Methodology
The study was conducted at Minilik II Comprehensive Specialized Hospital and Yekatit 12 Hospital Medical College, which are located in the capital city of Ethiopia, Addis Ababa. The hospitals provide service for wide scale of population from the city and for those who are from all corner regions with diverse socio-demographic background resident society.
Minilik II Comprehensive Specialized Hospital has various professionals that included more than 85 sub specialized and medical physicians, 203 nurses, 123 other health professionals, and 250 administrative staff, making nearly 700 staffs.
Yekatit 12 Hospital Medical College was build and starts to serve for the last 100 years which makes the hospital one of the few oldest institutes in Ethiopia. It is supposed to serve more than 5 million patients who are from the city and away from the catchment area. It has six major departments. In the hospital estimated 200 to 250 patients are being served per day through six units of the OPD.
A hospital based cross-sectional study was conducted from February to June, 2024.
All patients who were attended Minilik II Comprehensive Specialized Hospital and Yekatit 12 Medical College Hospital.
All patients who were attended Minilik II Comprehensive Specialized Hospital and Yekatit 12 Medical College Hospital for diagnosis of SBF infection and ordered SBF analysis for microbiology laboratory study during the study period.
All suspected patients for bacterial SBF infection that were cooperative to provide the clinical specimen and other required information.
Patients who were on antibiotic treatment in the last two weeks prior to the data collection period and those patients who had follow up at the same time at Minilik II Comprehensive Specialized Hospital and Yekatit 12 Hospital Medical College during the study period.
The sample size was calculated based on single population proportion formula as described below.
The value of p is taken as 14.1% (0.14) referring from a study conducted at Tikur Anbessa Specialized hospital [
2].
Where, n = sample size,
Z = Z-score, confidence interval (95% confidence interval, 1.96) P = 0.141 (Prevalence from previous study)
P = expected prevalence or proportion (P = 0.5)
d = precision (d = 0.05) and
(1.96)² X 0.141(1-0.14) =185.011~186
0.05²
Therefore, a total of 186 study participants were recruited using convenient sampling technique with equal proportion of participants from Minilik II Comprehensive Specialized Hospital and Yekatit 12 Hospital Medical College.
Socio-demographic characteristics such as, age, gender, residence, educational background, occupational status, marital status, monthly income, and clinical related data such as, specimen type, previous SBF infection history, ward unit, other known medical disease, and appearance of specimen.
Clinical and socio-demographic data were collected by assigned nurses at the study site hospital using a pretested structured questionnaire. The pretest was done at Kebena Health Centre, Arada Kifle Ketema, Addis Abeba.
All specimens, such as pleural fluid, peritoneal fluid, CSF, synovial, and pericardial fluids were collected and transported to the microbiology laboratory and processed within two (02), hours of collection. Simple and précised checklist was designed to collect participant’s information like participants code number, age, sex, date, medical record number, inpatient, and outpatient.
Following a signed agreement, a doctor used a needle and syringe to collect samples aseptically. After being collected, the samples were sent to a lab for gram staining, culture, and WBC count. Every collected fluid's physical characteristics including whether it is clear, bloody or traumatized, turbid, or straw-like was recorded.
Microbiology analysis was performed at Yekatit 12 Hospital Medical College laboratory, microbiology department for the isolation and determination of bacteria. The collected specimens were prepared in accordance with established procedures for gram staining, culture and biochemical testing followed to inoculating on blood agar, chocolate agar and MacConkey agar plates. Inoculated medium were aerobically incubated for 24 to 72 hours at 35 to 37°C. In order to create a microareophilic state for picky bacteria, the chocolate agar plates were specially incubated in a candle jar with a CO2 concentration of 5–10%.
Plates were investigated for the growth of bacterial colonies and those which had culture growths were taken for gram stain and biochemical tests based on the standards of the laboratory procedures. Identification of isolates was applied based on standard bacteriological techniques, such as colony morphology, gram staining and biochemical test.
Biochemical characteristics of gram-positive bacteria were determined by carrying out catalase test, coagulase test, optochin sensitivity test, bacitracin sensitivity test, mannitol test, and hemolytic activity on blood agar. While indole production test, citrate utilization test, kliger’s iron test, urease test, oxidase test, lysine decarboxylase (LDC) test, methyl red/voges proskauer (MR/VP) test, motility test, and hydrogen sulfide gas production test were carried out for the identification of gram-negative bacteria.
