Introduction
Plant leaf epidermis consists of several differentiated cell types, such as stomatal guard cells for gas exchange and trichomes that may serve protective roles for herbicides. Pavement cells (PCs) are most frequent cell types on leaf epidermis in model plant Arabidopsis [
1,
2]. They form characteristic intercalating structure with multiple lobes and indentations. PCs in adaxial side of leaves show distinctively different shape with PCs in abaxial sides, and those 2 types of cells are separated by a few layers of marginal cells with elongated cylindrical shape [
3]. Marginal cells have roles beyond physical distinction of adaxial/abaxial cells, since they are suggested to transport essential hormone auxin [
4,
5]. Moreover, recent report showed that marginal cells have meristematic activity specifically at the early stage of cotyledon/leaf development, which only maintains for a few days after germination of cotyledon or emergence of leaves [
6]. WUSCEL (WUS) and related homeobox transcription factors are regulators of stem cell activity in different tissues such as shoot apical meristem (SAM), root apical meristem (RAM) and vascular cambium [
7]. Among
WUSCHEL-RELATED HOMEOBOX (WOX) genes,
WOX1 and
PRESSED FLOWER (PRS)/WOX3 are involved in the development of marginal cells. Plants lacking function of those genes have multiple defects in leaves and floral organs, most notably defects in blade outgrowth[
8]. The double mutant is reported to lose marginal cells in leaves as well as sepals [
9]. Both WOX1 and PRS are expressed in the middle domain of leaf primordium and marginal cells [
10], supporting their role in maintaining stem cell activity of those cells. Further support of their roles in maintaining stem cell activity of marginal cells came from studies on orthologous genes in other plant species that have similar functions [
11,
12]. Adaxial-expressed MONOPTEROS (MP), abaxial-enriched auxin and abaxial expressed auxin response factor (ARF) repressors together act as positional cues for defining the
WOX expression in margin cells. MP regulates expression of
WOX genes by directly binding to the
WOX1 and
PRS promoters. Furthermore, the ARF2, ARF3, ARF4 repressors suppress
WOX1 and
PRS expression, also through direct binding [
10]. This work suggested that the nuclear auxin signal pathway is involved in WOX regulated leaf margin development. However, molecular mechanisms underlying acquisition of meristematic activity as well as developmental phase shift to lose such activity in marginal cells are unknown.
Phosphorylation of proteins has important biological significance not only by altering properties of phosphorylated protein itself, but also for inducing signaling cascades to enable dynamic regulation of downstream processes. In plants, receptor-like kinases (RLKs) mediated signaling plays key roles in multiple developmental events [
13]. Small peptides as well as other small molecule ligands such as plant hormones are perceived by RLKs for activating downstream signaling [
14]. Signaling mediated by CLAVATA3 (CLV3) peptide-CLAVATA1 (CLV1) RLK pair is involved in stem cell maintenance in the SAM and one of the most extensively characterized example of small peptide mediated signaling in plants [
15]. CLAVATA3/ESR (CLE) peptides act as mobile ligand in apoplast to activate signaling in neighboring cells [
16], and they typically involve in regulation of stem cells in plants [
17]. Direct binding of CLV3 as well as other CLE peptides to several closely related RLKs including CLV1, BAM1 and BAM2 has been detected [
18], and the signaling module is proposed to restrict meristematic activity of SAM stem cells by repressing WUS expression in the central zone of the SAM [
19]. Hormonal signaling is believed to cross-talk with WUS mediated signaling in both upstream and downstream of WUS [
20,
21,
22]. Although CLV1-related RLKs are proposed to have function beyond SAM based on severe abnormalities induced by loss of function of multiple
BAM genes [
23], exact function of these proteins in other tissues as well as their relationship to WOXs have not been investigated in detail yet.
In root tip, the auxin maximum is considered as a polarity signal for the cell layer patterning [
24,
25]. Besides auxin maximum, low concentration auxin also plays critical roles for plant development. For example, during leaf adaxial-abaxial patterning, it has been shown that a transient low auxin zone in the adaxial domain of early leaf primordia worked as a signal to promotes leaf adaxial-abaxial patterning [
26]. The formation of a local auxin minimum is necessary for the specification of the valve margin separation layer where Arabidopsis fruit opening takes place [
27]. It is reported that auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root [
28].
In this work, we used cotyledon margin cells as a new system to study meristematic activity of plant cells through analysis of cell elongation and cell division. In root, transition of the cell division stage to the cell elongation stage occurs successively along the root growth axis determined by the position of cells and thus regulated by spatial positional cues. In contrast, switching of cell division phase to cell division with cell elongation phase in margin cells seems to be regulated in a temporal and spatial manner. By utilizing this beneficial feature of margin cells for analysis, we demonstrated that stem cell factors BAM1 and WOX1 are involved in the suppression of cell division of margin cells evoked by low concentration auxin, which may connect auxin and stem cell factors in regulating meristematic activity of stem cells in plants.
