Submitted:
27 March 2025
Posted:
31 March 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Material and Methods
Plant Material
Phenotyping Data
SNP Selection and Genotyping
Genome-Wide Association (GWAS)
Results and Discussion
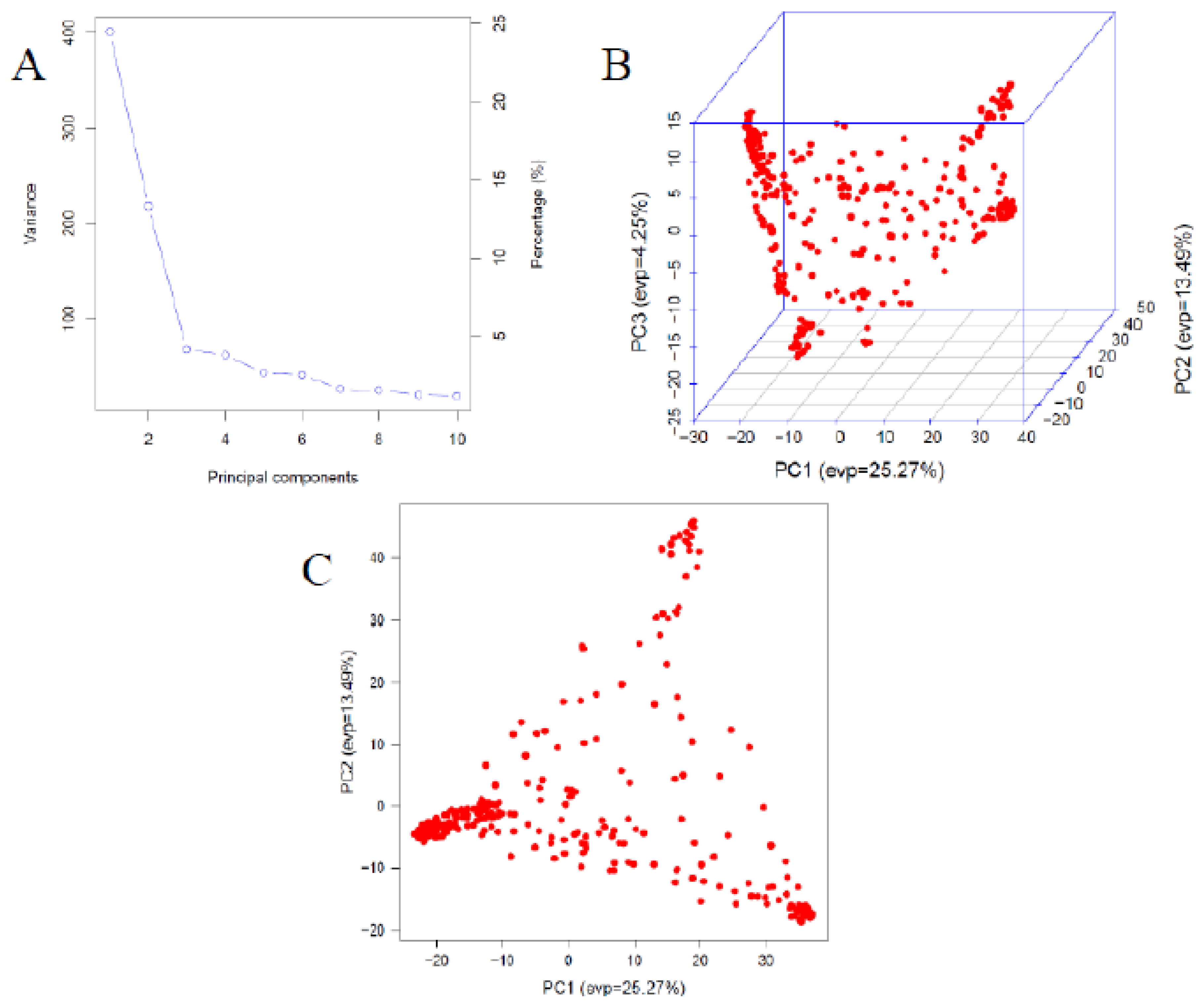
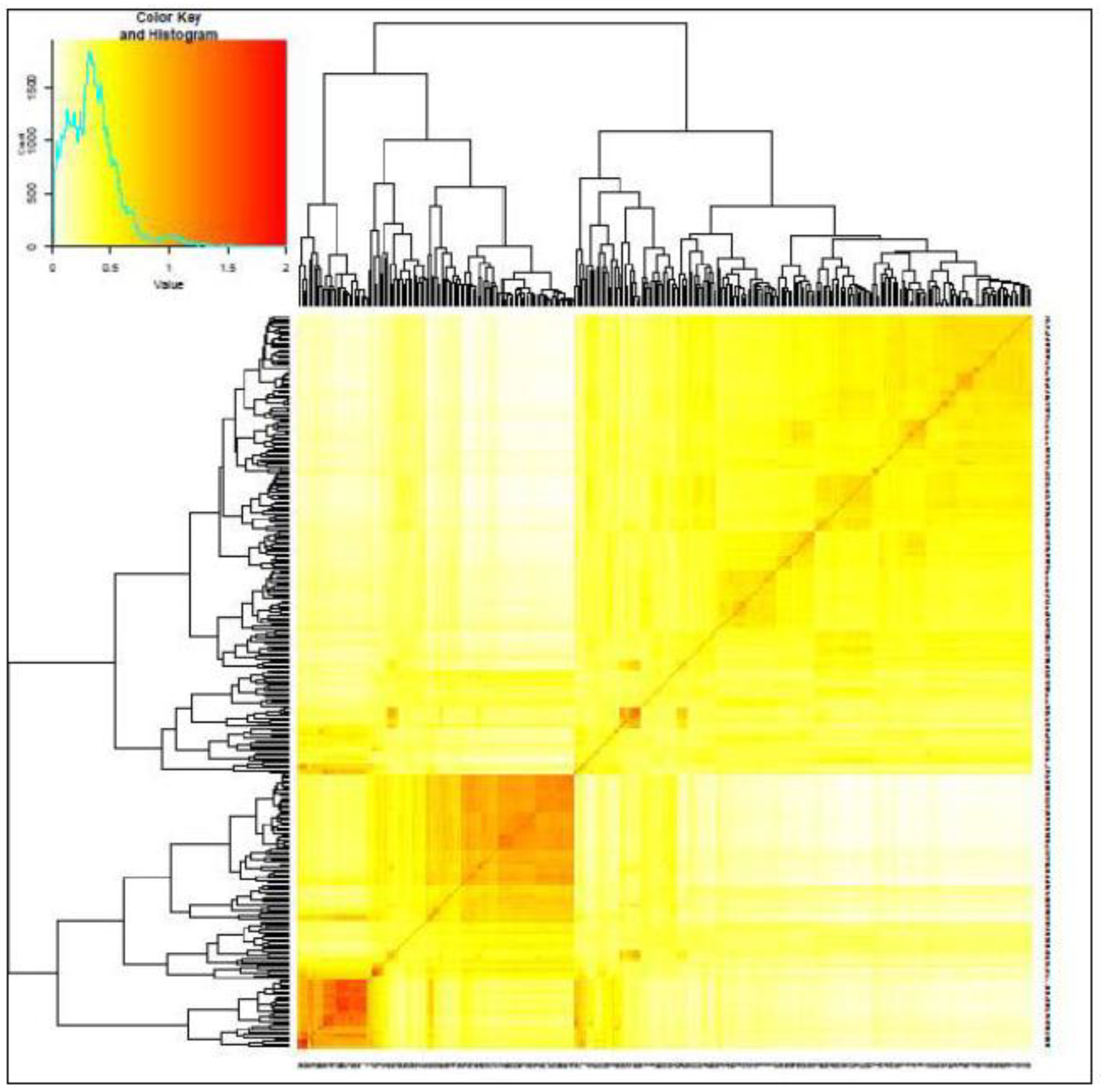
Principal Components (PCA)
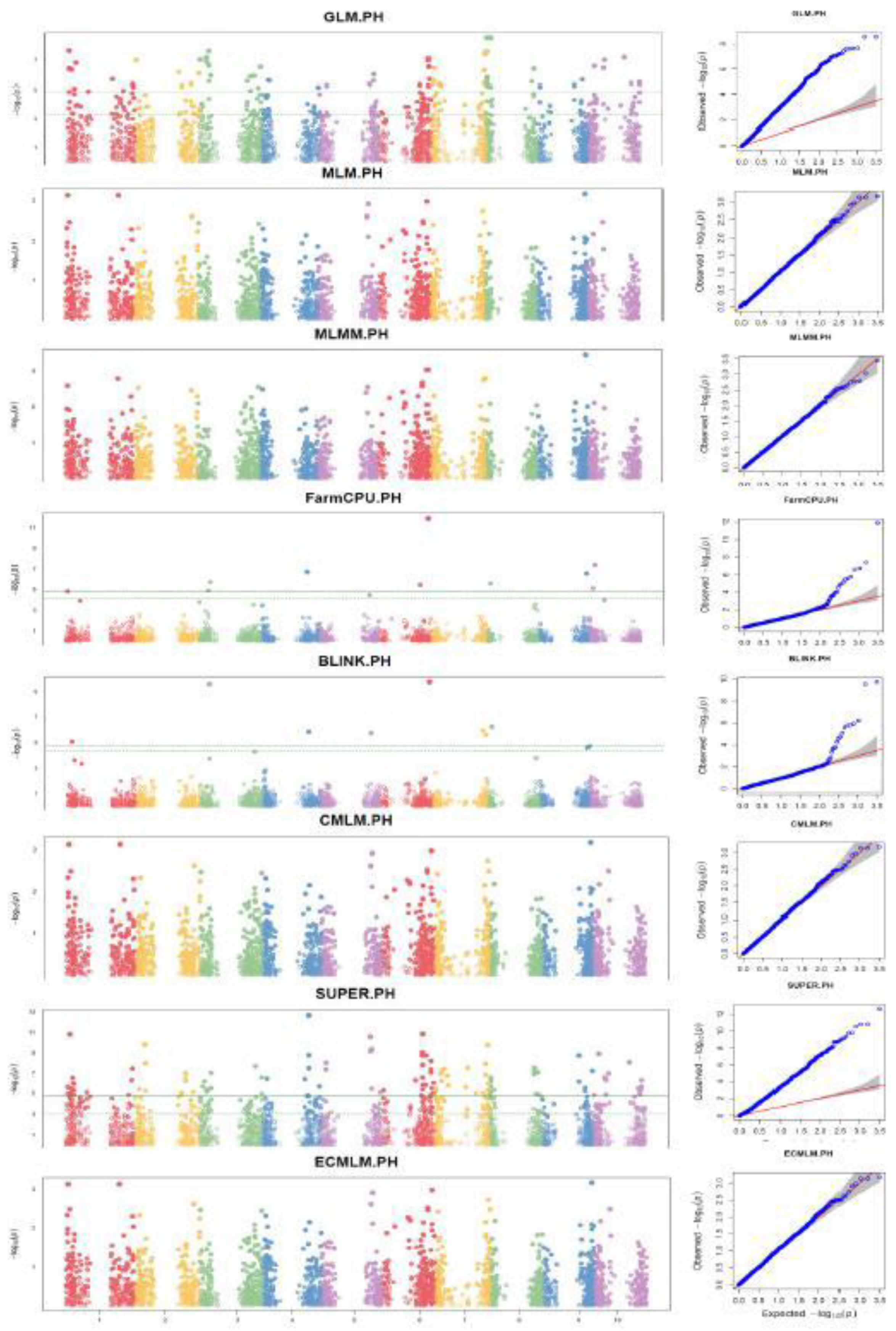
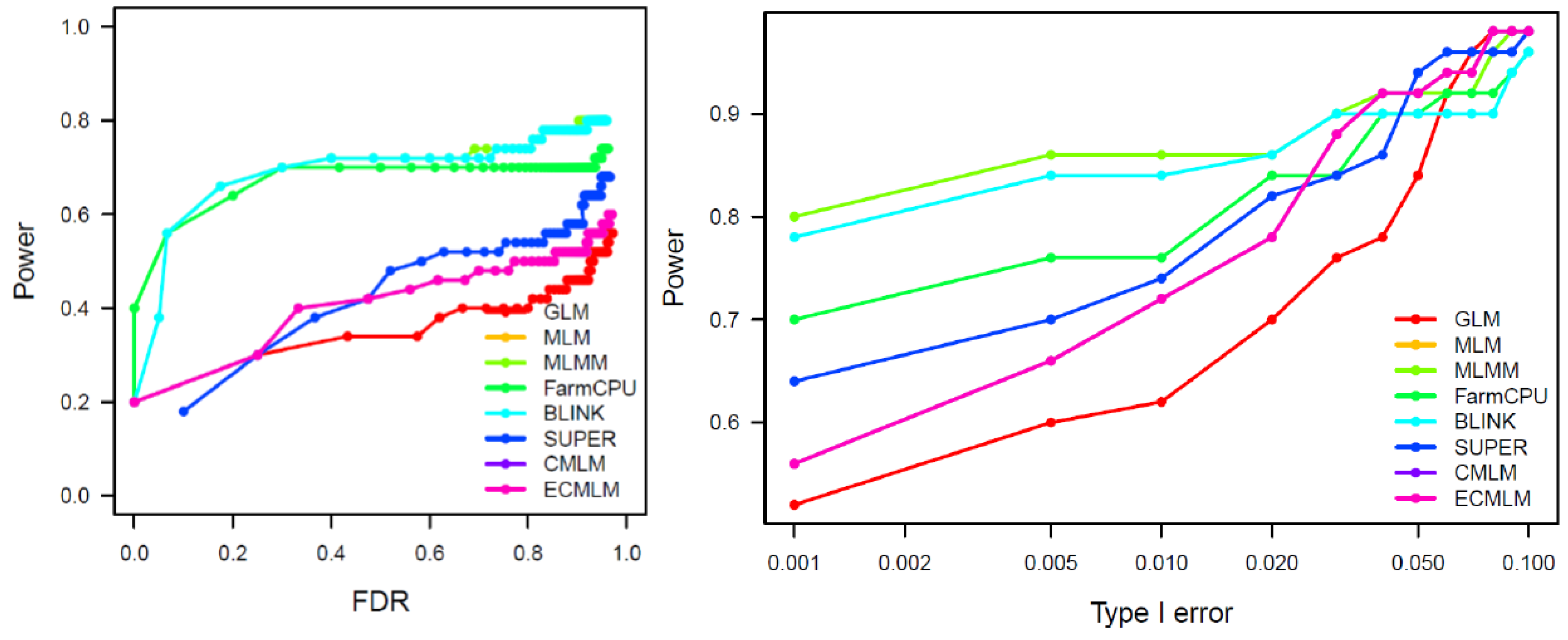
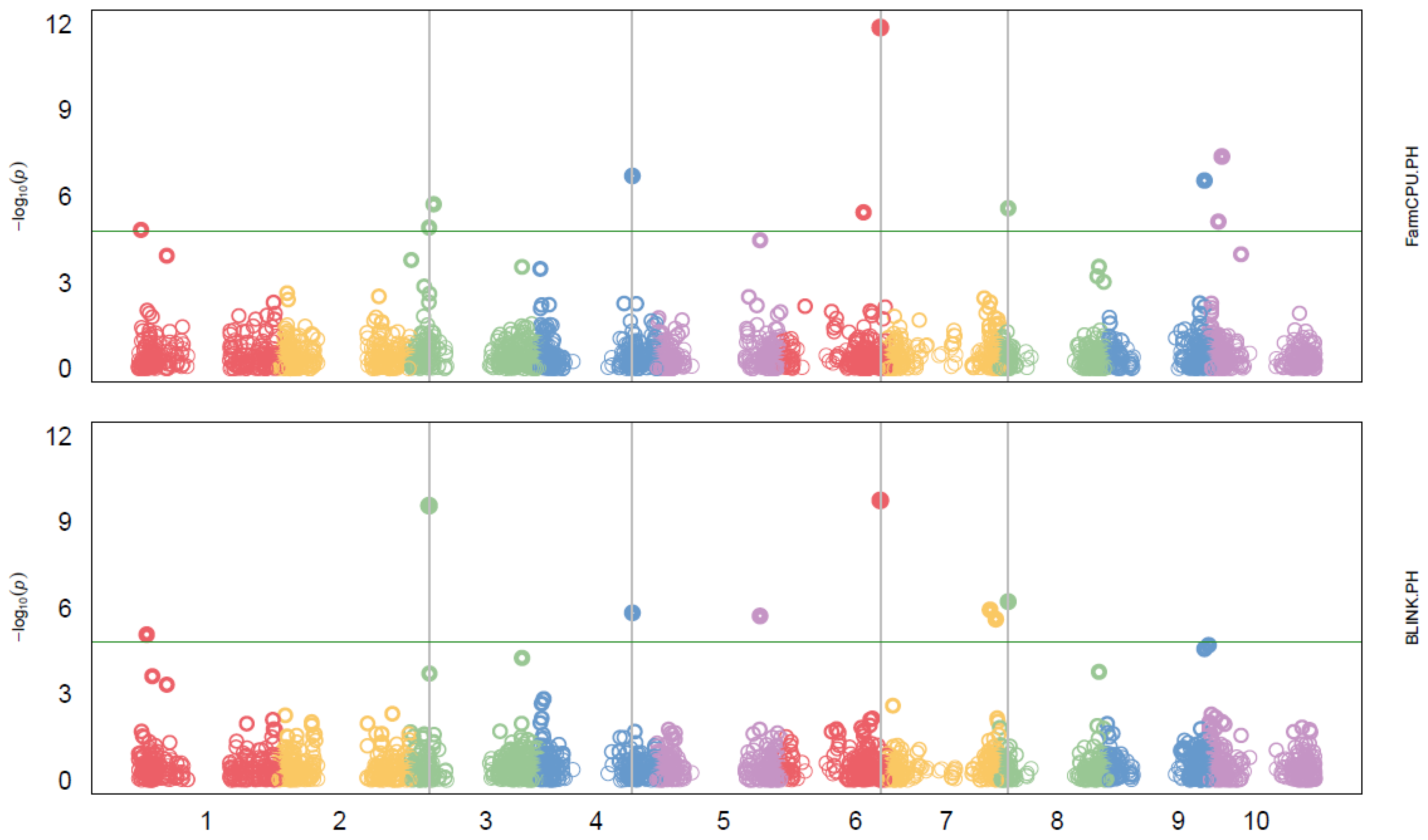
Markers-Trait Associated with Plant Height (PH)
Conclusions
References
- Enyew, M.; Feyissa, T.; Carlsson, A.S.; Tesfaye, K.; Hammenhag, C.; Seyoum, A.; Geleta, M. Genome-wide analyses using multi-locus models revealed marker-trait associations for major agronomic traits in Sorghum bicolor. Frontiers in Plant Science 2022, 13, 999692. [Google Scholar] [CrossRef] [PubMed]
- Enyew, M.; Geleta, M.; Tesfaye, K.; Seyoum, A.; Feyissa, T.; Alemu, A.; Hammenhag, C.; Carlsson, A.S. Genome-wide association study and genomic prediction of root system architecture traits in Sorghum (Sorghum bicolor (L.) Moench) at the seedling stage. BMC plant biology 2025, 25, 69. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOSTAT statistical database. Rome, Italy. Accessed on August 5. 2023. Available online: https://www.fao.org/faostat/en/#data/FS.
- FAO, IFAD, UNICEF, WFP, & WHO. (2024). The State of Food Security and Nutrition in the World 2024 – Financing to end hunger, food insecurity and malnutrition in all its forms. [CrossRef]
- Hossain, M.S.; Islam, M.N.; Rahman, M.M.; Mostofa, M.G.; Khan, M.A.R. Sorghum: A prospective crop for climatic vulnerability, food and nutritional security. Journal of Agriculture and Food Research 2022, 8, 100300. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Ulhassan, Z.; Brestic, M.; Zivcak, M.; Weijun, Z.; Allakhverdiev, S.I.; Yang, X.; Safdar, M.E.; Yang, W.; Liu, W. Photosynthesis research under climate change. Photosynthesis Research 2021, 150, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.; Fridman, E.; Tesso, T.T.; Yu, J. Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proceedings of the National Academy of Sciences – PNAS 2015, 112, 11823–11828. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS genetics 2016, 12(2), e1005767. [Google Scholar] [CrossRef] [PubMed]
