Submitted:
05 April 2025
Posted:
07 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and Establishment of Stable Transfectants
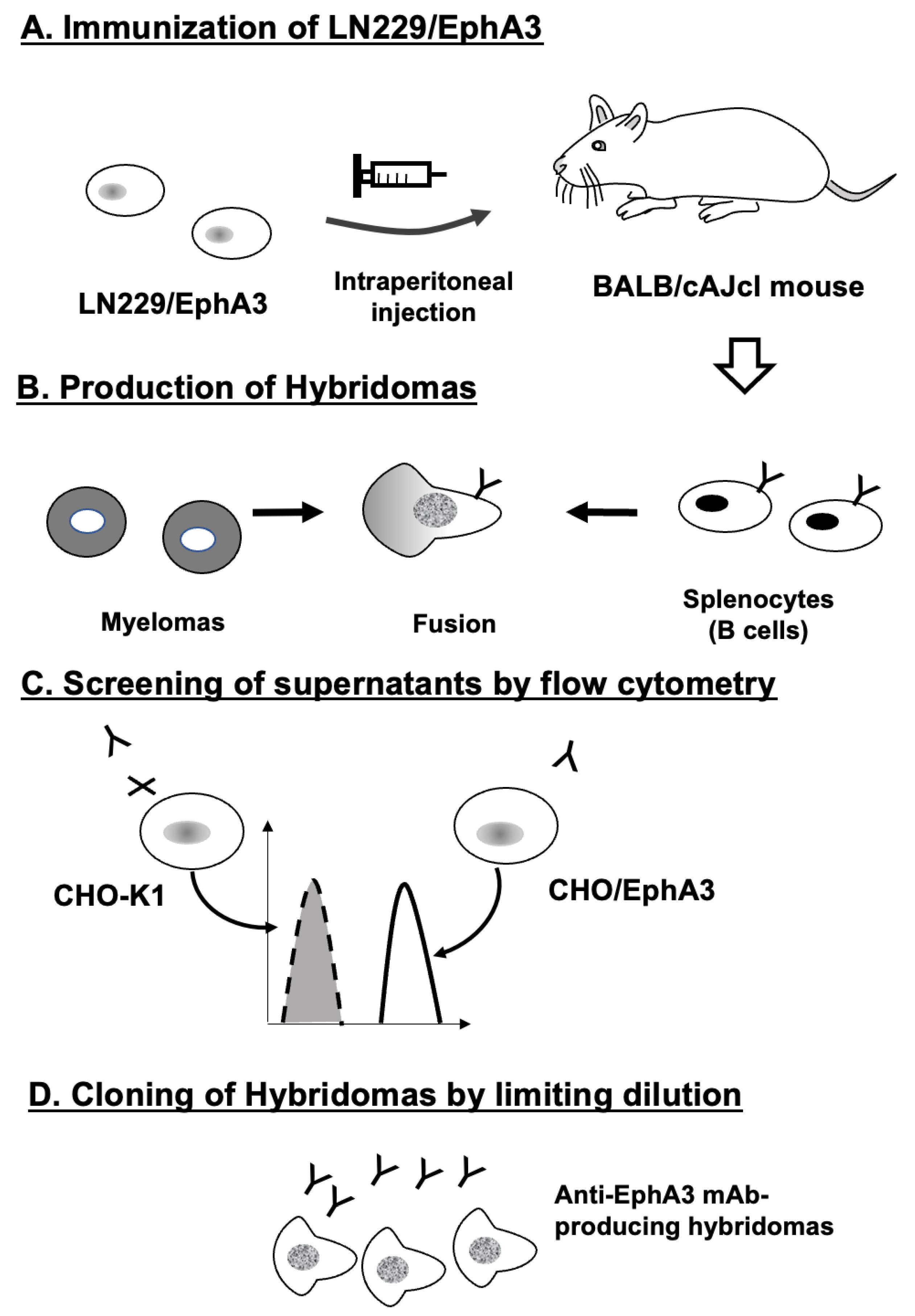
2.2. Development of Hybridomas
2.3. Flow Cytometry
2.4. Determination of Dissociation Constant Values Using Flow Cytometry