Susceptibility of bacterial isolate to antimicrobial agents of different classes were assessed by the Kirby-Bauer disk diffusion method in compliance with a commercially prepared antibiotic discs of known concentration on Mueller–Hinton agar(MHA) standard media per Clinical Laboratory Standard Institute (CLSI) guidelines [
10].
The MHA were applied with inoculums prepared with 0.5 McFarland turbidity standard following antimicrobial discs were applied on to the plate. Antimicrobial discs such as: Amikacin (30 ug Oxoid),Ceftazidime (30ug Oxoid), Ceftriaxone (30ug BD), Gentamycin (10ug Oxoid), TMP-SXT (1.25+23.75ug BD), Clindamycin(2μg BD), cefoxitin (10 ug BD), Erythromycin (15ug Oxoid), Ciprofloxacillin (5ug BD), Vancomycin (30ug BD), and amoxa-clavulanic acid (10ug Oxoid) were used.
The QC was performed to check the quality of the medium. Each new lot of stock reagents which were used for preparation of media was passed through quality control panel before used by testing with known standard strains. To observe the potency of performance of reagents, methods and techniques, QC was applied on staining reagents with known organisms.
Standard Operating Procedures (SOPs) were strictly applied, and verified if the media met an expiration date and quality control parameters per CLSI. Data collectors were trained before the data collection procedure and 5% of total prepared questionnaires were pre-tested before the data collection.
The Quality control of culture medias was assured by concurrent testing with the American Type Culture Collection (ATCC) strains including E. coli (ATCC 25922), P. aeruginosa (ATCC 27852), and S. aureus (ATCC 25923). Also visual inspections of cracks in media or plastic petri-dishes, unequal fill, hemolysis, evidence of freezing, bubbles, and contamination were performed.
The data entry was performed by EPI INFO version 3.1 statistics software and the entered data was double-checked before the analysis. The descriptive statistics (means, percentages or frequency) and the bi-variant logistic regression analysis were performed. The final results are presented in different forms of descriptive charts, graphs and tables.
Laboratory results were reported for respective physician/nurses using a cell phone and written report for better patient management. The findings of the study were re-reported to the hospital`s research director office. The study abstract submitted to Addis Ababa health bureau in order to use the findings as a baseline for further related studies and build general conceptual understanding about the issue also, to assist the responsible bodies to practice and thereby maximize the benefits of the patients and health services. The final report is submitted and presented to Wollo University, Department of Medical Laboratory Science.
The study was approved by the Department of Medical Laboratory Science, Wollo University. The proposal document was re-evaluated and represented to the Addis Ababa Health Bureau research directorate office and acknowledged the topic to accomplish the study at Minilik II Comprehensive Specialized Hospital and Yekatit 12 Hospital Medical College. The final permission letter was received from the hospital research and social service office with letter of assistance to the researcher after presentation and explanation of the purpose and procedures of the study. All information of patients remained as confidential as national protocol suggests. The necessary information about the purpose and procedures of the study was explained to the study participants and attendants and for professionals who has participated in the data collecting. Each laboratory result of study participants was reported to the requesting physicians on time.
Sterile body fluids: biological fluids found in human body which do not contain any microorganisms at normal state such as, cerebrospinal fluid, pleural fluid, pericardial fluid, peritoneal or ascitic fluid, synovial fluid [
9].
Sterile body fluids infection: microbial invasion and physiochemical change in the SBF site [
11].
Gross appearance: naked eye observable physical status of sample if it`s clear, turbid, bloody, yellowish, or purulent.
Types of specimens: referencing the site of the sterile fluid sample collected to classify as cerebrospinal fluid, pleural fluid, pericardial fluid, peritoneal or ascitic fluid, synovial fluid etc.
Multidrug resistance (MDR): resistant at least one agent in three or more antimicrobial categories [
12].