- Murchie, E.H.; Pinto, M.; Horton, P. Agriculture and the new challenges for photosynthesis research. The New phytologist 2009, 181, 532–552. [Google Scholar] [CrossRef] [PubMed]
- Nida, H.; Girma, G.; Mekonen, M.; Tirfessa, A.; Seyoum, A.; Bejiga, T.; Birhanu, C.; Dessalegn, K.; Senbetay, T.; Ayana, G.; Tesso, T.; Ejeta, G.; Mengiste, T. Genome-wide association analysis reveals seed protein loci as determinants of variations in grain mold resistance in sorghum. Theoretical and Applied Genetics 2021, 134(4), 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Sales, C.R.G.; Wang, Y.; Evers, J.B.; Kromdijk, J. Improving C4 photosynthesis to increase productivity under optimal and suboptimal conditions. Journal of Experimental Botany 2021, 72(17), 5942–5960. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genomics, proteomics & bioinformatics 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Zou, G.; Ding, Y.; Xu, J.; Feng, Z.; Cao, N.; Chen, H.; Liu, H.; Zheng, X.; Liu, X.; Zhang, L. Genome-wide dissection of genes shaping inflorescence morphology in 242 Chinese south–north sorghum accessions. Scientific Reports 2024, 14(1), 25828. [Google Scholar] [CrossRef]
| Trait | GWAS model | SNP ID | Chromosome number | Position | MAF1 | P-value | PVE (%)2 |
|---|---|---|---|---|---|---|---|
| PH3 | GLM | snp_sb001000003033 | 1 | 1803311 | 0.11650485 | 2.10E-08 | 0 |
| snp_sb001000003043 | 1 | 1803552 | 0.06796117 | 3.59E-07 | 1.17 | ||
| snp_sb042060173374 | 1 | 2053226 | 0.20711974 | 1.30E-05 | 3.06 | ||
| snp_sb042060176498 | 1 | 4583054 | 0.12297735 | 4.58E-07 | 3.30 | ||
| snp_sb042060176733 | 1 | 4778713 | 0.40453074 | 4.12E-07 | 0 | ||
| snp_sb042060183185 | 1 | 8066218 | 0.26213592 | 7.89E-06 | 4.04 | ||
| snp_sb040057635750 | 1 | 9985873 | 0.0987055 | 1.46E-07 | 7.01 | ||
| snp_sb001000040602 | 1 | 24769165 | 0.38673139 | 1.23E-05 | 0 | ||
| snp_sb042060233820 | 1 | 53456591 | 0.44012945 | 2.01E-06 | 0 | ||
| snp_sb042060260112 | 1 | 66258089 | 0.25080906 | 1.09E-05 | 0.75 | ||
| snp_sb042060273650 | 1 | 77744466 | 0.06148867 | 4.51E-06 | 0 | ||
| snp_sb042060274847 | 1 | 78464288 | 0.32524272 | 6.09E-06 | 0 | ||