Results
Cell Elongation of Margin Cells Precedes Cell Division in Wild Type Cotyledons During Germination
Epidermal cells locate at the marginal region in cotyledons and leaves play important roles in leaf shape formation [
29,
30] and plants adaptation to the environment [
2]. Although margin cells have been recognized as specialized cells with distinct shape compared to other cell types in leaf epidermis such as pavement cells or stomatal lineage cells, the developmental process of margin cells has not been characterized in detail [
6,
31]. There is no explicit definition on margin cells in Arabidopsis cotyledon. In this study, we only focus attention on the rectangle cells located at the outermost adaxial side and use them to represent margin cells while the small round cells besides the rectangle cells and the rectangle cells located in the abaxial side were not counted. We use mPS-PI staining method [
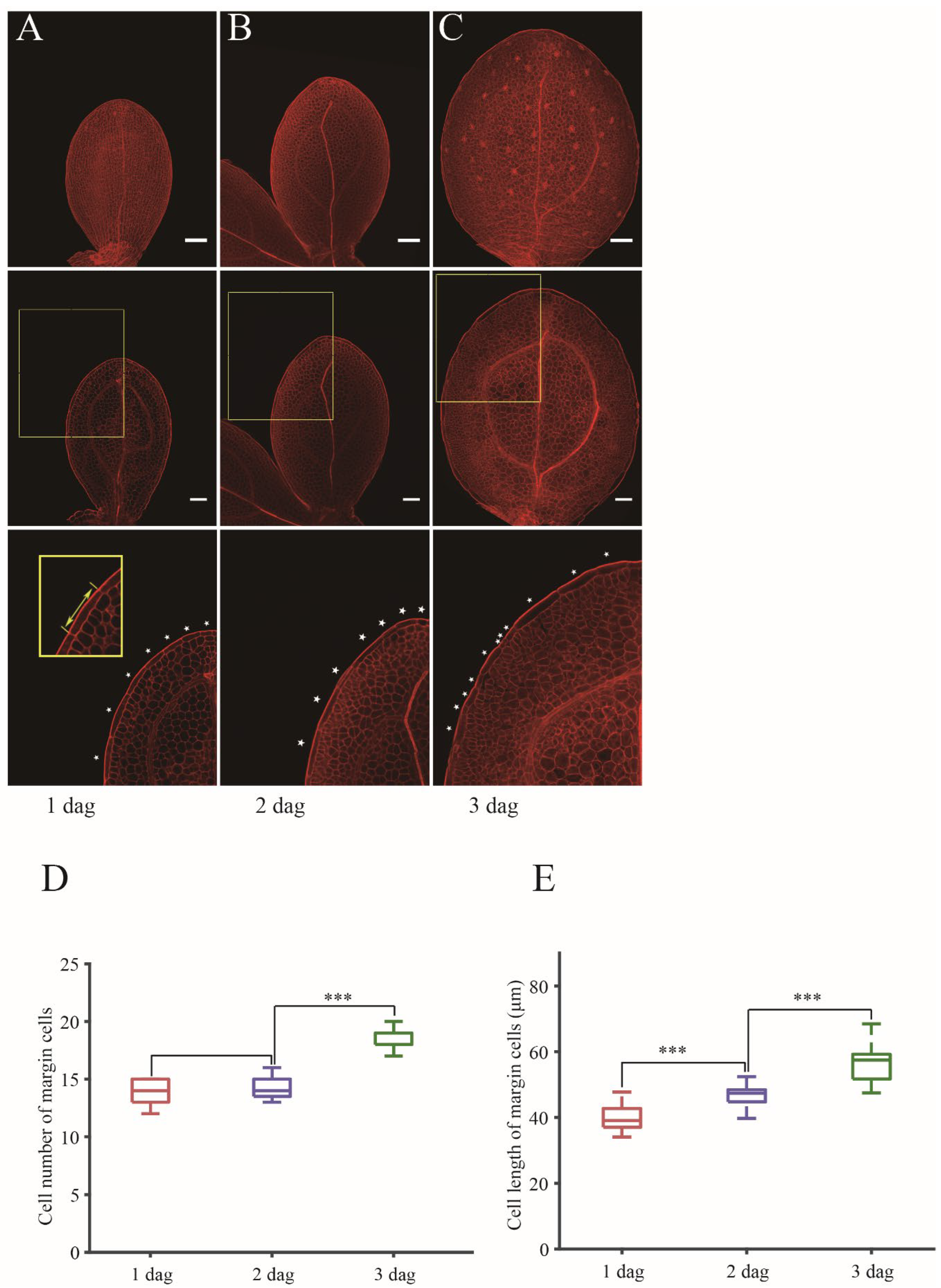
32] to visualize cell walls and measure cell number and longitudinal cell length. Since cotyledon is plane symmetric, we analyzed one side of cotyledon. Through counting the rectangle cells located at the outermost region of adaxial epidermis of wild type Col-0 (Col) plants at 1 dag (day after growth condition) to 3 dag (
Figure 1A–C), we found that the cell number of margin cells of 1 dag cotyledons has little variation ranging from 13 cells to 15 cells. The cell number of margin cells at 2 dag did not show significant change compare to 1 dag (
Figure 1D). These results indicated that margin cells do not endure cell division at 1 dag and 2 dag. In contrast, we found that the cell number increased at 3 dag (
Figure 1D). Then we measured the longitudinal cell length from the top of the margin cell to the bottom of the margin cell. The quantification of cell length in 1 dag, 2 dag and 3 dag showed that margin cells already increased in cell length at 2 dag, and thus they endured cell elongation without division before 2 dag (
Figure 1E).
Thus, we concluded that the cell elongation of margin cells precedes the cell division in developing wild type cotyledons, and the developmental phase transition from the “elongation” phase to the “elongation and division” phase occurs in cotyledons right after germination.
BAM1 Regulates Margin Cell Developmental Phase Transition
Our data showed that the cell elongation of margin cells precedes the cell division in developing wild type cotyledons, and we hypothesized that there is a signaling mechanisms to regulate such developmental phase transition. We have previously found that
