patients, stratified into training (n=725) and testing (n=311) sets at a 7:3 ratio. Clinicopathological characteristics were balanced between sets (
Tables S2 and S3). The cohort comprised 98.8% females (n=1,024) and 1.2% males (n=12), with 30.5% elderly patients (≥65 years) and 24.5% stage III/IV cases. Molecular subtypes included Luminal A (46.5%, n=482), Luminal B (18.2%, n=189), HER2+ (6.9%, n=72), and TNBC (15.4%, n=160). Postoperative adjuvant chemotherapy, radiotherapy, and trastuzumab therapy were documented for 49.1% (n=509), 47.5% (n=492), and 6.9% (n=71) of patients, respectively. The external GSE72245 validation cohort (n=118) comprised Luminal A (21.2%, n=25), Luminal B (27.1%, n=32), HER2+ (25.4%, n=30), and TNBC (26.3%, n=31) cases (
Table S4).
3.2. FAMOUS Construction and Validation
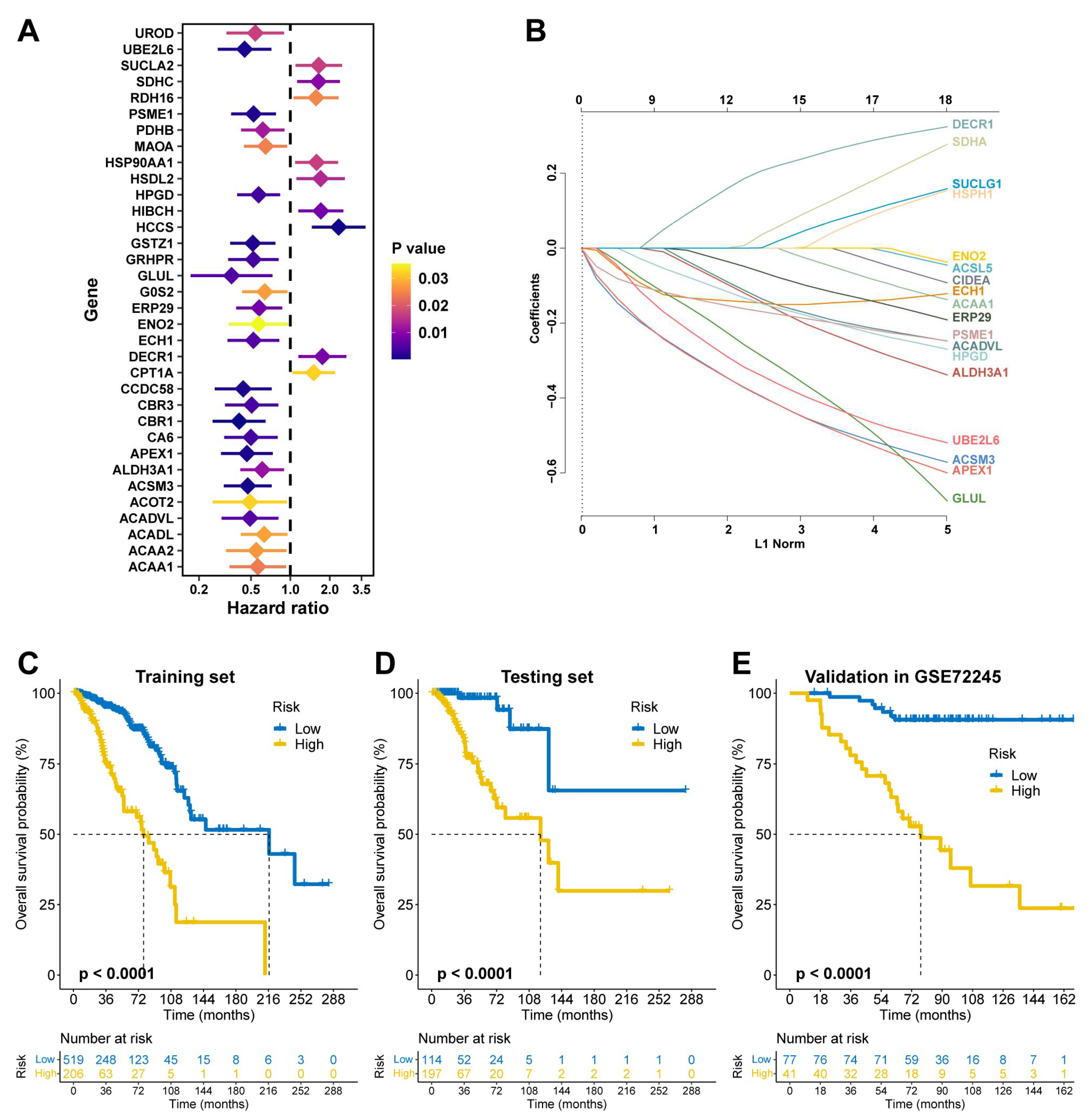
In the TCGA training set, univariate Cox regression identified 34 prognostic fatty acid metabolism-related genes (
Figure 1A), of which 15 were selected via LASSO regression to construct FAMOUS (
Figure 1B and
Table S5). Using the optimal OS-related cutoff, FAMOUS stratified patients into high- and low-risk groups. High-risk patients exhibited significantly worse OS in the TCGA training set (
Figure 1C) and testing set (
Figure 1D). These findings were validated in the GSE72245 cohort (
Figure 1E).