2.5. Immunohistochemical Analysis
3. Results
3.1. Development of Anti-Human EphA3 mAbs
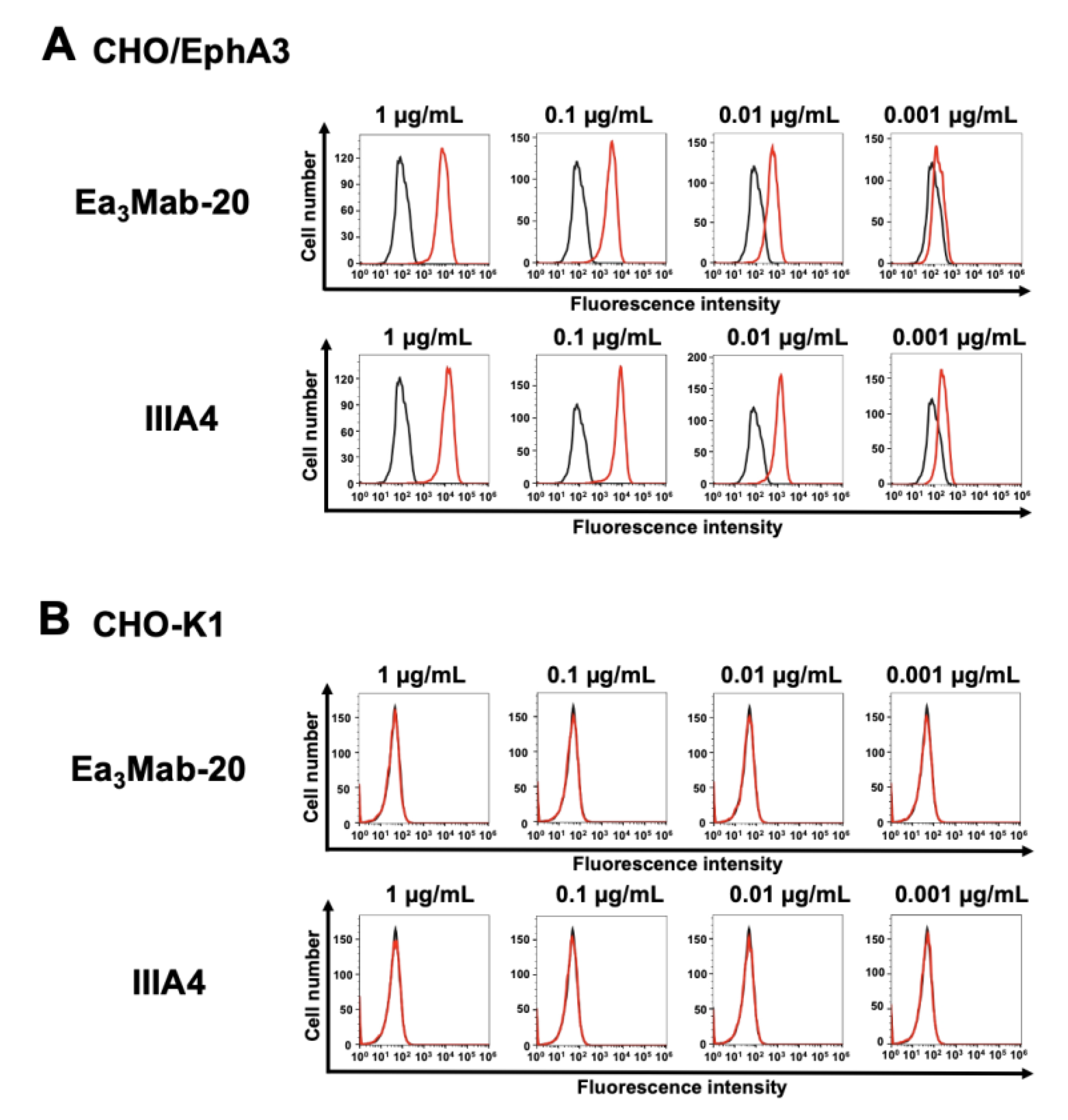
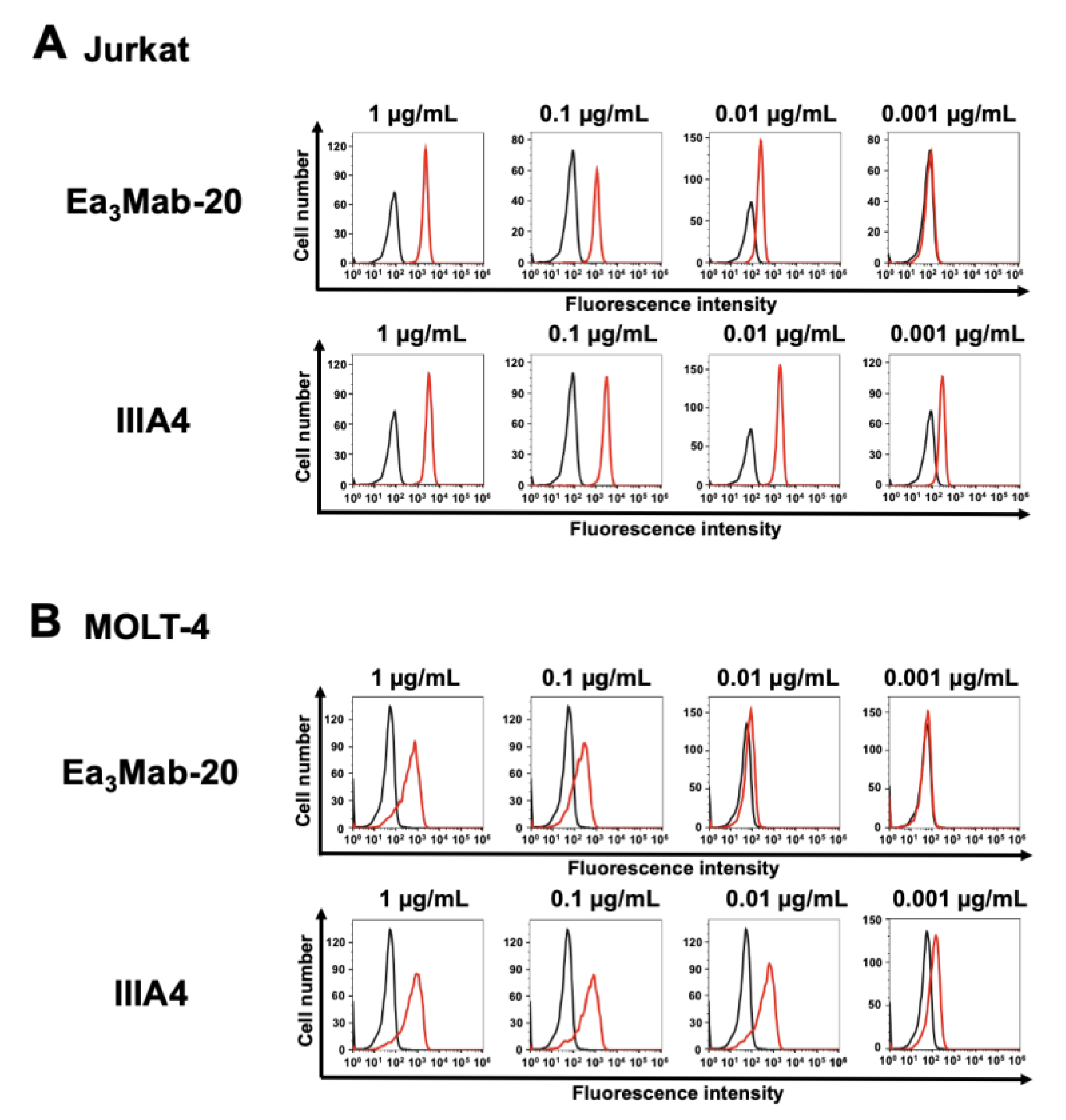
3.2. Flow Cytometry Using Ea3Mab-20 and IIIA4
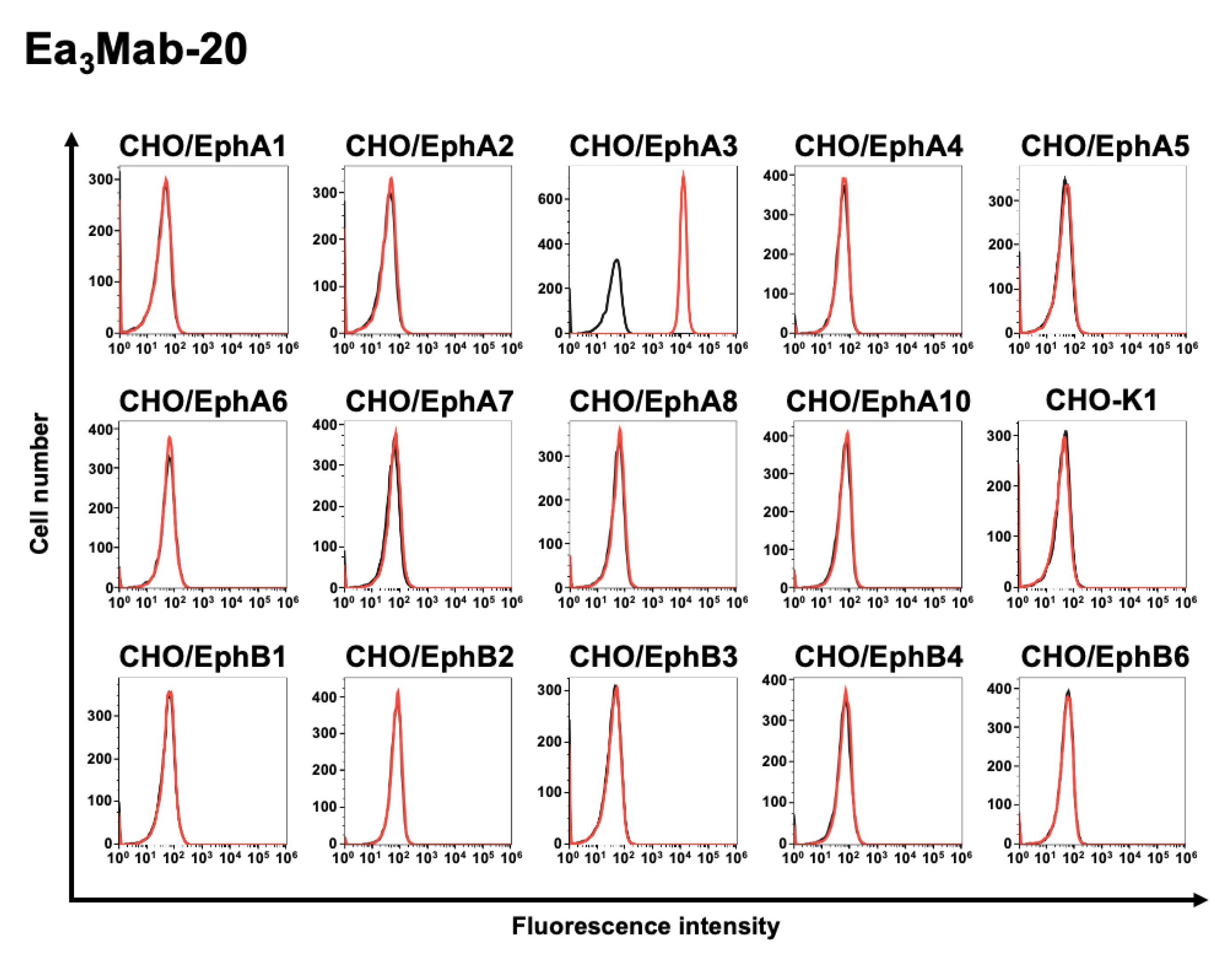