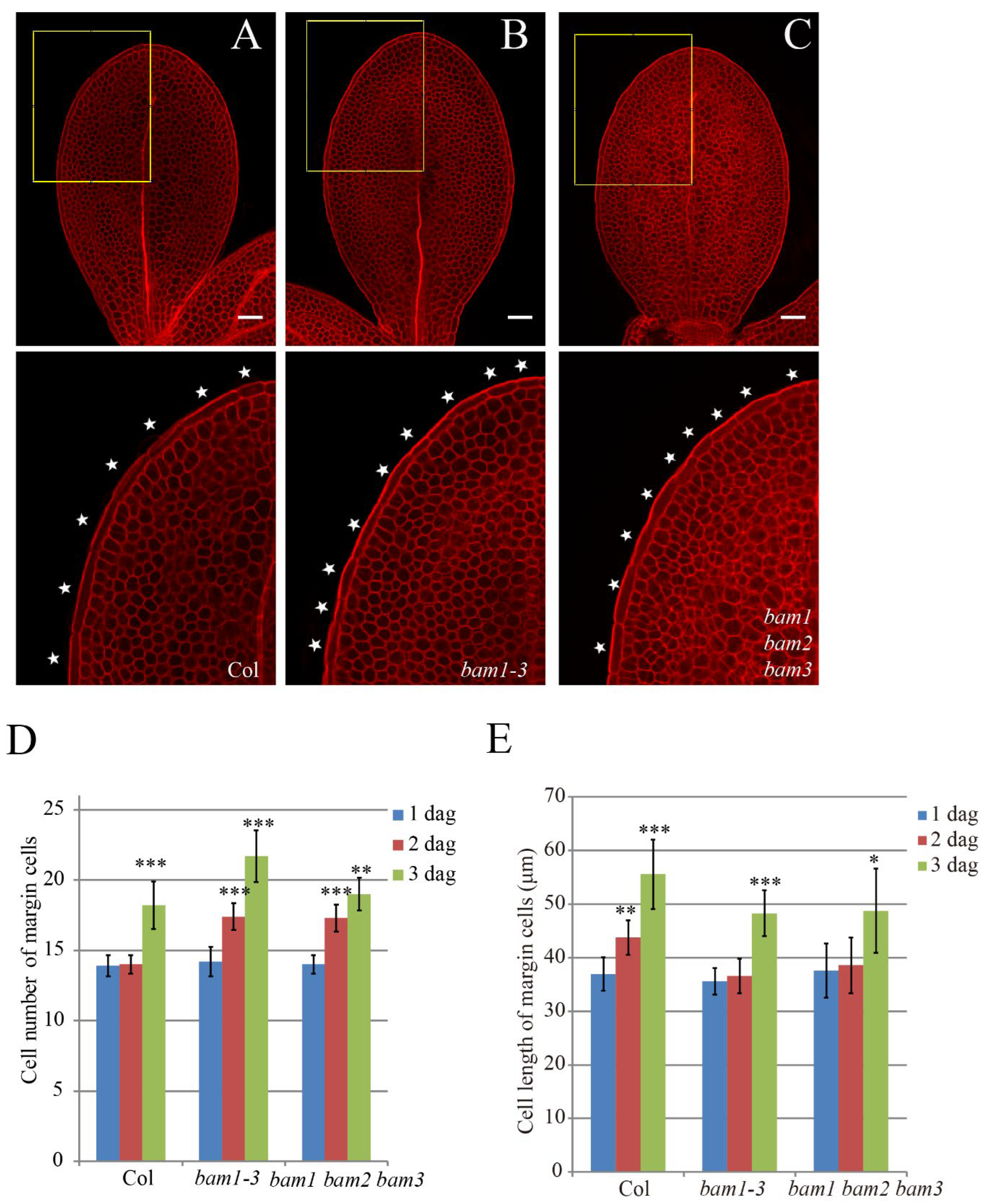
bam1-3 mutant (SALK_015302; Supplementary information, Figure S1A) has increased number of margin cells at 2 dag [
33]. Thus, we decided to check if
bam1-3 shows abnormality in the margin cell developmental phase transition (
Figure 2A–E). At 1dag, the cell number of margin cells in both Col and
bam1-3 cotyledons showed little variation ranging from 13 cells to 15 cells. However, unlike Col cotyledons, the average cell number of margin cells of 2 dag cotyledons in
bam1-3 increased up to 17 cells indicating that the cell division occurred between 1 dag to 2 dag (
Figure 2A, 2B and 2D). In contrast, unlike Col plants, the cell length of
bam1-3 did not increase from 1dag to 2dag (Figure2E). Similar to our previous analysis [
33], the cell number of palisade cells (Supplementary information, Figure S1B and S1C) and cotyledon size (Supplementary information, Figure S1D) of Col and
bam1-3 showed no significant difference at 2 dag, suggesting that the defect of
bam1-3 is specific to margin cells.
To test whether other BAM protein members, BAM2 and BAM3 are involved in regulating margin cells developmental phase transition, we observed margin cell development of bam1 bam2 bam3 triple mutant and found that bam1 bam2 bam3 displayed margin cell developmental phase transition defect as bam1-3, neither reinforcing nor alleviating margin cell developmental phase transition defect of bam1-3 mutant (
Figure 2C-2E). This data suggested that there is no genetic redundancy between BAM1 and BAM2/BAM3 in regulating margin cell developmental phase transition.
Next, to further confirm increase of cell division in
bam1-3, we applied a pulse-chase strategy for EdU labelling assay [
34] to quantify cell division frequency of margin cells of 1 dag, 1.5 dag and 2 dag in Col and
bam1-3 mutant. The EdU assay can monitor the cell division occurrs during the EdU treatment. Pulse-chasing allowed us to monitor the cell division orientation by identifying daughter cell pairs. We found that
bam1-3 displayed more longitudinal cell divisions than Col, from 32% to 16% of 1 dag and from 44% to 30% of 1.5 dag respectively (Table 1). These data confirmed that longitudinal cell division is increased during 1 dag to 2 dag in
bam1-3 compared to Col.
Margin Cells of bam1-3 Mutants Show Abnormal Auxin Response Monitored by an Auxin Response Marker
Auxin regulates timing of cell division or cell differentiation in different tissues [
35,
36,
37]. Since the developmental phase transition of margin cells is altered in