Results
In this study, 186 SBF samples were collected from patients attending at Minilik II Comprehensive Specialized Hospital and Yekatit 12 Medical College Hospital from February - June, 2024. From total participants, 110 (59.1%) were female and 54 (29.0%) were over 50 years old. Correspondingly, government employees took 48 (25.7%) while, based on participant`s monthly income, 69 (36.9%) had in a range 10001-20000 ETB. Similarly, 120 (64.5%) were above college level in their education status (
Table 1).
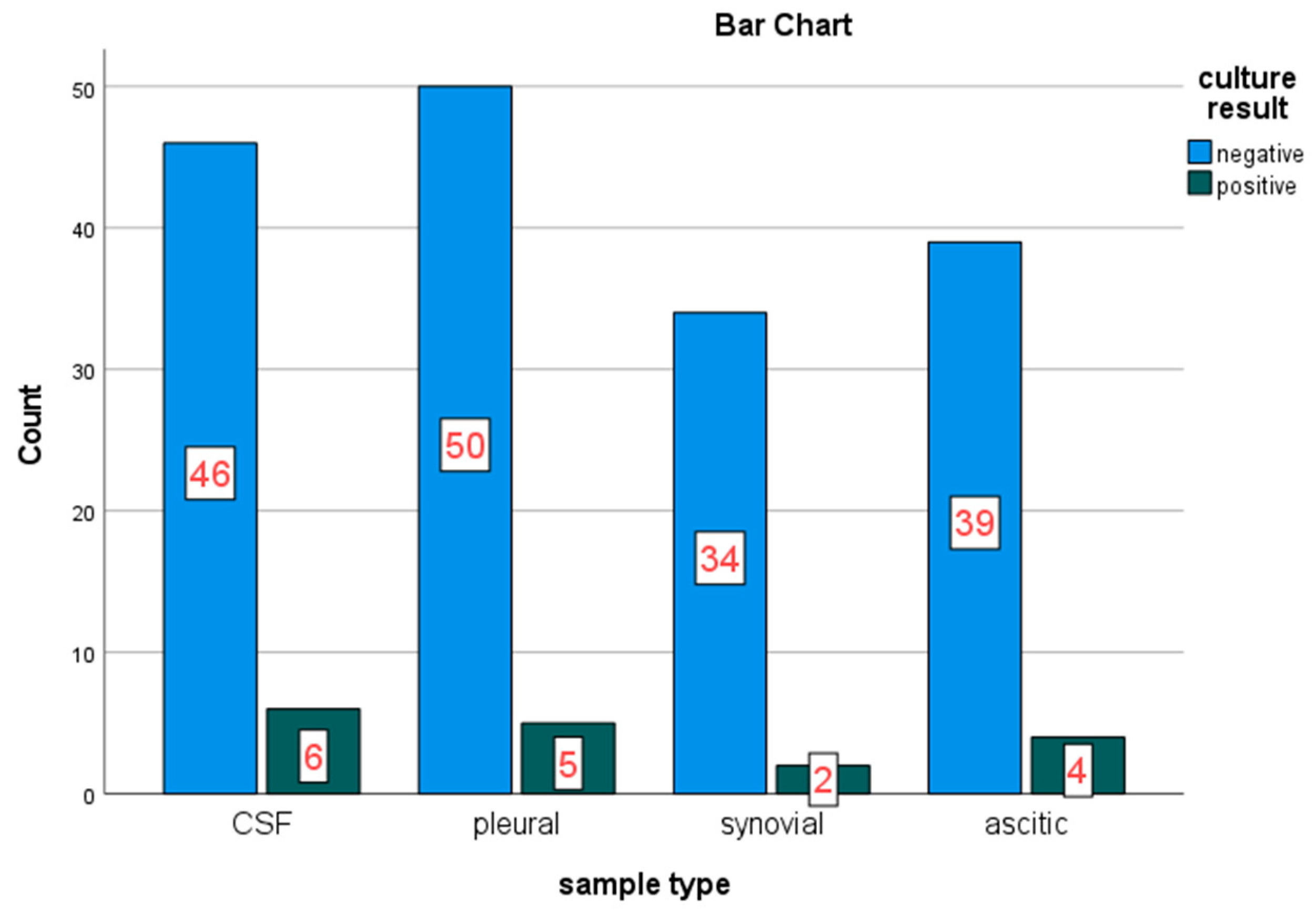
Sterile samples were collected aseptically from four (04) different sites secreting SBFs. In this study; CSF, pleural, synovial and peritoneal fluids were collected from participants. From these collected samples, Pleural fluid 55 (29.6%), Cerebrospinal fluids 52 (28.0%), Ascitic/peritoneal fluid 43 (23.1%) and synovial fluid 36 (19.4%) were encountered in the study. Among these samples types collected and investigated for bacterial growth on culture, CSF found with the highest burden among the collected SBF sample followed by pleural fluid, 6/17 (35.3%) and 5/17 (29.4%) respectively (
Figure 2).