3.3. FAMOUS Score Is an Independent Prognostic Predictor
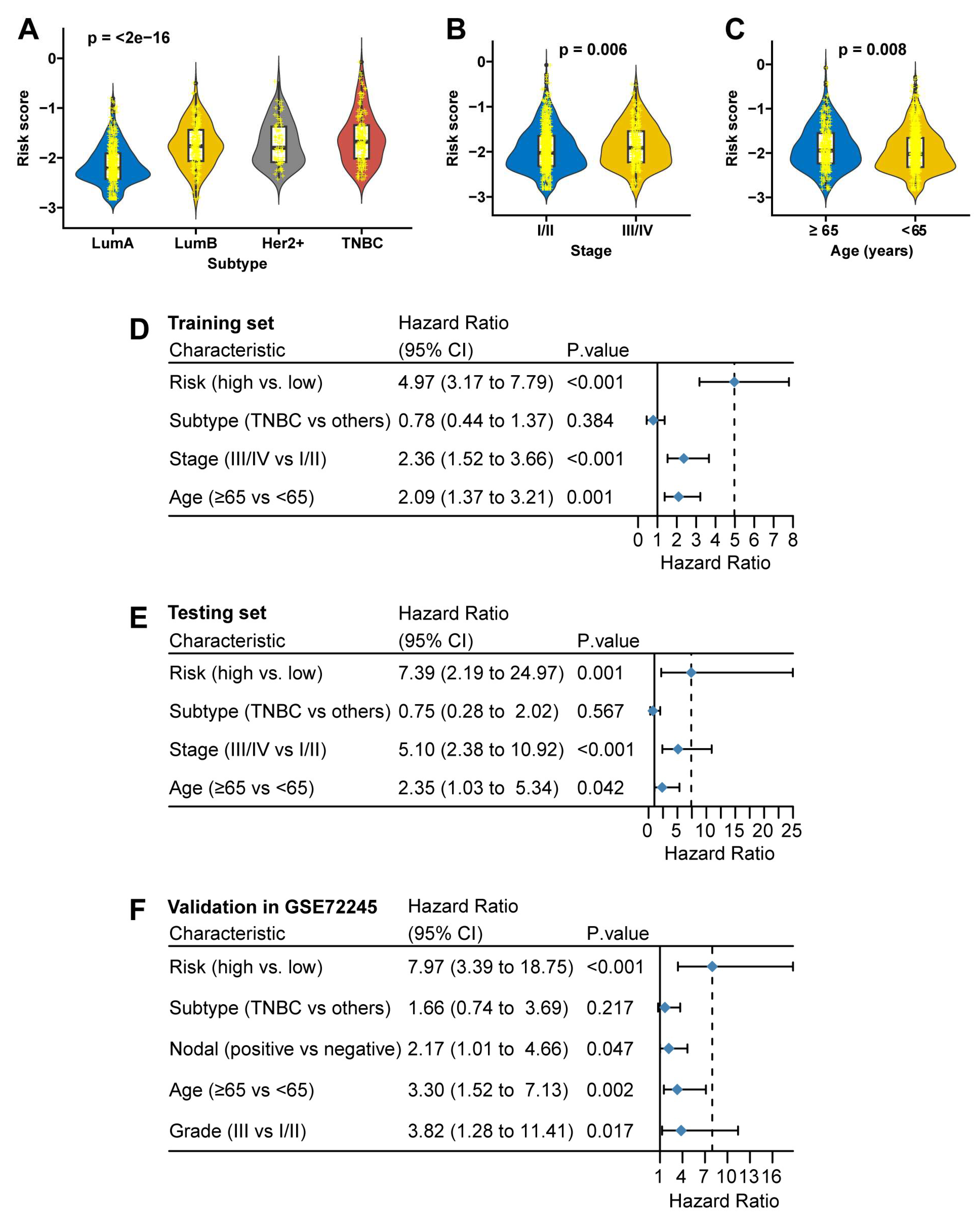
FAMOUS scores increased incrementally across subtypes (Luminal A < Luminal B < HER2+ < TNBC;
Figure 2A) and correlated with advanced stage (stage III/IV vs. I/II: p=0.006;
Figure 2B) and older age (≥65 vs. <65 years: p=0.008;
Figure 2C) in TCGA. In the GSE72245 cohort, FAMOUS score was also markedly higher in HER2-positive and TNBC compared to the other two subtypes (
Table S6). In multivariate analysis, FAMOUS remained an independent OS predictor in the TCGA training (HR=4.97, 95% CI: 3.17 to 7.79;
Figure 2D) and testing sets (HR=7.39, 95% CI: 2.19 to 24.97;
Figure 2E), validated in GSE72245 (HR=7.97, 95% CI: 3.39 to 18.75;
Figure 2F).