bam1-3 mutant, it is necessary to check whether the auxin response changed in
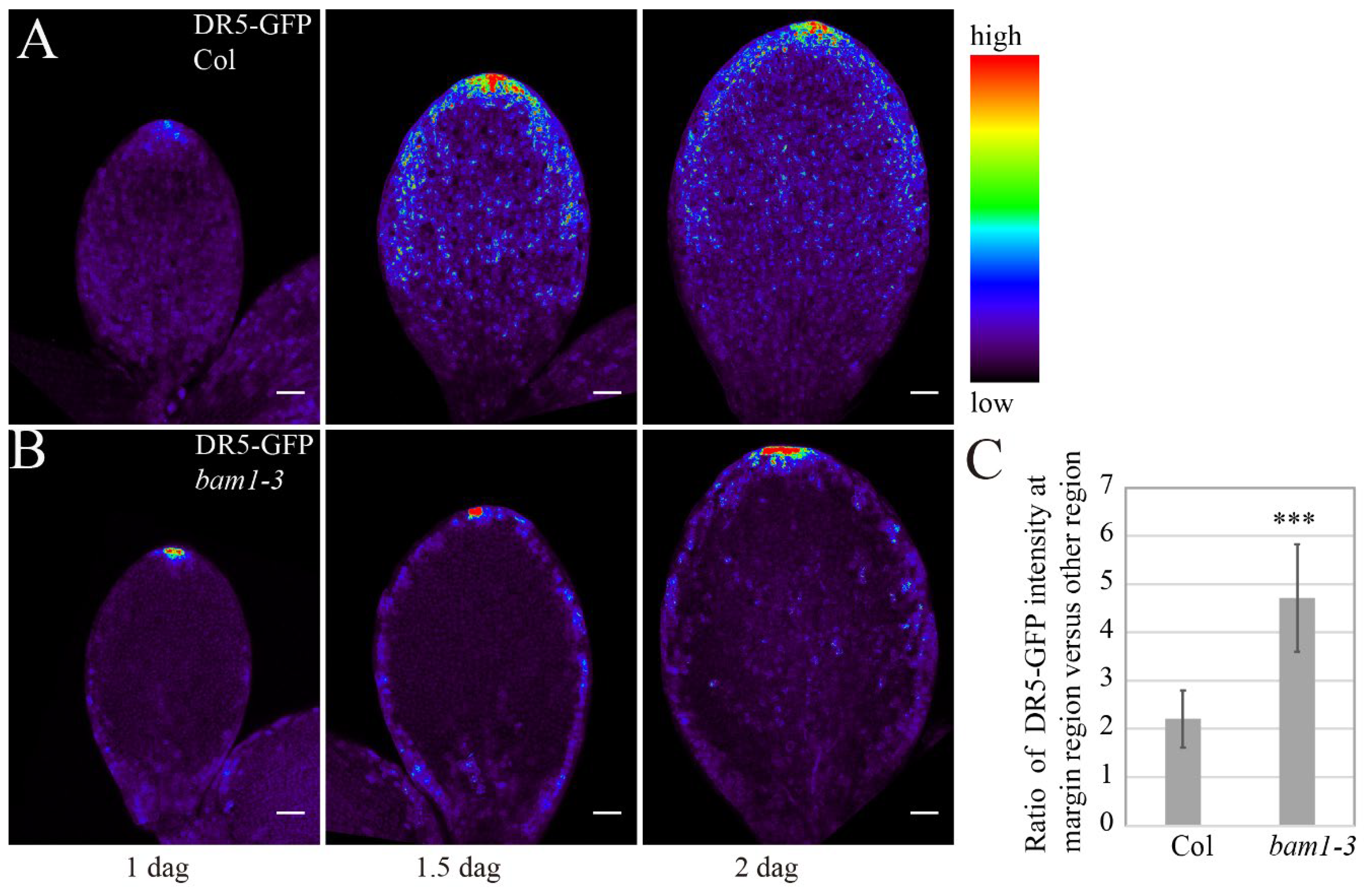
bam1-3 mutant. We checked GFP signal in pDR5::GFP-ER and found that the GFP intensity increased from 1 dag to 2 dag with maximum intensity mainly around the tip area (
Figure 3A). The maximum GFP intensity displayed in the cotyledon tip in
bam1-3 at 1.5 dag, demonstrating that the auxin accumulation at the tip is not affected in
bam1-3 (
Figure 3B). However, the GFP signal is much higher in margin cells compared to other epidermal cells in
bam1-3 at 1.5 dag and 2 dag (
Figure 3B). In Col cotyledons,the margin regions show only a slight increase in GFP intensity compared to other regions. However, we detected significant increase in GFP intensity in the margin region of
bam1-3 cotyledons (
Figure 3C). These results suggested that margin cells may endure higher auxin response compare to other cell types in
bam1-3.
WOX1 Coordinately Regulates Margin Cell Development with BAM1
Previous reports showed that WUS like transcription factors WOX1 and PRS are involved in the development of marginal cells in various shoot organs [
38,
39,
40], and
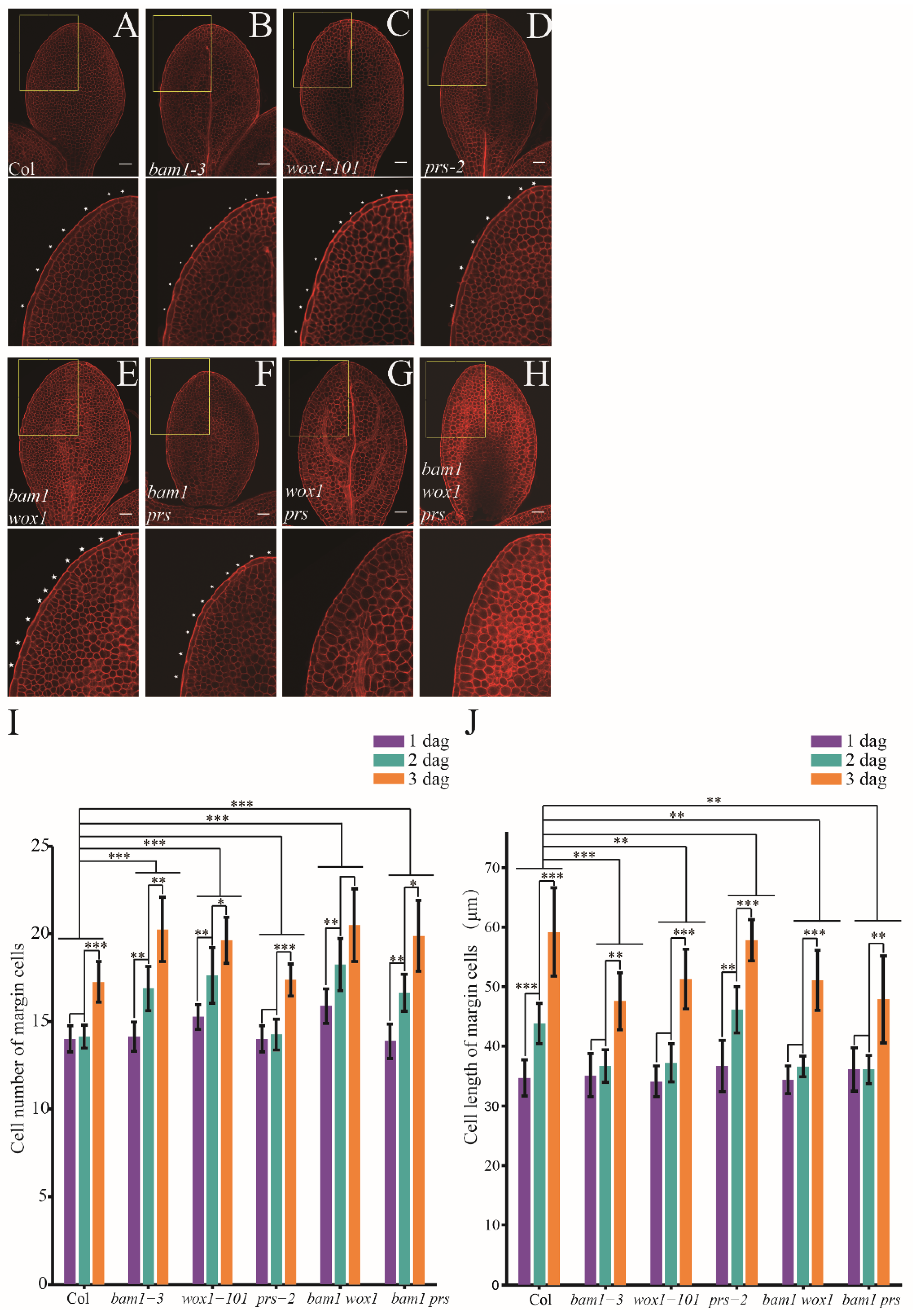
wox1 prs lacks leaf margin cells [
9]. We checked the cotyledon of
wox1 prs and found that the long, rectangular margin cells, were also absent in
wox1 prs cotyledons (
Figure 4A,B). Then, we checked single loss of function alleles for
WOX1 and
PRS on developmental defects in margin cells. Unlike margin cells of Col cotyledons which did not divide before 2 dag, margin cells of the
wox1-101 cotyledons divided from 1 dag similar to
bam1-3 (
Figure 4C,D). In contrast,
prs-2 cotyledons did not show alteration in margin cell development (
Figure 4E). Neither
wox1 bam1-3 nor
prs-2 bam1-3 showed enhancement of margin cell developmental defects from defects observed for
bam1-3 (
Figure 4F,G). We crossed
bam1-3 with
wox1 prs, and found that the
bam1 wox1 prs triple mutants lacks margin cell as in
wox1 prs mutant (
Figure 4H). Quantification of cell division and cell elongation in mutants confirmed that bam1-3 and wox1-101 show similar alteration in developmental phase transition of margin cells (
Figure 4I,J). However, in
wox1-101 and
wox1 bam1, the cell number of margin cells increased up to 15 and 16 respectively at 1 dag (
Figure 4I). Given that in both Col and
bam1-3 the cell number of margin cells at 1dag is constant around 14, the results suggested that WOX1 may regulate margin cells development in an early stage before 1dag.