According to the present study, the overall prevalence of SBF bacterial infection among participants is 17 /186 (9.1%) (CI=95%, 3.8-14.5).
Regarding to the gram stain characteristics of the isolated bacteria, 12/17 (70.6%) were gram-negative bacteria.
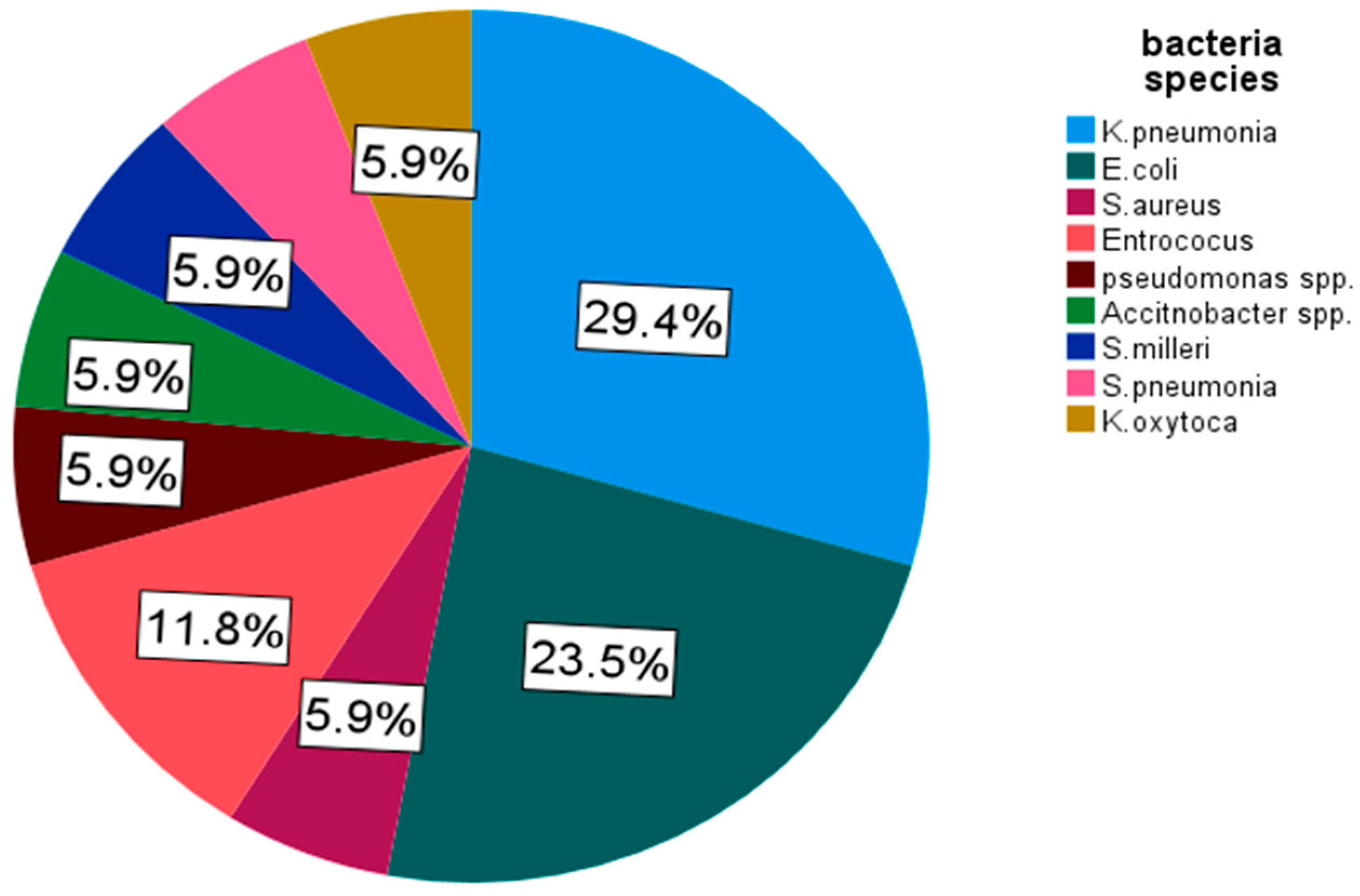
From the culture growth isolated bacteria,
K. pneumoniae was found as the most predominant one 29.4% (5/17) and followed by
E. coli attaining 23.5% (4/17) for SBF infection (
Figure 3).