| snp_sb042060279054 | 2 | 1028568 | 0.18770227 | 9.40E-08 | 0 | ||
| snp_sb042060334327 | 2 | 51764695 | 0.15048544 | 6.59E-07 | 3.89 | ||
| snp_sb042060339135 | 2 | 55665363 | 0.34951456 | 5.71E-06 | 0.51 | ||
| snp_sb042060355039 | 2 | 66604865 | 0.12459547 | 3.85E-06 | 0 | ||
| snp_sb042060363157 | 2 | 75994759 | 0.12621359 | 7.87E-07 | 0 | ||
| snp_sb042060365193 | 3 | 542076 | 0.26213592 | 8.95E-08 | 1.09 | ||
| snp_sb042060375446 | 3 | 8002244 | 0.1828479 | 6.13E-08 | 3.06 | ||
| snp_sb042060375491 | 3 | 8012102 | 0.22168285 | 1.16E-07 | 0.42 | ||
| snp_sb042060379604 | 3 | 10734118 | 0.06148867 | 2.10E-07 | 0 | ||
| snp_sb042060379940 | 3 | 10836535 | 0.0776699 | 2.23E-08 | 0 | ||
| snp_sb042060380150 | 3 | 10921954 | 0.08090615 | 2.30E-07 | 0 | ||
| snp_sb042060380199 | 3 | 10932720 | 0.07281553 | 2.71E-06 | 5.54 | ||
| snp_sb042060384040 | 3 | 12866372 | 0.24433657 | 1.10E-06 | 0.21 | ||
| snp_sb042060421455 | 3 | 56322246 | 0.28964401 | 2.65E-06 | 0 | ||
| snp_sb042060428418 | 3 | 60472707 | 0.0566343 | 1.07E-05 | 1.77 | ||
| snp_sb042060428585 | 3 | 60501386 | 0.05339806 | 1.29E-06 | 0 | ||
| snp_sb042060429232 | 3 | 60811994 | 0.0566343 | 1.24E-06 | 0 | ||
| snp_sb042060434280 | 3 | 64830003 | 0.06148867 | 1.19E-06 | 0 | ||
| snp_sb042060439477 | 3 | 69764576 | 0.2038835 | 6.96E-06 | 0 | ||
| snp_sb042060445645 | 3 | 72284237 | 0.05825243 | 2.79E-07 | 0 | ||
| snp_sb042060463777 | 4 | 7424990 | 0.06148867 | 2.38E-06 | 0 | ||
| snp_sb042060526464 | 4 | 8890484 | 0.08252427 | 2.51E-06 | 1.30 | ||
| snp_sb042060523231 | 4 | 66858877 | 0.25889968 | 6.67E-06 | 0.11 | ||
| snp_sb042060545830 | 5 | 8563133 | 0.05177994 | 5.31E-06 | 0 | ||
| snp_sb042060589862 | 5 | 59844243 | 0.41585761 | 4.13E-06 | 4.45 | ||
| snp_sb042060600784 | 5 | 64447085 | 0.24919094 | 2.52E-06 | 0 | ||
| snp_sb042060601544 | 5 | 64669481 | 0.06634304 | 9.37E-07 | 0 | ||
| snp_sb001000702141 | 6 | 47599757 | 0.35598706 | 4.80E-06 | 0 | ||
| snp_sb001000703068 | 6 | 47897247 | 0.42394822 | 1.42E-05 | 0.68 | ||
| snp_sb001000719123 | 6 | 57647043 | 0.39320388 | 7.47E-08 | 0 | ||
| snp_sb001000719127 | 6 | 57647287 | 0.3802589 | 1.01E-07 | 0.43 | ||
| snp_sb001000719254 | 6 | 57704110 | 0.31067961 | 5.42E-06 | 4.23 | ||