3.3. Specificity of Ea3Mab-20 Using CHO-K1 Cells Overexpressed Various Eph Receptors
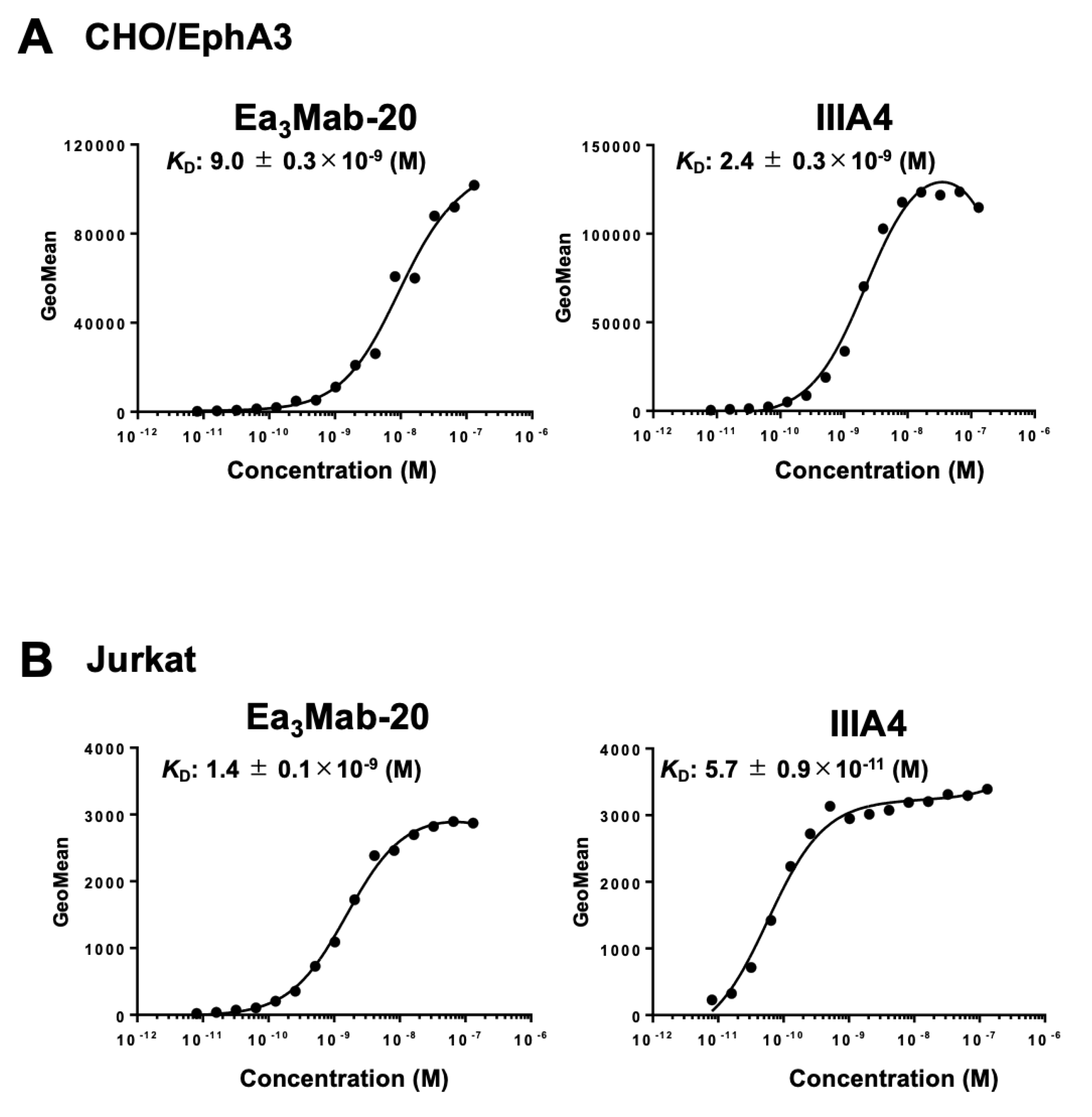
3.4. Determination of Binding Affinity of Ea3Mab-20 and IIIA4 Using Flow Cytometry
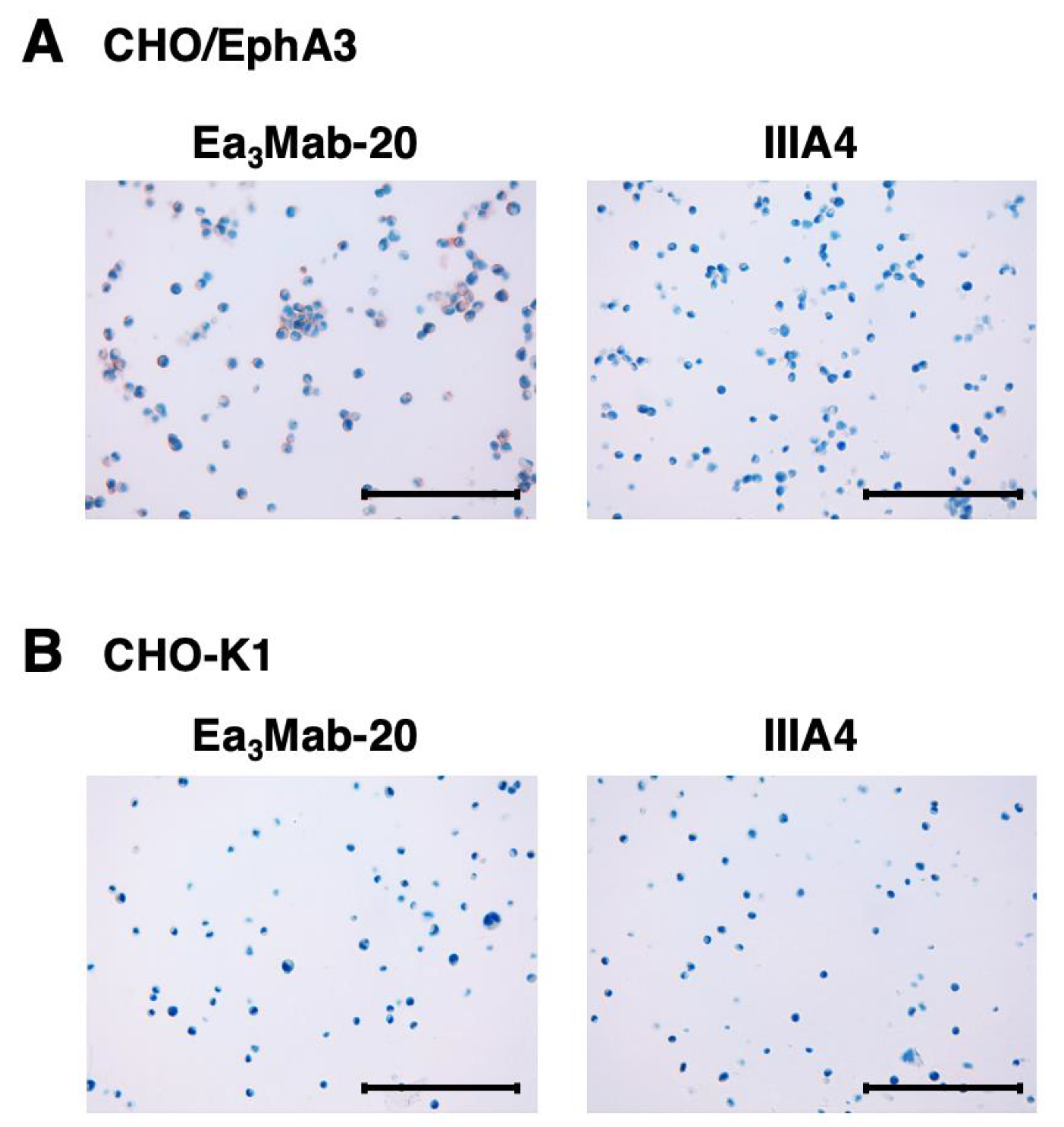
3.5. Immunohistochemistry Using Anti-EphA3 mAbs
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barquilla, A.; Pasquale, E.B. Eph receptors and ephrins: therapeutic opportunities. Annu Rev Pharmacol Toxicol 2015;55: 465-487. [CrossRef]
- Darling, T.K.; Lamb, T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front Immunol 2019;10: 1473.
- Liang, L.Y.; Patel, O.; Janes, P.W.; Murphy, J.M.; Lucet, I.S. Eph receptor signalling: from catalytic to non-catalytic functions. Oncogene 2019;38(39): 6567-6584.
- Pasquale, E.B. Eph receptors and ephrins in cancer progression. Nat Rev Cancer 2024;24(1): 5-27. [CrossRef]
- Buckens, O.J.; El Hassouni, B.; Giovannetti, E.; Peters, G.J. The role of Eph receptors in cancer and how to target them: novel approaches in cancer treatment. Expert Opin Investig Drugs 2020;29(6): 567-582.