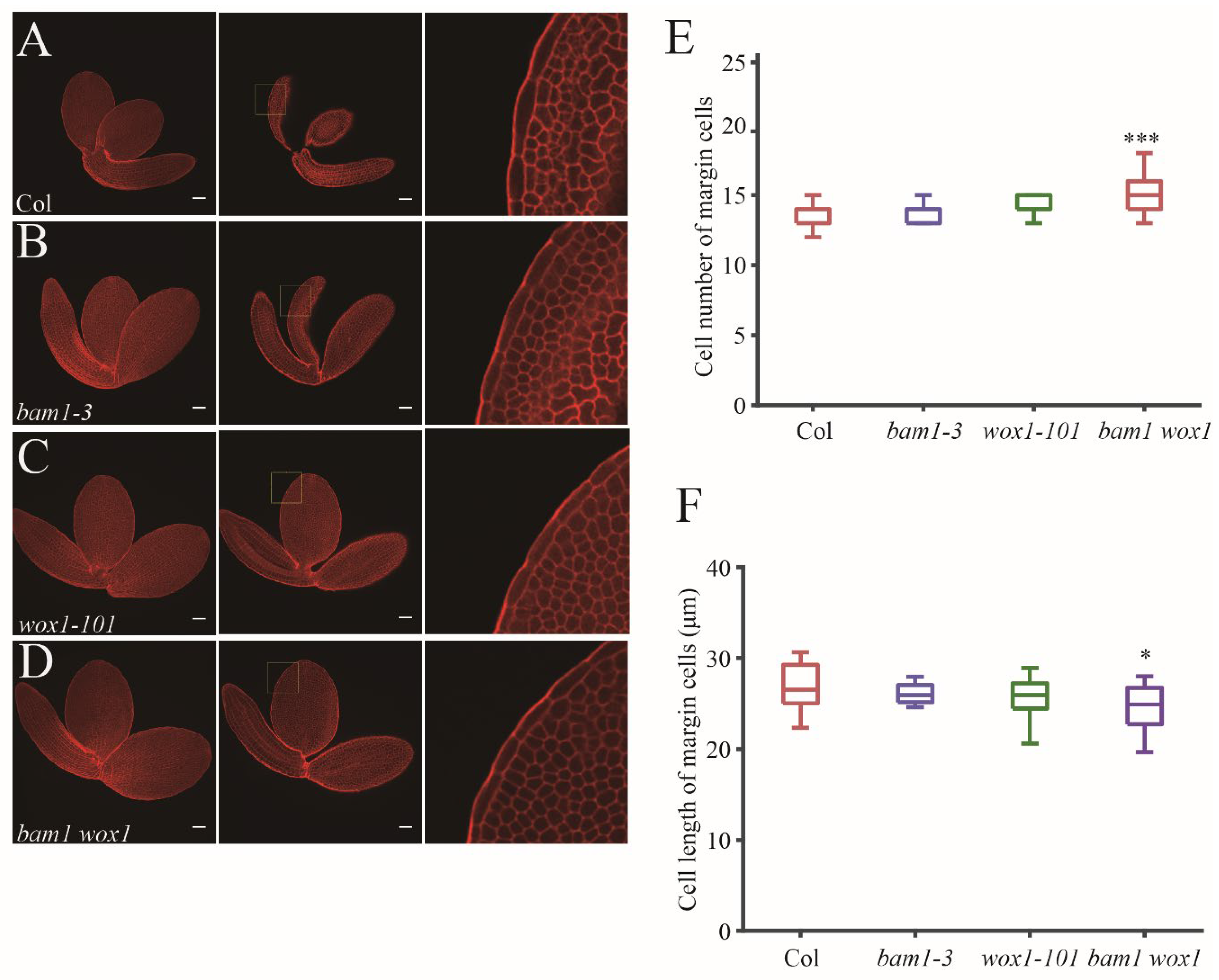
Next, in order to detect early developmental defects before germination, we checked mature embryo of Col,
bam1-3, wox1-101 and
wox1 bam1 (
Figure 5A–D). We found that the margin cell number of mature embryo in
bam1-3 and
wox1-101 are similar to Col. The cell number of mature embryo margin cells in
wox1 bam1 is increased slightly to around 15 (
Figure 5E) and the cell length of this mutant is decreased slightly (
Figure 5F), suggesting that the BAM1 has synergistic function with WOX1 in regulating margin cell numbers in embryo stage.