| snp_sb001000720111 | 6 | 58537465 | 0.20226537 | 2.75E-07 | 0 | ||
| snp_sb001000721097 | 6 | 59276261 | 0.1407767 | 2.04E-06 | 0 | ||
| snp_sb042060760081 | 7 | 2622240 | 0.10032362 | 2.83E-06 | 0 | ||
| snp_sb042060760098 | 7 | 2623348 | 0.10355987 | 2.29E-06 | 0 | ||
| snp_sb042060760099 | 7 | 2623635 | 0.0987055 | 3.48E-06 | 0 | ||
| snp_sb042060761490 | 7 | 3776173 | 0.07119741 | 3.33E-07 | 7.40 | ||
| snp_sb042060766358 | 7 | 6455441 | 0.29449838 | 1.57E-05 | 0 | ||
| snp_sb042060772026 | 7 | 9428572 | 0.16504854 | 4.27E-06 | 0 | ||
| snp_sb042060774985 | 7 | 12018772 | 0.43042071 | 5.26E-06 | 0 | ||
| snp_sb042060850373 | 7 | 63227425 | 0.45631068 | 3.42E-06 | 0.50 | ||
| snp_sb042060850458 | 7 | 63300346 | 0.12783172 | 4.78E-07 | 5.09 | ||
| snp_sb042060852727 | 7 | 64318593 | 0.22330097 | 3.69E-08 | 0 | ||
| snp_sb042060852735 | 7 | 64319205 | 0.22491909 | 2.58E-08 | 0 | ||
| snp_sb042060856436 | 8 | 606304 | 0.197411 | 2.44E-08 | 0.13 | ||
| snp_sb042060857050 | 8 | 1016470 | 0.27346278 | 2.70E-09 | 0.27 | ||
| snp_sb042060857649 | 8 | 1241472 | 0.09223301 | 3.50E-06 | 0 | ||
| snp_sb042060857701 | 8 | 1243863 | 0.08090615 | 1.82E-06 | 3.31 | ||
| snp_sb042060858244 | 8 | 1459101 | 0.36893204 | 1.40E-06 | 0.16 | ||
| snp_sb042060858246 | 8 | 1462792 | 0.36731392 | 5.57E-06 | 0 | ||
| snp_sb042060867162 | 8 | 5053406 | 0.08576052 | 6.16E-06 | 1.54 | ||
| snp_sb042060867731 | 8 | 5247147 | 0.23300971 | 2.68E-09 | 0 | ||
| snp_sb042060939820 | 8 | 54615437 | 0.19417476 | 2.82E-06 | 2.26 | ||
| snp_sb042060945149 | 8 | 56847745 | 0.09385113 | 3.71E-07 | 0 | ||
| snp_sb042060958548 | 9 | 2005306 | 0.15372168 | 5.86E-06 | 0 | ||
| snp_sb042061005134 | 9 | 42849238 | 0.08252427 | 5.11E-06 | 0 | ||
| snp_sb042061021148 | 9 | 52373447 | 0.2605178 | 2.16E-06 | 0 | ||
| snp_sb001001049696 | 10 | 7303004 | 0.05987055 | 2.49E-07 | 8.78 | ||
| snp_sb001001075487 | 10 | 18388160 | 0.23300971 | 3.25E-07 | 0 | ||
| snp_sb042061062036 | 10 | 19451315 | 0.19417476 | 9.11E-08 | 3.57 | ||
| snp_sb001001125741 | 10 | 42872791 | 0.40291262 | 5.98E-08 | 0 | ||
| snp_sb001001143490 | 10 | 51455048 | 0.2394822 | 3.66E-06 | 1.23 | ||
| snp_sb001001159813 | 10 | 60977250 | 0.4012945 | 1.39E-05 | 0.12 | ||