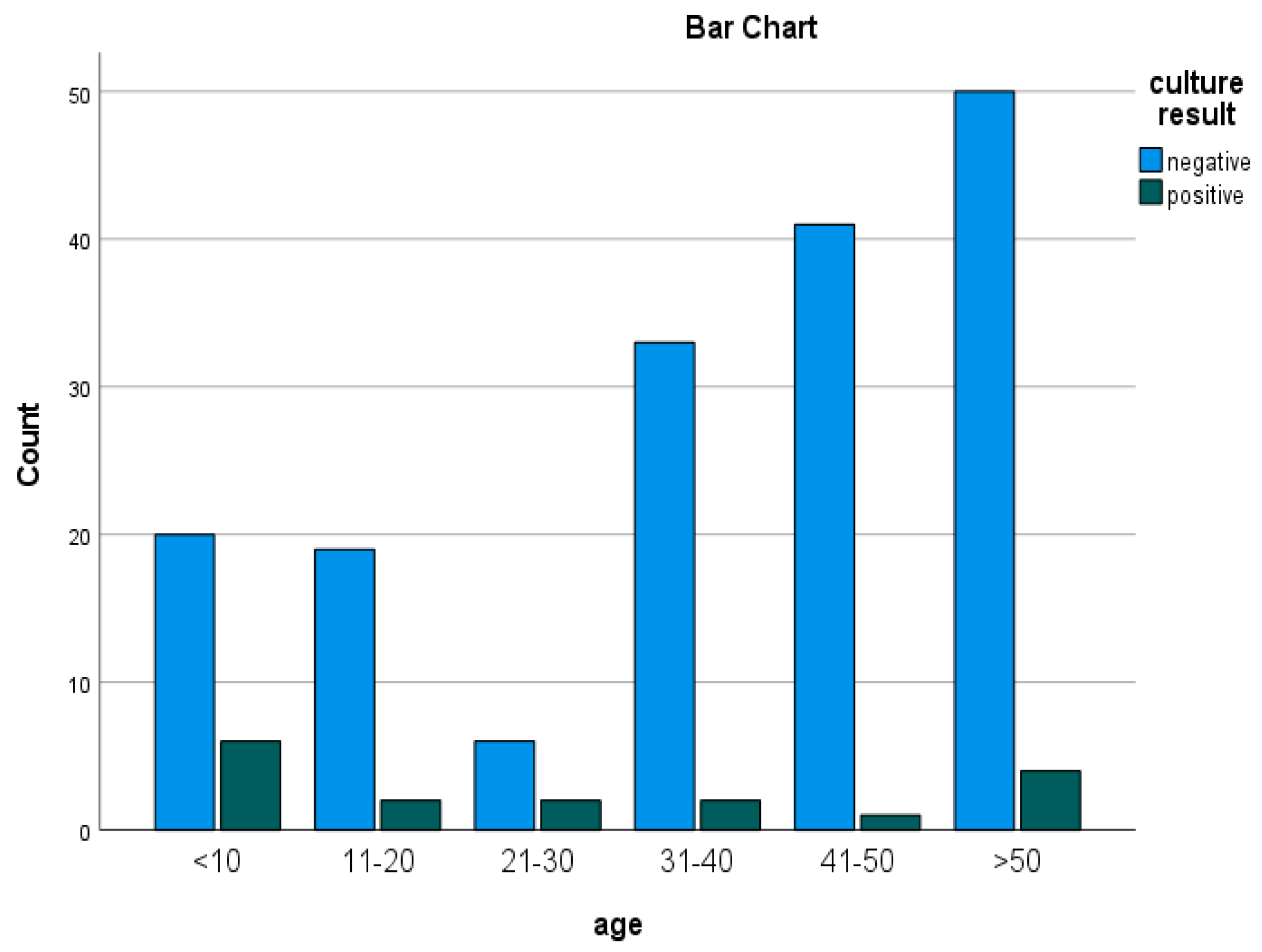
In this study, the incidence of bacterial SBF infection was higher in females when compared to males. From the isolated bacteria, 11/17 (64.7%) were from female study participants.