The pulse-chase EdU labelling assay showed that at 1 dag the frequency of longitudinal cell divisions in wox1-101 and wox1 bam1 increased, from 16% to 34% and 16% to 26% respectively. However, at 2 dag, the frequency of longitudinal cell divisions in wox1-101 and wox1 bam1 decreased, from 16% to 4% and 16% to 8% respectively. Thus, synergistic effect of combining wox1-101 and bam1-3 is limited to embryo development stage, while it shows no enhancement of abnormality in post-embryonic development.
To check the expression pattern of BAM1 and WOX1 in young cotyledons, we constructed the promoter reporter line for BAM1 and WOX1 which contained YFP fused with nucleus localization signal downstream of promoter sequence. Consistent with the defects at the germinating cotyledons, the transgenic line proBAM1::YFP-NLS started to show YFP signal from the middle region of cotyledon at 1dag and the signal expanded to appear on cells over the whole cotyledon after 2 dag (Supplementary information, Figure S2A–C). The transgenic line proWOX1::YFP-NLS started to show YFP signal from the margin cells of cotyledons at 1 dag and the signal was restricted to the margin region till later stage (Supplementary information, Figure S2D–F). These results suggested that the margin cell development in cotyledon is regulated by BAM1 and WOX1 at different stages. WOX1 works from the mature embryo to 2dag while BAM1 acts at a relatively later stage, from 1 dag to 2 dag. However, BAM1 seems to play roles in margin cell development in embryo stage with WOX1 which may be hindered by genetic redundancy with other BAMs/RLKs.
Low Concentration Auxin Suppresses Longitudinal Cell Division of Margin Cells
Next, to gain further insights into the effect of auxin on margin cell development, we examined whether different concentrations of auxin has differential effect on cell division of margin cells in Col by applying EdU labelling assay to monitor cell division. Seedlings were divided into 5 groups and cultured in 1/2 MS with 0.1 nM NAA, 1 nM NAA, 10 nM NAA and EtOH as control respectively before EdU staining assay at 1.5 dag. The results demonstrated that with 1 nM NAA treatment, the frequency of transversal and longitudinal cell divisions increased, from 23% to 30% and from 20% to 46% respectively (Table 2). With 100 nM NAA treatment, the frequency of transversal and longitudinal cell divisions decreased, from 23% to 10% and from 20% to 6% respectively (Table 2). These results suggested that 1 nM NAA treatment accelerate longitudinal cell division while 100 nM NAA depress both transversal and longitudinal cell division.
Then we tested potential negative contribution of BAM1 and WOX1 in the low concentration auxin response, since bam1-3 and wox1-1 showed similar alteration of cell division frequency to the low concentration auxin treatment. When we applied pulse-chase EdU labelling assay with 1 nM NAA treatment in bam1-3 and wox1-101 at 1.5 dag, we found that neither bam1-3 nor wox1-101 showed increased cell division frequency compared to Col (Table 3). All together, these data suggest that BAM1 and WOX1 act as negative regulators of longitudinal cell division induced by low concentration auxin treatment.
Discussion
Our results show that margin cells elongated before 2 dag and then started to divide along with the growth of cotyledon. Suppression of margin cell division before 2 dag is dependent on BAM1. Importantly, initiation of cell elongation of margin cells is retarded in
bam1-3. This indicates that defects we observed for
bam1-3 are not caused by simple acceleration of developmental program inducing a precocious developmental phase shift, but caused by alteration of developmental program to prevent cell division. Similar to
bam1-3, margin cells developmental phase transition occurs at earlier timing in
wox1-101 (
Figure 4I). Phenotype analysis of mature embryo margin cells in
wox1-101,
bam1-3 and
bam1 wox1 demonstrated that BAM1 and WOX1 work synergistically to control embryo margin cells development (
Figure 5E,F). Unlike opposite phenotype of
clv1 and
wus exhibited in the CLV3-WUS feedback loop of controlling SAM maintenance,
wox1-101 and
bam1-3 seem to have abnormal phenotype in similar direction. Increased division of stem cells may result in loss of stem cell activity, which in turn leads to consumption of proliferating cells in meristem. Detail analysis on mutants with weaker phenotype due to reduction of
WUS genes and genetic analysis among multiple mutants in the CLV-WUS pathway using such weak mutants may reveal whether this idea is correct or not.
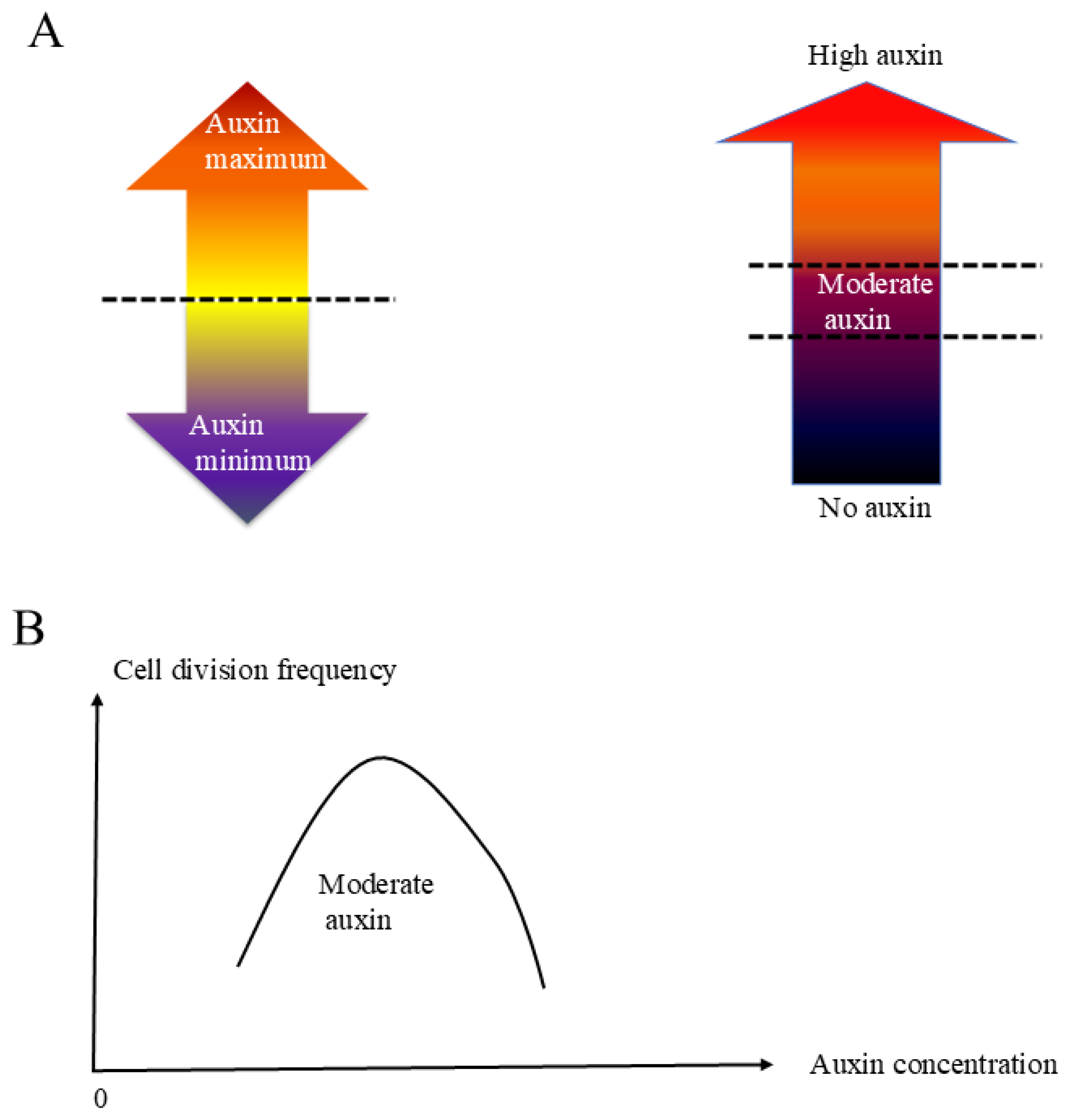
As
Figure 3A shows, gradient of auxin response is formed over cotyledon surface with pronounced accumulation of signal along margin cells by 2 dag. Thus, it is reasonable to assume that increased auxin evokes the cell division after 2 dag. Interestingly, exogenously applied low concentration NAA promotes cell division while high concentration NAA application inhibits cell division. These data suggests that a moderate auxin concentration is necessary for margin cells to transit from “elongation” phase to the “elongation and division” phase, and higher or lower concentration of auxin out of that range keeps cells to be elongated. Auxin concentration is controlled by auxin biosynthesis, auxin transport and auxin degradation. Since our data was obtained from exogenously applied auxin, exact concentration range of auxin to induce margin cell division is currently unclear. Further experiments such as characterizing development of margin cells in auxin synthesis mutant lines or testing the effect of blocking auxin transportation or inhibiting auxin degradation will be done in the future. Different from previous hypothesis that the auxin maximum or auxin minimum triggers the developmental switch[