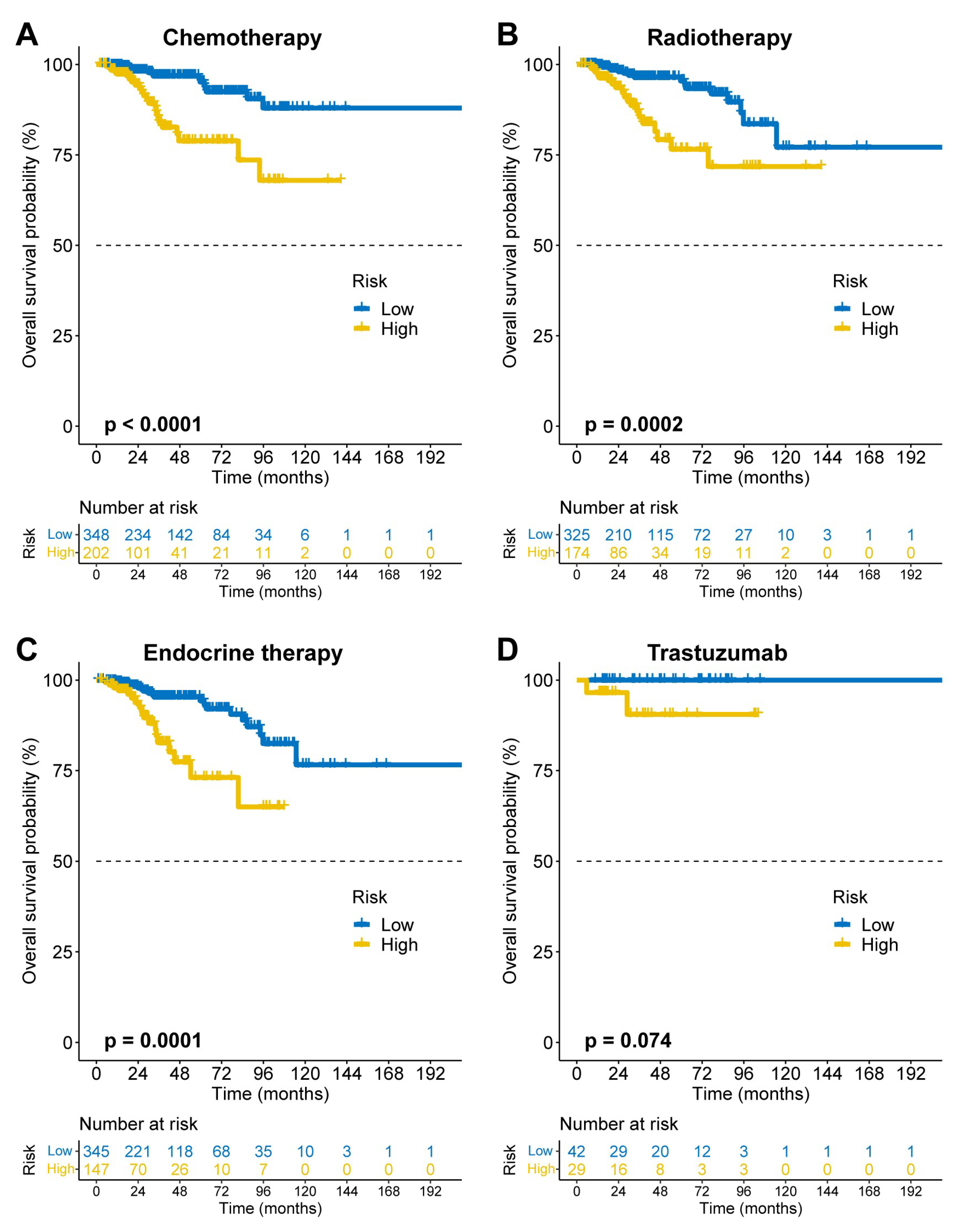
3.4. Pan-Subtype and Pan-Therapeutic Applicability of FAMOUS
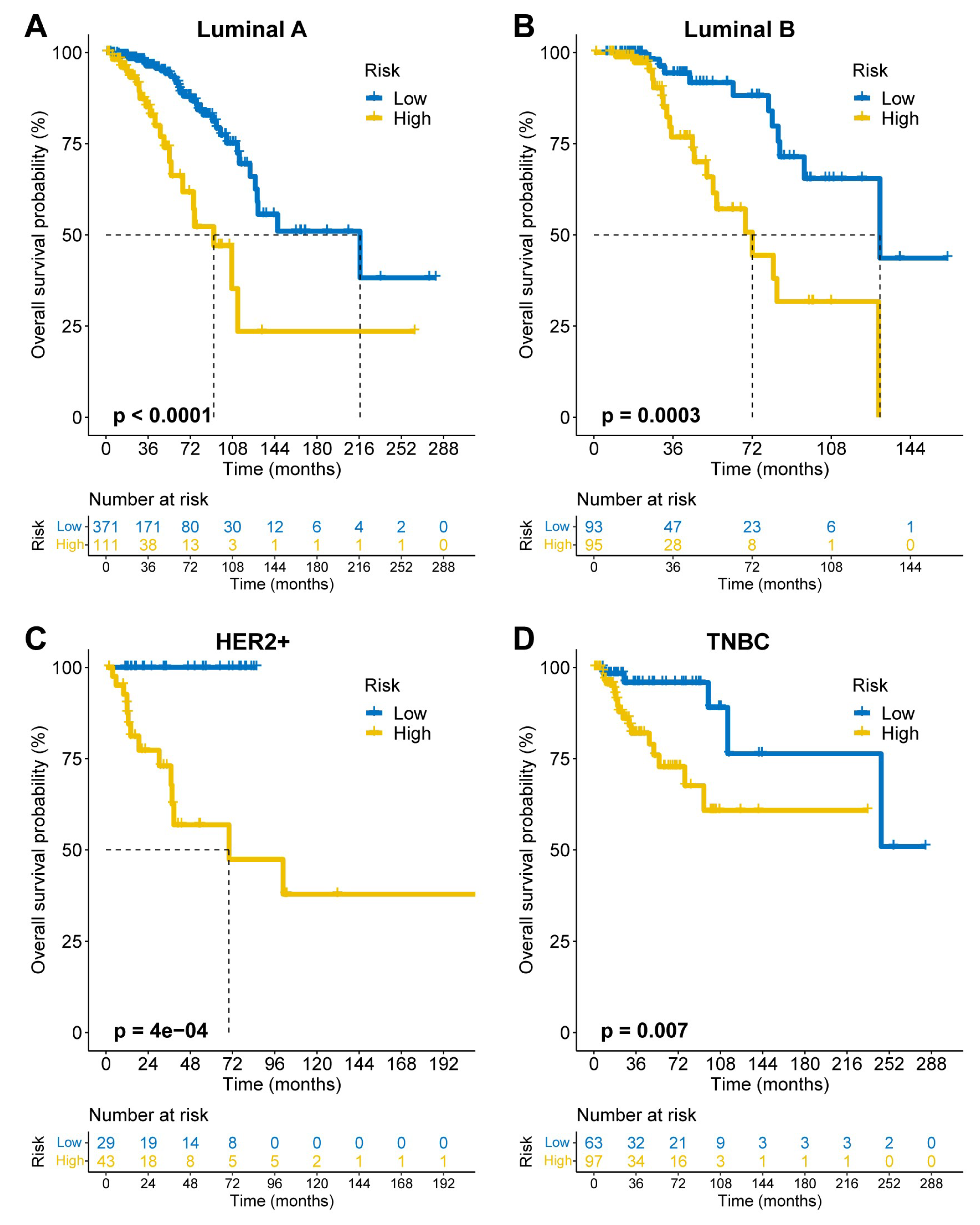
FAMOUS significantly stratified OS risk across all molecular subtypes (Luminal A/B, HER2+, TNBC; p<0.05;
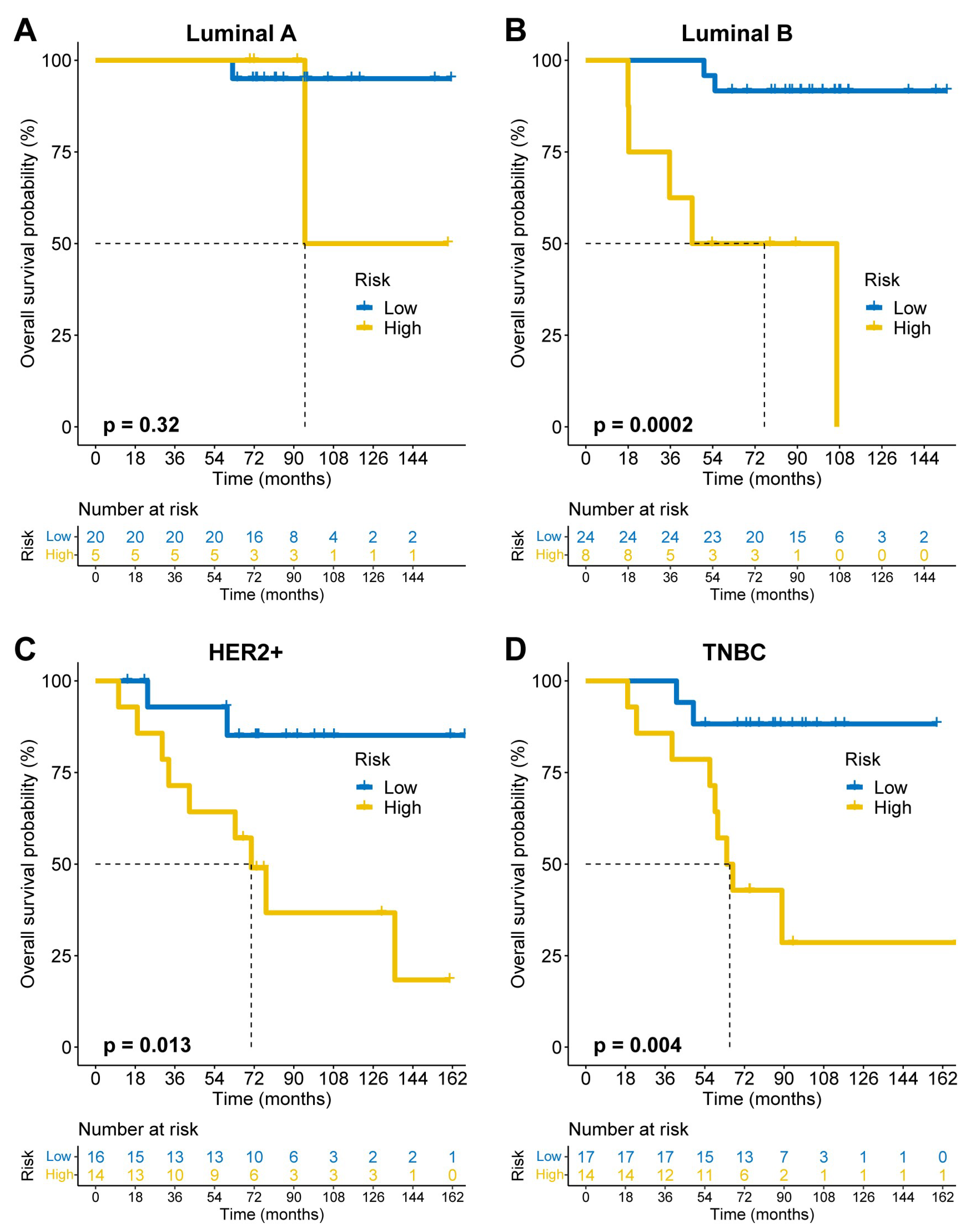
Figure 3A–D) in TCGA. These results were validated in the GSE72245 cohort (
Figure 4A–D), though statistical significance was not reached in Luminal A due to limited sample size (
Figure 4A). Besides, FAMOUS-based risk stratification significantly predicted prognosis across therapies (chemotherapy, radiotherapy, endocrine therapy; p<0.05;
Figure 5A–C) in TCGA. In trastuzumab-treated patients, a clinically relevant trend emerged despite limited statistical power (
Figure 5D).