| snp_sb001001159934 | 10 | 61069500 | 0.13915858 | 2.99E-06 | 0 | ||
| snp_sb001001160062 | 10 | 61144250 | 0.20064725 | 3.98E-06 | 0 | ||
| SUPER | snp_sb042060173350 | 1 | 2036942 | 0.15534 | 1.10E-05 | - | |
| snp_sb042060173374 | 1 | 2053226 | 0.20712 | 2.30E-06 | - | ||
| snp_sb042060176733 | 1 | 4778713 | 0.404531 | 1.60E-11 | - | ||
| snp_sb042060177238 | 1 | 5073279 | 0.493528 | 1.14E-05 | - | ||
| snp_sb042060177747 | 1 | 5705669 | 0.461165 | 9.13E-06 | - | ||
| snp_sb042060177833 | 1 | 5736900 | 0.307443 | 3.18E-06 | - | ||
| snp_sb042060183033 | 1 | 8034619 | 0.210356 | 3.49E-06 | - | ||
| snp_sb042060183185 | 1 | 8066218 | 0.262136 | 2.99E-07 | - | ||
| snp_sb001000014533 | 1 | 8870319 | 0.423948 | 9.51E-07 | - | ||
| snp_sb042060184103 | 1 | 9022825 | 0.315534 | 5.03E-06 | - | ||
| snp_sb042060184571 | 1 | 9291528 | 0.457929 | 1.08E-06 | - | ||
| snp_sb001000015314 | 1 | 9412963 | 0.449838 | 1.40E-05 | - | ||
| snp_sb001000040602 | 1 | 24769165 | 0.386731 | 7.69E-06 | - | ||
| snp_sb042060267164 | 1 | 73737253 | 0.349515 | 1.36E-05 | - | ||
| snp_sb042060273269 | 1 | 77432019 | 0.199029 | 4.72E-07 | - | ||
| snp_sb042060274847 | 1 | 78464288 | 0.325243 | 3.62E-08 | - | ||
| snp_sb042060287884 | 2 | 5838445 | 0.459547 | 3.69E-06 | - | ||
| snp_sb042060302008 | 2 | 12243572 | 0.331715 | 1.62E-10 | - | ||
| snp_sb042060304434 | 2 | 13177574 | 0.18123 | 5.33E-06 | - | ||
| snp_sb042060304638 | 2 | 13293723 | 0.427184 | 1.07E-08 | - | ||
| snp_sb042060338476 | 2 | 55292382 | 0.31877 | 1.32E-05 | - | ||
| snp_sb042060339135 | 2 | 55665363 | 0.349515 | 8.25E-06 | - | ||
| snp_sb042060347981 | 2 | 59810106 | 0.480583 | 3.55E-07 | - | ||
| snp_sb042060348862 | 2 | 60374738 | 0.451456 | 6.24E-07 | - | ||
| snp_sb042060349202 | 2 | 60415546 | 0.470874 | 9.69E-08 | - | ||
| snp_sb042060349216 | 2 | 60416638 | 0.438511 | 1.16E-07 | - | ||
| snp_sb042060358990 | 2 | 69586319 | 0.072816 | 6.97E-06 | - | ||
| snp_sb042060368691 | 3 | 3720735 | 0.364078 | 2.09E-06 | - | ||
| snp_sb042060370968 | 3 | 5535752 | 0.375405 | 6.31E-07 | - | ||
| snp_sb042060375446 | 3 | 8002244 | 0.182848 | 3.91E-06 | - | ||
| snp_sb042060375491 | 3 | 8012102 | 0.221683 | 6.55E-06 | - | ||