41], our work suggested that an appropriate moderate auxin concentration triggers the transition of margin cells from the cell elongation to the cell division. Thus, we would like to introduce a new “moderate auxin concentration model”, which proposes the existence of mechanisms to achieve switching of developmental program specifically by a certain auxin concentration range. The major differences between the auxin maximum or minimum model and the moderate auxin concentration model are summarized in
Figure 6A. Auxin maximum or minimum model emphasizes that achieving high or low auxin concentration works as a trigger to developmental regulation. The moderate auxin concentration model proposes that there is a moderate range for auxin to trigger developmental regulation. Too high or too low auxin concentration will not trigger the relevant developmental regulation (
Figure 6B). It is possible that the reported auxin minimum is actually a moderate auxin concentration instead of minimum auxin concentration since the auxin markers used in those studies, DR5-GFP or DII-VENUS are both based on TIR mediated auxin response expected to reflect only certain range of high auxin concentration. To confirm this idea, a new auxin marker to detect low auxin concentration at a few nM range need to be developed and investigated in each developmental context where auxin minimum is reported to be playing important role. Experiments using auxin inducible lines and auxin markers in auxin biosynthesis mutants to control auxin concentration artificially should be done. Plant development is complicated, and auxin concentration is not the only factor determines margin cells fate. It is reported that cytokinin and brassinosteroid can affect cell elongation and cell division together with or without auxin as well [
42,
43]. Whether the two hormones also participate in the margin cell development and whether they have crosstalk with auxin need to be clarified in the future. Besides, cell division is a result from cell cycle. Whether the cell division increase triggered by moderate auxin concentration changes the expression of some key regulators in cell cycle, for example, translation elongation factor 2 (EF2) needs to be investigated in the future studies [
44].
Figure 1.
Cell elongation of margin cells preceding cell division in wild type cotyledons during germination process. (A-C) Images of cotyledons with mPS-PI staining at 1 dag (A), 2 dag (B) and 3 dag (C) in Col. Maximum projection images of Z-stacks are showed in upper panel. Single slice of optimal sections of margin cells are showed in middle panel. Bottom panels show magnified images of middle panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (D, E) Quantitative analysis of cell number (D) and cell length (E) of margin cells in (A-C). Cell lengths are measured by drawing straight line in longitudinal axis along a cell as indicated in the bottom left panel in (A). Data are presented as mean ± SD. Student’s t-tests were performed based on the differences of 2 dag to 1 dag and 3 dag to 2 dag (***p < 0.001, n = 16).
Figure 1.
Cell elongation of margin cells preceding cell division in wild type cotyledons during germination process. (A-C) Images of cotyledons with mPS-PI staining at 1 dag (A), 2 dag (B) and 3 dag (C) in Col. Maximum projection images of Z-stacks are showed in upper panel. Single slice of optimal sections of margin cells are showed in middle panel. Bottom panels show magnified images of middle panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (D, E) Quantitative analysis of cell number (D) and cell length (E) of margin cells in (A-C). Cell lengths are measured by drawing straight line in longitudinal axis along a cell as indicated in the bottom left panel in (A). Data are presented as mean ± SD. Student’s t-tests were performed based on the differences of 2 dag to 1 dag and 3 dag to 2 dag (***p < 0.001, n = 16).
Figure 2.
BAM1 regulating margin cell developmental phase transition. (A-C) mPS-PI stained 2 dag cotyledons of Col (A), bam1-3 (B) and bam1, 2, 3 (C). Single optimal section of margin cells is showed in upper panel. Bottom panels show magnified images of upper panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (D, E) Quantitative analysis of cell number (D) and cell length (E) of margin cells in Col, bam1-3 and bam1, 2, 3. Data are presented as mean ± SD. Student’s t-test was performed to assess significance of differences of between 2 dag to 1 dag and 3 dag to 2 dag in each group. The difference between each mutant line and Col was determined by two-way ANOVA. (*, ** and *** denote p < 0.05, p < 0.01 and p < 0.001, respectively. n = 8).