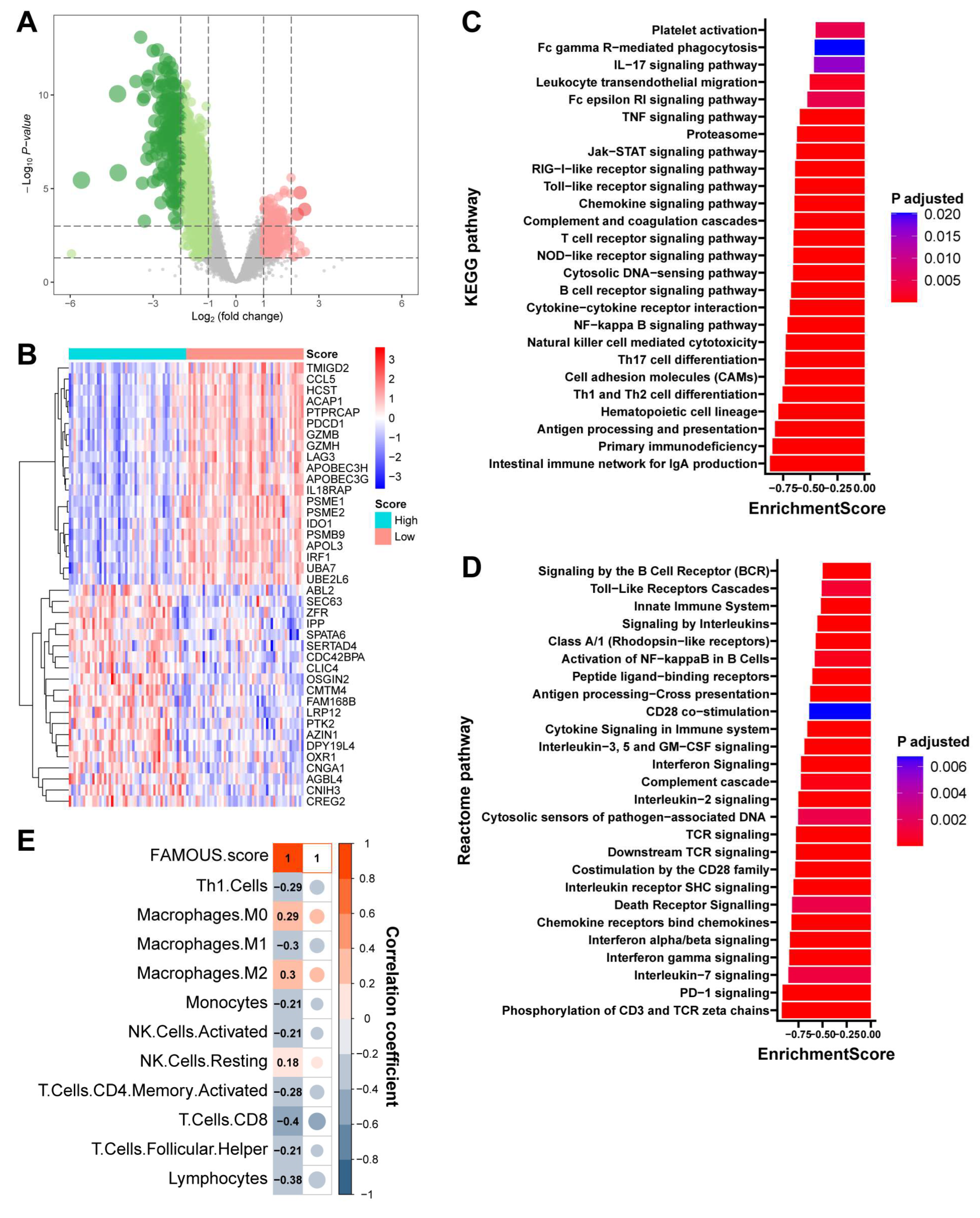
3.5. Immune Relevance of FAMOUS
In TNBC from TCGA, 1079 DEGs distinguished high- and low-FAMOUS groups (
Figure 6A and
Table S7). Among the top 40 most significantly DEGs, immune-related genes such as PDCD1 (PD-1), LAG3, and GZMB were observed (
Figure 6B). GSEA based on KEGG (
Figure 6C) and Reactome (
Figure 6D) revealed enrichment of immune activation pathways (e.g., antigen processing and presentation, T cell receptor signaling pathway, and interferon gamma signaling) in low-FAMOUS groups, while high-FAMOUS groups exhibited immune suppression (
Table S8). Similar results were obtained from the biological process analysis of GO (
Table S8). Consistent with these findings, FAMOUS score inversely correlated with immune-activated cells such as CD8+ T cells and positively correlated with immune-suppressive cells like M2 macrophages (
Figure 6E). Similar immune trends were observed across all subtypes (
Tables S9–S11), indicating FAMOUS reflects tumor immune microenvironment (“hotness”) polarization.