Similarly, the highest prevalence was recorded from under ten (<10) years study participants with burden of 6/17 (35.5%). (
Figure 4).
A drug sensitivity test was performed for all (17) bacterial isolates. These isolates had shown various resistance and susceptibility patterns against different antibiotics that were tested on MHA. The AST pattern of gram-negative bacteria was observed that antibiotics drugs of Meropenem and Amikacin had showed the most effectiveness against gram-negative isolates both with 83.3% effective action on the isolates consequently also, Amoxa-clavlanic acid at 75% effect over the isolated bacteria. However, highest resistance rate was observed on third-generation cephalosporin, Ceftazidime and Ceftraxone with 58.3% weak effect on gram-negative isolates also; Trimethoprim-Sulfamethoxazole had expelled 41.7% resistance to isolated gram negatives. Most effective antibiotics against K. pneumoniae isolates (n=5) on AST were Amikacin, Meropenem and Amoxa-clavlanic acid which showed 80% effectiveness. In contrast, the highest resistance was observed for Ceftraxone and Ceftazidime each with 60% résistance rate. All the E. coli isolates (n=4) were susceptible to Amikacin, Meropium and Amoxa-clavulanic acid 75% for each isolates. But they were 50% resistant to Gentamycin, Trimethoprim-Sulfamethoxazole and third-generation cephalosporins such as Ceftazidime and Ceftriaxone.
The overall resistance rate of Gram positive bacteria had showed that, Trimptoprin-sulfamethoxazole 60% (3/5), ceftriaxone 60% (3/5) and ciprofloxacilin 40% (2/5). However, the gram positive isolates showed no resistance for Cefoxtin and Vancomycin that isolates were 100% sensitive and Erythromycin showed 80% effective action on isolates (
Table 2).
According to the present study, 12 (70.5%) bacteria isolated were MDR. From these, 66.7% (8/12) MDR isolates were gram-negative isolates and 80% (4/5) of isolates of Gram-positive isolates. From the frequently isolated bacteria, 75% (3/4) of
E. coli and 2/5 (40%) of
K. Pneumoniae isolates were found as MDR (
Table 3).
Discussion
Biological fluids collected from sterile bodily areas are anticipated to be free from both pathogenic and commensal bacteria. Any pathological agents or environmental skin pollutants could be the source of infection in these SBFs. Although SBFs, such as pleural fluid, ascitic fluid, and CSF, are typically sterile, they may become contaminated by various microorganisms and develop potentially fatal diseases. Microbial invasion of normally sterile parts of the body can cause systemic illness. Meningitis, pericarditis, pleural infection either complicated parapneumonic effusion or empyema and septic arthritis cover major forms of SBFs infections [
13,
14].
In the present study, the overall prevalence of bacterial SBF infection was 9.1% (17/186). This prevalence finding in some extent is close with most previous studies, while offset with few studies.
Accordingly, the prevalence of bacterial SBF infection in this study found similar to the study conducted in Turkey in 2021, which was 9.7%, and Nepal in 2019, 10.7% [
14,
15]. In advance, studies from Addis Ababa, Ethiopia, at EPHI and TASH, reported a prevalence of bacterial SBF infection as 11.5% and 14.1%, respectively [
2,
4]. These results tell that the prevalence of bacteria causing SBF infection in studies done in Addis Ababa in some extent is similar. Although, for the least contradictions observed, study population socio-economic background, study methods and sample sizes are noted as contributing factors.