| snp_sb042060378852 | 3 | 10370193 | 0.122977 | 3.87E-06 | - | ||
| snp_sb042060384040 | 3 | 12866372 | 0.244337 | 3.43E-06 | - | ||
| snp_sb042060385209 | 3 | 13494213 | 0.380259 | 2.88E-06 | - | ||
| snp_sb042060385211 | 3 | 13494361 | 0.496764 | 9.35E-08 | - | ||
| snp_sb042060434769 | 3 | 65706545 | 0.472492 | 2.15E-08 | - | ||
| snp_sb042060446486 | 3 | 72742644 | 0.343042 | 2.67E-07 | - | ||
| snp_sb042060447019 | 3 | 73010484 | 0.187702 | 1.08E-05 | - | ||
| snp_sb042060523179 | 4 | 4755657 | 0.158576 | 3.40E-07 | - | ||
| snp_sb042060527001 | 4 | 49335014 | 0.302589 | 3.70E-07 | - | ||
| snp_sb042060525591 | 4 | 52459901 | 0.478964 | 1.44E-05 | - | ||
| snp_sb042060524523 | 4 | 54044421 | 0.446602 | 2.22E-13 | - | ||
| snp_sb001000469722 | 4 | 54108806 | 0.18932 | 3.37E-08 | - | ||
| snp_sb042060524643 | 4 | 54165068 | 0.383495 | 1.76E-09 | - | ||
| snp_sb042060526825 | 4 | 66343002 | 0.496764 | 2.07E-06 | - | ||
| snp_sb042060539991 | 5 | 5752947 | 0.453074 | 4.32E-08 | - | ||
| snp_sb042060541082 | 5 | 6039745 | 0.346278 | 9.84E-09 | - | ||
| snp_sb042060587348 | 5 | 57995949 | 0.443366 | 2.81E-11 | - | ||
| snp_sb042060587478 | 5 | 58078499 | 0.365696 | 6.82E-10 | - | ||
| snp_sb042060589677 | 5 | 59776476 | 0.482201 | 3.34E-06 | - | ||
| snp_sb042060589862 | 5 | 59844243 | 0.415858 | 4.54E-10 | - | ||
| snp_sb042060613793 | 5 | 69766819 | 0.299353 | 7.77E-06 | - | ||
| snp_sb001000591841 | 6 | 1893544 | 0.067961 | 4.97E-06 | - | ||
| snp_sb001000685308 | 6 | 39650537 | 0.433657 | 4.84E-07 | - | ||
| snp_sb001000699642 | 6 | 46593261 | 0.067961 | 1.52E-05 | - | ||
| snp_sb001000702141 | 6 | 47599757 | 0.355987 | 9.67E-10 | - | ||
| snp_sb001000702204 | 6 | 47614312 | 0.415858 | 1.83E-09 | - | ||
| snp_sb001000702655 | 6 | 47747134 | 0.389968 | 2.63E-07 | - | ||
| snp_sb001000703068 | 6 | 47897247 | 0.423948 | 1.61E-08 | - | ||
| snp_sb001000703373 | 6 | 48007508 | 0.126214 | 3.89E-06 | - | ||
| snp_sb001000703397 | 6 | 48019015 | 0.386731 | 1.50E-11 | - | ||
| snp_sb001000703435 | 6 | 48022337 | 0.326861 | 1.39E-06 | - | ||
| snp_sb001000710036 | 6 | 51604058 | 0.391586 | 1.49E-06 | - | ||
| snp_sb001000710785 | 6 | 51877933 | 0.404531 | 1.99E-09 | - | ||