4. Discussion
Lipids serve as essential macromolecules, functioning as energy reservoirs, signaling mediators, and structural components of cellular membranes. Dysregulated lipid homeostasis promotes tumor progression by modulating therapeutic responses and metastatic potential (Wang et al., 2025). Cancer cells exploit lipid metabolic pathways to rewire tumor-stromal interactions and evade immune surveillance, fostering a treatment-resistant TME (Wan et al., 2025). Here, we developed the FAMOUS score to quantify fatty acid metabolic activity in breast cancer. FAMOUS independently stratifies prognosis across molecular subtypes and therapies while revealing immune-suppressive TME features, positioning it as a pan-subtype prognostic tool with implications for immunotherapy.
The mammary stroma is uniquely enriched with lipid-laden adipocytes that secrete bioavailable fatty acids and adipocytokines. These peritumoral adipocytes drive metastatic competence via lipidomic remodeling, epithelial-mesenchymal transition activation, and pro-survival signaling (Chang et al., 2024). While breast cancer subtypes exhibit distinct metabolic profiles, all rely on fatty acid metabolism for progression (He et al., 2023). This convergence on lipid dysregulation—a shared mechanism of therapeutic resistance—underpins FAMOUS’s ability to transcend molecular heterogeneity and predict outcomes across treatment paradigms.
Notably, FAMOUS captures the interplay between fatty acid metabolism and immune evasion. High FAMOUS scores correlate with depleted CD8+ T cells, enriched M2 macrophages, and suppressed immune activation pathways (e.g., antigen presentation, interferon signaling). These findings align with the metabolic competition model, wherein tumor cells outcompete immune cells for nutrients, reshaping the TME into an immunosuppressive niche (Zhang et al., 2024). Thus, FAMOUS may identify patients likely to benefit from immunotherapy, particularly in aggressive subtypes like TNBC.
Immunotherapy has emerged as a cornerstone of breast cancer management, particularly following the approval of pembrolizumab combined with chemotherapy for TNBC. This regimen is now approved for both neoadjuvant and advanced settings, broadening therapeutic avenues for this aggressive subtype (Cortes et al., 2022; Schmid et al., 2022, 2024). Recent trials, including KEYNOTE-756 (Cardoso et al., 2025) and CheckMate 7FL (Loi et al., 2025), demonstrated that adding nivolumab or pembrolizumab to neoadjuvant chemotherapy significantly enhances pathological complete response rates in high-risk, early-stage ER+/HER2− patients, particularly those with elevated PD-L1 expression. Nevertheless, immunotherapy efficacy remains suboptimal, with pronounced response heterogeneity across molecular subtypes (Onkar et al., 2023; Heater et al., 2024). The FAMOUS score, which reflects fatty acid metabolism-driven immune modulation, exhibits pan-subtype relevance and may refine patient selection for immunotherapy by identifying subtype-specific populations poised to benefit.
This study has several limitations. First, FAMOUS was constructed using public datasets; validation in proprietary cohorts is needed. Second, population heterogeneity may affect generalizability despite a large sample size in the construction of FAMOUS. Moreover, the molecular mechanisms by which the 15 genes comprising FAMOUS affect breast cancer are not fully understood, with some genes not yet reported in the context of breast cancer. Finally, clinical correlations between FAMOUS and immunotherapy outcomes remain untested. The strengths of this study lie in the development of a universal breast cancer prognostic evaluation system. FAMOUS surpasses existing models in prognostic power (Qian et al., 2023; Tang et al., 2023; Yang et al., 2023), likely due to its focused gene selection and categorical expression thresholds optimized for clinical relevance.
5. Conclusions
In summary, our findings underscore the pan-subtype influence of fatty acid metabolism on breast cancer progression and therapeutic responses, as quantified by the FAMOUS score. Future investigations should prioritize elucidating the mechanistic underpinnings of FAMOUS-associated pathways, identifying druggable targets within this metabolic axis, developing multi-omics-integrated prognostic models, and advancing translational studies to bridge preclinical insights with clinical applications.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org, Table S1. Genes included for analysis. Table S2. TCGA cohort. Table S3. Clinicopathological features between the testing and training sets. Table S4. GSE72245 cohort. Table S5. Regression coefficients of the genes in FAMOUS. Table S6. Clinicopathological features and FAMOUS score in GSE72245. Table S7. DEGs between the high and low FAMOUS score groups in TNBC. Table S8. GSEA results in TNBC. Table S9. GSEA results in Luminal A subtype. Table S10. GSEA results in Luminal B subtype. Table S11. GSEA results in HER2+ subtype.