In contrast, the prevalence of this study is found higher than study from Debremarkos, North-western Ethiopia, (7.5%), and lower to the study from Mekele, Ethiopia, (20.2%) [
16,
17]. For the variations of the prevalence’s among studies can be attributed to differences in sample processing/laboratory technique, environmental status of the study area and the practice of infection control. Accordingly, the appropriate management of patient, early detection and identification of organism with the results of AST is crucial. Positive cultures are usually low because of less number of pathogens and prior administration of empirical antibiotics in patients [
14,
18,
19,
20].
In the present study, the most frequently isolated bacteria are gram-negative bacteria (70.6%) that`s identical with the study conducted at HFSH, Harar, Ethiopia. Similarly, study from India, gram negative isolates were 71% likely, from EPHI, Addis Ababa, Ethiopia, 74.6% of total isolates were gram negative [
4,
21,
22].
In contrast, clashing with this study, studies conducted in Mekelle, Ethiopia and India, gram positive bacteria were the dominant isolates as 52.3% and 85.2% of total isolates respectively [
16,
23]. This obverse finding on the bacterial characteristics in these studies, can possibly be due to differences on sample sizes, study areas, hospital-acquired infections, and different epidemiological factors also standard infection control precautions applied on the sites [
24].
In general, from isolated bacteria in this study,
K. pneumoniae (5/17, 29.4%)
, E.coli (4/17, 23.5%), and
S. aureus (2/17, 11.8%) were the most frequently isolates causing SBF infection. This data record is similar with earlier studies done in Ethiopia (HFSH and TASH), Turkey, India, and Kenya. This uniform data pattern observed could be explained by their distinctive structure of gram-negative bacteria, are their increasingly resistant presentation to most available antibiotics or opportunistic nature of organisms [
9,
14,
22,
25].
In the present study, antibiotics, Meropenem (83.3%) and Amikacin (83.3%) found with the most effective action against gram-negative isolates. Whereas, cefraxone and cefepime found as the least effective antibiotics against gram -negatives in which 58.3% of each antibiotics were resisted by isolates. This result is complementary with the studies carried out from HFSH, Harar, tertiary care hospital, India, Accra, Gana, and with Debremarkos, North west Ethiopia [
9,
17,
22,
26].
In contrast to the Gram positives bacteria isolates of this study, 100% of the isolates were noted as sensitive to Cefoxin and vancomycin, and 80% of the isolates were found sensitive to Erytromycin. Contrarily, 60% of gram positives executed resistance to ceftriaxone and Sulfamethoxazole-trimethoprim. In advance,
S. pneumoniae and
Entrococcus were found resistant for most classes of antibiotics such as Sulfamethoxazole-trimethoprim, ciprofloxacilin and ceftriaxone. This record of data is homogenous with study from Turkey that showed both isolates,
S. pneumoniae and
Entrococcus were the most frequent isolate with the most problematic resistance patterns [
14].
From this study, the increased rate of phenomenon of beta-lactam including third generation cephalosporins drug resistance may be linked to various factors, including but not limited to the frequent use of antibiotics, their easy availability, and the practice of self-medication, the scarcity of diagnostic facilities, the inappropriate use of antibiotics and the tradition of manipulation of prescriptions without susceptibility data. Also a survey in Sub-Saharan Africa and Asia reveal that affordable first line agents such as ampicillin and gentamicin are unlikely to be clinically efficacious in a substantial proportion of infections. This results in increasing reliance on the third generation cephalosporins for empirical treatment of serious infections. However, the spread of extended-spectrum beta-lactamase producing strains into the community, probably accelerated by this increased consumption, is eroding the usefulness of these drugs. Alternative agents for treating multi-resistant coliform infections, such as the carbapenems, are unaffordable for treatment of community-acquired infections in low-income countries. In addition, these organisms have a range of mechanisms to prevent the action of many antimicrobials used in clinical medicine such as efflux pumps, alteration of the drug binding site and membrane permeability, degradation enzymes [
27,
28].