- Poliakov, A.; Cotrina, M.; Wilkinson, D.G. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell 2004;7(4): 465-480. [CrossRef]
- Pasquale, E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol 2005;6(6): 462-475.
- Beckmann, M.P.; Cerretti, D.P.; Baum, P.; et al. Molecular characterization of a family of ligands for eph-related tyrosine kinase receptors. EMBO J 1994;13(16): 3757-3762.
- Hirai, H.; Maru, Y.; Hagiwara, K.; Nishida, J.; Takaku, F. A novel putative tyrosine kinase receptor encoded by the eph gene. Science 1987;238(4834): 1717-1720.
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol 2013;5(9).
- Batlle, E.; Wilkinson, D.G. Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harb Perspect Biol 2012;4(1): a008227. [CrossRef]
- Pasquale, E.B. Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008;133(1): 38-52.
- Hanover, G.; Vizeacoumar, F.S.; Banerjee, S.L.; et al. Integration of cancer-related genetic landscape of Eph receptors and ephrins with proteomics identifies a crosstalk between EPHB6 and EGFR. Cell Rep 2023;42(7): 112670.
- Xi, H.Q.; Wu, X.S.; Wei, B.; Chen, L. Eph receptors and ephrins as targets for cancer therapy. J Cell Mol Med 2012;16(12): 2894-2909. [CrossRef]
- London, M.; Gallo, E. Critical role of EphA3 in cancer and current state of EphA3 drug therapeutics. Mol Biol Rep 2020;47(7): 5523-5533.
- Charmsaz, S.; Al-Ejeh, F.; Yeadon, T.M.; et al. EphA3 as a target for antibody immunotherapy in acute lymphoblastic leukemia. Leukemia 2017;31(8): 1779-1787. [CrossRef]
- Day, B.W.; Stringer, B.W.; Al-Ejeh, F.; et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell 2013;23(2): 238-248.
- Nasri, B.; Inokuchi, M.; Ishikawa, T.; et al. High expression of EphA3 (erythropoietin-producing hepatocellular A3) in gastric cancer is associated with metastasis and poor survival. BMC Clin Pathol 2017;17: 8.
- Kim, S.H.; Lee, W.H.; Kim, S.W.; et al. EphA3 maintains radioresistance in head and neck cancers through epithelial mesenchymal transition. Cell Signal 2018;47: 122-130.
- Wu, R.; Wang, H.; Wang, J.; et al. EphA3, induced by PC-1/PrLZ, contributes to the malignant progression of prostate cancer. Oncol Rep 2014;32(6): 2657-2665.
- Peng, J.; Wang, Q.; Liu, H.; et al. EPHA3 regulates the multidrug resistance of small cell lung cancer via the PI3K/BMX/STAT3 signaling pathway. Tumour Biol 2016;37(9): 11959-11971.
- Lisabeth, E.M.; Fernandez, C.; Pasquale, E.B. Cancer somatic mutations disrupt functions of the EphA3 receptor tyrosine kinase through multiple mechanisms. Biochemistry 2012;51(7): 1464-1475.
- Vail, M.E.; Murone, C.; Tan, A.; et al. Targeting EphA3 inhibits cancer growth by disrupting the tumor stromal microenvironment. Cancer Res 2014;74(16): 4470-4481.
- Swords, R.T.; Greenberg, P.L.; Wei, A.H.; et al. KB004, a first in class monoclonal antibody targeting the receptor tyrosine kinase EphA3, in patients with advanced hematologic malignancies: Results from a phase 1 study. Leuk Res 2016;50: 123-131.
- Tomasevic, N.; Luehrsen, K.; Baer, M.; et al. A high affinity recombinant antibody to the human EphA3 receptor with enhanced ADCC activity. Growth Factors 2014;32(6): 223-235. [CrossRef]
- Lv, X.Y.; Wang, J.; Huang, F.; et al. EphA3 contributes to tumor growth and angiogenesis in human gastric cancer cells. Oncol Rep 2018;40(4): 2408-2416.
- Lertsumitkul, L.; Iliopoulos, M.; Wang, S.S.; et al. EphA3-targeted chimeric antigen receptor T cells are effective in glioma and generate curative memory T cell responses. J Immunother Cancer 2024;12(8).