Author Contributions
Conceptualization, Y.S., J.L., D.W., Z.S., L.O. and L.Y.; methodology, Y.S., J.L., D.W., Z.S., L.O. and L.Y.; software, D.W., Z.S., L.O. and L.Y.; validation, D.W., Z.S., L.O. and L.Y.; formal analysis, Y.S., J.L., D.W., Z.S., L.O. and L.Y.; investigation, Y.S., J.L., M.Z., Y.X., D.W., Z.S., L.O. and L.Y.; resources, Y.S., J.L., M.Z., Y.X., D.W., Z.S., L.O. and L.Y; data curation, D.W., Z.S., L.O. and L.Y.; writing—original draft preparation, D.W., Z.S., L.O. and L.Y.; writing—review and editing, L.Y.; supervision, L.Y.; project administration, L.Y.. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by The Natural Science Foundation of Jiangsu Province, grant number BK20231252 and the Key Medical Research Projects of Jiangsu Provincial Health Commission, grant number K2023026.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Affiliated Hospital of Jiangsu University (protocol code KY2021K0908 and September 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the TCGA and GSE72245 cohorts according to related literature.
Data Availability Statement
All data relevant to the study that are not in the article and
supplementary material are available from the corresponding author on reasonable request.
Acknowledgments
We thank all the patients, their families, and the institutions for supporting this Study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| FAMOUS |
The Fatty Acid Metabolic Signature for Prognosis |
| TNBC |
Triple-negative breast cancer |
| TCGA |
The Cancer Genome Atlas |
| GEO |
Gene Expression Omnibus |
| OS |
Overall survival |
| LASSO |
The least absolute shrinkage and selection operator |
| DEGs |
Differentially expressed genes |
| GO |
Gene Ontology |
| KEGG |
Kyoto Encyclopedia of Genes and Genomes |
| TME |
Tumor microenvironment |
| HRs |
Hazard ratios |
| CIs |
Confidence intervals |
References
- Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R.L., Soerjomataram, I., Jemal, A., 2024. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 74 (3), 229-263. [CrossRef]
- Cardoso, F., O’Shaughnessy, J., Liu, Z., McArthur, H., Schmid, P., Cortes, J., Harbeck, N., Telli, M.L., Cescon, D.W., Fasching, P.A., Shao, Z., Loirat, D., Park, Y.H., Fernandez, M.G., Rubovszky, G., Spring, L., Im, S.A., Hui, R., Takano, T., André, F., Yasojima, H., Ding, Y., Jia, L., Karantza, V., Tryfonidis, K., Bardia, A., 2025. Pembrolizumab and chemotherapy in high-risk, early-stage, ER+/HER2- breast cancer: a randomized phase 3 trial. Nat. Med. 31 (2), 442-448. [CrossRef]
- Chang, Y., Du, R., Xia, F., Xu, X., Wang, H., Chen, X., 2024. Dysregulation of Fatty Acid Metabolism in Breast Cancer and Its Targeted Therapy. Breast Cancer (Dove Med Press) 16, 825-844. [CrossRef]
- Cortes, J., Rugo, H.S., Cescon, D.W., Im, S.A., Yusof, M.M., Gallardo, C., Lipatov, O., Barrios, C.H., Perez-Garcia, J., Iwata, H., Masuda, N., Torregroza Otero, M., Gokmen, E., Loi, S., Guo, Z., Zhou, X., Karantza, V., Pan, W., Schmid, P., KEYNOTE-355 Investigators, 2022. Pembrolizumab plus Chemotherapy in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 387 (3), 217-226. [CrossRef]
- Giaquinto, A.N., Sung, H., Newman, L.A., Freedman, R.A., Smith, R.A., Star, J., Jemal, A., Siegel, R.L., 2024. Breast cancer statistics 2024. CA Cancer J Clin 74 (6), 477-495. [CrossRef]
- He, M., Xu, S., Yan, F., Ruan, J., Zhang, X., 2023. Fatty Acid Metabolism: A New Perspective in Breast Cancer Precision Therapy. Front Biosci (Landmark Ed) 28 (12), 348. [CrossRef]
- Heater, N.K., Warrior, S., Lu, J., 2024. Current and future immunotherapy for breast cancer. J Hematol Oncol 17 (1), 131. [CrossRef]
- Hu, Y., Wang, C., Liang, H., Li, J., Yang, Q., 2024. The treatment landscape of triple-negative breast cancer. Med. Oncol. 41 (10), 236. [CrossRef]
- Jeschke, J., Bizet, M., Desmedt, C., Calonne, E., Dedeurwaerder, S., Garaud, S., Koch, A., Larsimont, D., Salgado, R., Van den Eynden, G., Willard Gallo, K., Bontempi, G., Defrance, M., Sotiriou, C., Fuks, F., 2017. DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J. Clin. Invest. 127 (8), 3090-3102. [CrossRef]