Figure 2.
BAM1 regulating margin cell developmental phase transition. (A-C) mPS-PI stained 2 dag cotyledons of Col (A), bam1-3 (B) and bam1, 2, 3 (C). Single optimal section of margin cells is showed in upper panel. Bottom panels show magnified images of upper panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (D, E) Quantitative analysis of cell number (D) and cell length (E) of margin cells in Col, bam1-3 and bam1, 2, 3. Data are presented as mean ± SD. Student’s t-test was performed to assess significance of differences of between 2 dag to 1 dag and 3 dag to 2 dag in each group. The difference between each mutant line and Col was determined by two-way ANOVA. (*, ** and *** denote p < 0.05, p < 0.01 and p < 0.001, respectively. n = 8).
Figure 3.
Margin cells of bam1-3 mutant show abnormal accumulation of GFP signal from DR5-GFP. (A, B) DR5-GFP expression pattern in Col (A) and bam1-3 (B) cotyledons at early stage of germination. From left to right, 1 dag, 1.5 dag. 2 dag. Bars = 50 μm. Signal intensities are coded from violet to red corresponding to increasing intensity levels (see colour scale). (C) Mean ratio of DR5-GFP abundance at the margin cells versus other epidermal cells in Col and bam1-3 at 1.5 dag. n = 8 with 20 cells for each plants were analyzed. Error bars represent SD, P-value was calculated according to Student’s t-test. * denotes p < 0.05.
Figure 3.
Margin cells of bam1-3 mutant show abnormal accumulation of GFP signal from DR5-GFP. (A, B) DR5-GFP expression pattern in Col (A) and bam1-3 (B) cotyledons at early stage of germination. From left to right, 1 dag, 1.5 dag. 2 dag. Bars = 50 μm. Signal intensities are coded from violet to red corresponding to increasing intensity levels (see colour scale). (C) Mean ratio of DR5-GFP abundance at the margin cells versus other epidermal cells in Col and bam1-3 at 1.5 dag. n = 8 with 20 cells for each plants were analyzed. Error bars represent SD, P-value was calculated according to Student’s t-test. * denotes p < 0.05.
Figure 4.
The role of WOX in regulating margin cells development. (A-H) mPS-PI stained cotyledons in Col (A), wox1 prs (B), bam1-3 (C), wox1-101 (D), prs-2 (E), bam1 wox1 (F), bam1 prs (G) and bam1 wox1 prs (H) at 2 dag. Single slice of optimal sections of margin cells are showed in upper panel. Bottom panels show magnified images of upper panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (I, J) Quantitative analysis of cell number (I) and cell length (J) of margin cells in Col and the mutants. Data are presented as mean ± SD. Student’s t-tests were performed to assess significance of based on the differences between of 2 dag to 1 dag and 3 dag to 2 dag in each group. The significance of differences between each mutant line and Col was assessed by two-way ANOVA. (*, ** and *** denote p < 0.05, p < 0.01 and p < 0.001, respectively. n = 8).
Figure 4.
The role of WOX in regulating margin cells development. (A-H) mPS-PI stained cotyledons in Col (A), wox1 prs (B), bam1-3 (C), wox1-101 (D), prs-2 (E), bam1 wox1 (F), bam1 prs (G) and bam1 wox1 prs (H) at 2 dag. Single slice of optimal sections of margin cells are showed in upper panel. Bottom panels show magnified images of upper panels (outlined in yellow boxes). White stars indicate margin cells. Bars = 50 μm. (I, J) Quantitative analysis of cell number (I) and cell length (J) of margin cells in Col and the mutants. Data are presented as mean ± SD. Student’s t-tests were performed to assess significance of based on the differences between of 2 dag to 1 dag and 3 dag to 2 dag in each group. The significance of differences between each mutant line and Col was assessed by two-way ANOVA. (*, ** and *** denote p < 0.05, p < 0.01 and p < 0.001, respectively. n = 8).
Figure 5.
BAM1 and WOX1 synergistically regulate margin cell development in mature embryo. (A-D) mPS-PI stained mature embryos of in Col (A), bam1-3 (B), wox1-101 (C) and bam1 wox1 (D). Maximum projection images of Z-stacks are showed in left panels. Single slices of optimal section of margin cells are showed in middle panels. Right panels show magnified images of middle panels (outlined in yellow boxes). Bars = 50 μm. (E, F) Quantitative analysis of cell number (E) and cell length (F) of margin cells in Col and the mutants. Data are presented as mean ± SD. Student’s t-tests were performed to assess significance of the differences between the mutant lines and Col (* p < 0.05, *** p < 0.001, n = 14).
Figure 5.
BAM1 and WOX1 synergistically regulate margin cell development in mature embryo. (A-D) mPS-PI stained mature embryos of in Col (A), bam1-3 (B), wox1-101 (C) and bam1 wox1 (D). Maximum projection images of Z-stacks are showed in left panels. Single slices of optimal section of margin cells are showed in middle panels. Right panels show magnified images of middle panels (outlined in yellow boxes). Bars = 50 μm. (E, F) Quantitative analysis of cell number (E) and cell length (F) of margin cells in Col and the mutants. Data are presented as mean ± SD. Student’s t-tests were performed to assess significance of the differences between the mutant lines and Col (* p < 0.05, *** p < 0.001, n = 14).
Figure 6.
A moderate auxin concentration model for cell division control in margin cells. (A) Basic description of auxin minimum model (left panel) and moderate auxin concentration model (right panel). (B) The relationship of auxin concentration and cell division frequency. Our data suggest that there is a moderate auxin concentration range which promotes margin cell division. In normal condition without exogenous auxin, this moderate auxin concentration showed up around 2 dag. However, if we treat the seedlings with low concentration exogenous auxin at 1.5 dag, the moderate auxin concentration will be achieved earlier and additional cell division happen at this stage.
Figure 6.
A moderate auxin concentration model for cell division control in margin cells. (A) Basic description of auxin minimum model (left panel) and moderate auxin concentration model (right panel). (B) The relationship of auxin concentration and cell division frequency. Our data suggest that there is a moderate auxin concentration range which promotes margin cell division. In normal condition without exogenous auxin, this moderate auxin concentration showed up around 2 dag. However, if we treat the seedlings with low concentration exogenous auxin at 1.5 dag, the moderate auxin concentration will be achieved earlier and additional cell division happen at this stage.