- Martins, P.; D'Souza, R.C.J.; Skarne, N.; et al. EphA3 CAR T cells are effective against glioblastoma in preclinical models. J Immunother Cancer 2024;12(8).
- Ubukata, R.; Suzuki, H.; Hirose, M.; et al. Establishment of a highly sensitive and specific anti-EphB2 monoclonal antibody (Eb2Mab-12) for flow cytometry. MI 2025. [CrossRef]
- Tanaka, T.; Suzuki, H.; Li, G.; et al. Ea 1 Mab-30: A Novel Monoclonal Antibody Against Erythropoietin-Producing Hepatocellular Receptor A1 for Versatile Applications Preprint 2025.
- Tanaka, T.; Kaneko, Y.; Yamamoto, H.; et al. Development of a novel anti-erythropoietin-producing hepatocellular receptor B6 monoclonal antibody Eb(6)Mab-3 for flow cytometry. Biochem Biophys Rep 2025;41: 101960. [CrossRef]
- Satofuka, H.; Suzuki, H.; Tanaka, T.; et al. Development of an anti-human EphA2 monoclonal antibody Ea2Mab-7 for multiple applications. Biochemistry and Biophysics Reports 2025;42: 101998.
- Nanamiya, R.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Development of an Anti-EphB4 Monoclonal Antibody for Multiple Applications Against Breast Cancers. Monoclon Antib Immunodiagn Immunother 2023;42(5): 166-177.
- Fujii, Y.; Kaneko, M.; Neyazaki, M.; et al. PA tag: a versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr Purif 2014;95: 240-247.
- Fujii, Y.; Kaneko, M.K.; Kato, Y. MAP Tag: A Novel Tagging System for Protein Purification and Detection. Monoclon Antib Immunodiagn Immunother 2016;35(6): 293-299.
- Tanaka, T.; Suzuki, H.; Isoda, Y.; et al. Development of a Sensitive Anti-Human CCR9 Monoclonal Antibody (C(9)Mab-11) by N-Terminal Peptide Immunization. Monoclon Antib Immunodiagn Immunother 2022;41(6): 303-310.
- Ghorashian, S.; Kramer, A.M.; Onuoha, S.; et al. Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat Med 2019;25(9): 1408-1414.
- Okada, Y.; Suzuki, H.; Tanaka, T.; Kaneko, M.K.; Kato, Y. Epitope Mapping of an Anti-Mouse CD39 Monoclonal Antibody Using PA Scanning and RIEDL Scanning. Monoclon Antib Immunodiagn Immunother 2024;43(2): 44-52.
- Asano, T.; Kaneko, M.K.; Takei, J.; Tateyama, N.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C(44)Mab-46) Using the REMAP Method. Monoclon Antib Immunodiagn Immunother 2021;40(4): 156-161. [CrossRef]
- Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Epitope Mapping System: RIEDL Insertion for Epitope Mapping Method. Monoclon Antib Immunodiagn Immunother 2021;40(4): 162-167.
- Sano, M.; Kaneko, M.K.; Aasano, T.; Kato, Y. Epitope Mapping of an Antihuman EGFR Monoclonal Antibody (EMab-134) Using the REMAP Method. Monoclon Antib Immunodiagn Immunother 2021;40(4): 191-195.
- Nanamiya, R.; Sano, M.; Asano, T.; et al. Epitope Mapping of an Anti-Human Epidermal Growth Factor Receptor Monoclonal Antibody (EMab-51) Using the RIEDL Insertion for Epitope Mapping Method. Monoclon Antib Immunodiagn Immunother 2021;40(4): 149-155.
- Ishikawa, K.; Suzuki, H.; Ohishi, T.; et al. Antitumor activities of anti-CD44 monoclonal antibodies in mouse xenograft models of esophageal cancer. Oncol Rep 2024;52(5).
- Ishikawa, K.; Suzuki, H.; Ohishi, T.; et al. Anti-CD44 Variant 10 Monoclonal Antibody Exerts Antitumor Activity in Mouse Xenograft Models of Oral Squamous Cell Carcinomas. Int J Mol Sci 2024;25(17). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