- Liu, S., Zhang, X., Wang, W., Li, X., Sun, X., Zhao, Y., Wang, Q., Li, Y., Hu, F., Ren, H., 2024. Metabolic reprogramming and therapeutic resistance in primary and metastatic breast cancer. Mol. Cancer 23 (1), 261. [CrossRef]
- Loi, S., Salgado, R., Curigliano, G., Romero Díaz, R.I., Delaloge, S., Rojas García, C.I., Kok, M., Saura, C., Harbeck, N., Mittendorf, E.A., Yardley, D.A., Suárez Zaizar, A., Caminos, F.R., Ungureanu, A., Reinoso-Toledo, J.G., Guarneri, V., Egle, D., Ades, F., Pacius, M., Chhibber, A., Chandra, R., Nathani, R., Spires, T., Wu, J.Q., Pusztai, L., McArthur, H., 2025. Neoadjuvant nivolumab and chemotherapy in early estrogen receptor-positive breast cancer: a randomized phase 3 trial. Nat. Med. 31 (2), 433-441. [CrossRef]
- Onkar, S.S., Carleton, N.M., Lucas, P.C., Bruno, T.C., Lee, A.V., Vignali, D., Oesterreich, S., 2023. The Great Immune Escape: Understanding the Divergent Immune Response in Breast Cancer Subtypes. Cancer Discov 13 (1), 23-40. [CrossRef]
- Qian, L., Liu, Y.F., Lu, S.M., Yang, J.J., Miao, H.J., He, X., Huang, H., Zhang, J.G., 2023. Construction of a fatty acid metabolism-related gene signature for predicting prognosis and immune response in breast cancer. Front Genet 14, 1002157. [CrossRef]
- Schmid, P., Cortes, J., Dent, R., McArthur, H., Pusztai, L., Kümmel, S., Denkert, C., Park, Y.H., Hui, R., Harbeck, N., Takahashi, M., Im, S.A., Untch, M., Fasching, P.A., Mouret-Reynier, M.A., Foukakis, T., Ferreira, M., Cardoso, F., Zhou, X., Karantza, V., Tryfonidis, K., Aktan, G., O’Shaughnessy, J., KEYNOTE-522 Investigators, 2024. Overall Survival with Pembrolizumab in Early-Stage Triple-Negative Breast Cancer. N. Engl. J. Med. 391 (21), 1981-1991. [CrossRef]
- Schmid, P., Cortes, J., Dent, R., Pusztai, L., McArthur, H., Kümmel, S., Bergh, J., Denkert, C., Park, Y.H., Hui, R., Harbeck, N., Takahashi, M., Untch, M., Fasching, P.A., Cardoso, F., Andersen, J., Patt, D., Danso, M., Ferreira, M., Mouret-Reynier, M.A., Im, S.A., Ahn, J.H., Gion, M., Baron-Hay, S., Boileau, J.F., Ding, Y., Tryfonidis, K., Aktan, G., Karantza, V., O’Shaughnessy, J., KEYNOTE-522 Investigators, 2022. Event-free Survival with Pembrolizumab in Early Triple-Negative Breast Cancer. N. Engl. J. Med. 386 (6), 556-567. [CrossRef]
- Tang, L., Lei, X., Hu, H., Li, Z., Zhu, H., Zhan, W., Zhang, T., 2023. Investigation of fatty acid metabolism-related genes in breast cancer: Implications for Immunotherapy and clinical significance. Transl Oncol 34, 101700. [CrossRef]
- Wan, M., Pan, S., Shan, B., Diao, H., Jin, H., Wang, Z., Wang, W., Han, S., Liu, W., He, J., Zheng, Z., Pan, Y., Han, X., Zhang, J., 2025. Lipid metabolic reprograming: the unsung hero in breast cancer progression and tumor microenvironment. Mol. Cancer 24 (1), 61. [CrossRef]
- Wang, J., Wang, M., Zeng, X., Li, Y., Lei, L., Chen, C., Lin, X., Fang, P., Guo, Y., Jiang, X., Wang, Y., Chen, L., Long, J., 2025. Targeting membrane contact sites to mediate lipid dynamics: innovative cancer therapies. Cell Commun. Signal 23 (1), 89. [CrossRef]
- Xiong, X., Zheng, L.W., Ding, Y., Chen, Y.F., Cai, Y.W., Wang, L.P., Huang, L., Liu, C.C., Shao, Z.M., Yu, K.D., 2025. Breast cancer: pathogenesis and treatments. Signal Transduct Target Ther 10 (1), 49. [CrossRef]
- Yang, X., Tang, W., He, Y., An, H., Wang, J., 2023. A novel fatty-acid metabolism-based classification for triple negative breast cancer. Aging (Albany NY) 15 (4), 1177-1198. [CrossRef]
- Yang, Y., Ma, X., Li, Y., Jin, L., Zhou, X., 2024. The evolving tumor-associated adipose tissue microenvironment in breast cancer: from cancer initiation to metastatic outgrowth. Clin Transl Oncol. [CrossRef]
- Ying, L., Cheng, M., Lu, Y., Tao, Q., Chen, X., Shen, B., Xiong, F., Hu, Z., Wang, D., Li, X., 2021. Glutamine Metabolism Scoring Predicts Prognosis and Therapeutic Resistance in Hepatocellular Carcinoma. Pathol. Oncol. Res. 27, 1610075. [CrossRef]
- Zhang, S., Lv, K., Liu, Z., Zhao, R., Li, F., 2024. Fatty acid metabolism of immune cells: a new target of tumour immunotherapy. Cell Death Discov 10 (1), 39. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).