Multi drug resistance (MDR) pattern at 70.6% of isolates was recorded in this study. This result in fewer extents is similar with the studies from HFSH, TASH in Addis Ababa, and Debremarkos, north-west Ethiopia with 76.4, 75% and 78.6% respectively. However, this finding had higher MDR rate with the study from Mekele, Northern Ethiopia, which is 40.1% also, higher than study in Nepal that had 30% MDR rate. There are many possible explanations for this difference and increased prevalence among the studies conducted socio-demography and clinical determinants status including: Self-medication, poor adherence to complete antibiotic regimens and low quality, often counterfeit drugs are all common. The disease burden from bacterial pathogens is greatest different bacterial strains, geographic variation, patients’ awareness towards the use of the antimicrobials, the difference in infection control practice, the difference in antibiotic prescribing policies, easy availability of some drugs without a prescription, and indiscriminate/prolonged use of common antibiotics lead to rapid and extensive spread of antimicrobial resistance Also, the environmental persistence of resistance genes or drug residues. Antibiotic resistance spreads through a variety of environmental reservoirs, such as soil, water, hospitals, industries, farm waste, and other contaminated ecological niches [
1,
2,
17,
22,
29].
The etiology of bacteria for infections of SBF being at minimal dose, detection rate is reduced as of the growing inquiry on culture media also, the avoidance of anaerobic bacteria enrichment, which were excluded from this investigation due to the seldom availability of national laboratory setup, the overall culture positivity rate of SBF samples was relatively low. Since the diagnosis of anaerobic bacteria is expensive and requires special facilities and expertise to perform this study doesn`t include anaerobic bacteria. The risk of false-negative results in agar-based culture media is high because only a small number of microorganisms may be present in the specimens.
The present study, unlike most previous studies, it is conducted at the two of the largest highest hospitals, Minilik II Comprehensive Specialized Hospital and Yekatit 12 Hospital Medical College which brought a better diverse participant population that built the study finding represents a wide and diverse population so that it assure to develop a better generalization for further hypothetical study.
Conclusions
A variety of gram-positive, as well as gram-negative organisms, were isolated from collected SBF samples analyzed in the study, which yielded 17 of 186 (9.1%). Dominantly, K.pneumoniae and E.coli were the leading causative agents for SBF infection. Amikacin and meropenem were found to be the most effective drugs, both executing 83.3% effective action against gram-negative isolates. However, the gram negatives were 58.3% resistance to third-generation cephalosporin. Also the gram-positive isolates showed 60% resistant to trimethoprim-sulfamethoxazole and ceftriaxone. While, cefoxitin and erytromycin, were observed with effective action against gram- positive isolates 100% and 80%, respectively. The majority, (70.5%) of the isolates have shown multidrug resistance (MDR) pattern, especially to commonly in use and easily available drugs.
The following recommendations are made based on the findings of the present study.
For Yekatit 12 hospital medical college and Minilik II comprehensive hospital Hospital:
The hospital should acknowledge that Culture and susceptibility testing has to be an integral part of routine laboratory tests for the best management of infections in patients, and the choice of drugs should be based on results of sensitivity testing.
For Addis Ababa health bureau:
This data should be used as a baseline for development of better formulation of practical antibiotic policy and a regular surveillance of hospital-associated infections and monitoring of antibiotic susceptibility pattern based on the current study are suggested. In addition, advancement and technical support for laboratories found under AAHB on the bacteria growing and identification technique like molecular and automated machines is preferable for a better detection capability.
For health care workers:
The antibiotic sensitivity profile suggests that ciprofloxacin and amikacin are a choice of drugs for sterile body infections. However, Beta-lactam antibiotics such as third-generation cephalosporins, ampicillin, and penicillin should be used carefully unless tested.
The careful prescribing practice or rational use of antibiotics based on sensitivity testing result is needed to halt the trend of increasing antibiotic resistance. Besides, great care should be given to inpatients, as they are more vulnerable to bacterial infections in sterile body areas.
For future researchers:
Since this study was not conducted alongside the nation with well enough data. The present study was conducted on a limited number of samples; this may not reflect the entire scenarios of the study. This highlights the need for further studies to be conducted on larger sample sizes. Further, antimicrobial resistance is also gradually increasing, which needs much attention. Thus, continuous monitoring of susceptibility profile of the clinically important pathogens is of great importance to guide effective antimicrobial therapy.