| snp_sb001000719123 | 6 | 57647043 | 0.393204 | 4.25E-08 | - | ||
| snp_sb001000719127 | 6 | 57647287 | 0.380259 | 8.49E-08 | - | ||
| snp_sb001000719254 | 6 | 57704110 | 0.31068 | 6.61E-09 | - | ||
| snp_sb001000719606 | 6 | 57947690 | 0.393204 | 1.40E-05 | - | ||
| snp_sb001000720111 | 6 | 58537465 | 0.202265 | 2.82E-06 | - | ||
| snp_sb001000722757 | 6 | 60369078 | 0.050162 | 9.03E-06 | - | ||
| snp_sb001000723739 | 6 | 60908842 | 0.300971 | 8.88E-07 | - | ||
| snp_sb042060760081 | 7 | 2622240 | 0.100324 | 3.49E-06 | - | ||
| snp_sb042060760098 | 7 | 2623348 | 0.10356 | 5.02E-06 | - | ||
| snp_sb042060760099 | 7 | 2623635 | 0.098706 | 4.78E-06 | - | ||
| snp_sb042060770742 | 7 | 8646573 | 0.360841 | 8.44E-08 | - | ||
| snp_sb042060770748 | 7 | 8647644 | 0.415858 | 1.17E-07 | - | ||
| snp_sb042060771257 | 7 | 8852228 | 0.346278 | 5.59E-07 | - | ||
| snp_sb042060774985 | 7 | 12018772 | 0.430421 | 3.83E-08 | - | ||
| snp_sb042060777297 | 7 | 15926543 | 0.456311 | 1.70E-07 | - | ||
| snp_sb042060778451 | 7 | 17139586 | 0.381877 | 1.57E-06 | - | ||
| snp_sb042060810749 | 7 | 37587848 | 0.286408 | 2.36E-06 | - | ||
| snp_sb042060814271 | 7 | 39762751 | 0.441748 | 3.17E-08 | - | ||
| snp_sb042060829083 | 7 | 49513148 | 0.300971 | 9.34E-06 | - | ||
| snp_sb042060840008 | 7 | 57778908 | 0.425566 | 9.31E-06 | - | ||
| snp_sb042060844978 | 7 | 60737379 | 0.331715 | 1.46E-07 | - | ||
| snp_sb042060844980 | 7 | 60737490 | 0.33657 | 1.26E-08 | - | ||
| snp_sb042060850373 | 7 | 63227425 | 0.456311 | 1.77E-10 | - | ||
| snp_sb042060850888 | 7 | 63387510 | 0.482201 | 4.16E-06 | - | ||
| snp_sb042060851559 | 7 | 63687599 | 0.242718 | 9.21E-07 | - | ||
| snp_sb042060852735 | 7 | 64319205 | 0.224919 | 1.08E-05 | - | ||
| snp_sb042060856436 | 8 | 606304 | 0.197411 | 1.09E-05 | - | ||
| snp_sb042060857701 | 8 | 1243863 | 0.080906 | 6.59E-06 | - | ||
| snp_sb042060857985 | 8 | 1398622 | 0.228155 | 2.77E-07 | - | ||
| snp_sb042060858064 | 8 | 1417507 | 0.436893 | 6.98E-07 | - | ||
| snp_sb042060858244 | 8 | 1459101 | 0.368932 | 3.51E-07 | - | ||
| snp_sb042060858246 | 8 | 1462792 | 0.367314 | 2.15E-06 | - | ||
| snp_sb042060867731 | 8 | 5247147 | 0.23301 | 4.64E-06 | - | ||
| snp_sb042060935546 | 8 | 52669272 | 0.377023 | 1.27E-07 | - | ||
| snp_sb042060935548 | 8 | 52669338 | 0.36246 | 6.20E-08 | - | ||
| snp_sb042060935700 | 8 | 52708389 | 0.349515 | 3.18E-08 | - | ||
| snp_sb042060939820 | 8 | 54615437 | 0.194175 | 1.13E-05 | - | ||
| snp_sb042060944584 | 8 | 56617256 | 0.440129 | 8.10E-08 | - | ||
| snp_sb042060945149 | 8 | 56847745 | 0.093851 | 1.14E-05 | - | ||
| snp_sb042061005177 | 9 | 42889324 | 0.360841 | 1.89E-09 | - | ||
| snp_sb042061005325 | 9 | 42941172 | 0.430421 | 6.97E-07 | - | ||
| snp_sb042061016556 | 9 | 50101247 | 0.378641 | 2.71E-06 | - | ||
| snp_sb042061016557 | 9 | 50101376 | 0.372168 | 5.49E-06 | - | ||
| snp_sb042061032708 | 9 | 58880811 | 0.428803 | 6.21E-08 | - | ||
| snp_sb001001039940 | 10 | 1485692 | 0.320388 | 1.38E-05 | - | ||
| snp_sb001001039971 | 10 | 1488770 | 0.338188 | 1.46E-05 | - | ||
| snp_sb001001049162 | 10 | 7201807 | 0.359223 | 1.23E-09 | - | ||
| snp_sb001001066189 | 10 | 14137762 | 0.44822 | 2.53E-07 | - | ||
| snp_sb001001067710 | 10 | 15183661 | 0.47411 | 4.61E-07 | - | ||
| snp_sb001001068022 | 10 | 15305891 | 0.378641 | 1.23E-05 | - | ||
| snp_sb001001075487 | 10 | 18388160 | 0.23301 | 1.03E-07 | - | ||
| snp_sb001001125741 | 10 | 42872791 | 0.402913 | 8.92E-09 | - | ||
| snp_sb042061092070 | 10 | 53108112 | 0.050162 | 3.14E-06 | - | ||
| snp_sb001001150146 | 10 | 54629603 | 0.401295 | 1.02E-05 | - | ||
| snp_sb042061095323 | 10 | 55477852 | 0.338188 | 2.49E-06 | - | ||
| snp_sb001001151401 | 10 | 55478741 | 0.305825 | 1.33E-05 | - | ||
| snp_sb001001151403 | 10 | 55479226 | 0.309061 | 5.54E-06 | - | ||
| snp_sb001001153208 | 10 | 56339534 | 0.092233 | 4.37E-06 | - | ||
| snp_sb001001154375 | 10 | 56953779 | 0.490291 | 4.74E-07 | - | ||
| MLM | -4 | - | - | - | - | - | |
| MLMM | -5 | - | - | - | - | - | |
| CMLM | -6 | - | - | - | - | - | |
| ECMLM | -7 | - | - | - | - | - | |
| farmCPU | snp_sb001000002532 | 1 | 1495213 | 0.113269 | 1.45E-05 | 27.4 | |
| snp_sb042060379940 | 3 | 10836535 | 0.07767 | 1.20E-05 | 0.11 | ||
| snp_sb042060385209 | 3 | 13494213 | 0.380259 | 1.85E-06 | 8.23 | ||
| snp_sb001000469722 | 4 | 54108806 | 0.18932 | 1.91E-07 | 0 | ||
| snp_sb001000703373 | 6 | 48007508 | 0.126214 | 3.56E-06 | 0.65 | ||
| snp_sb001000719254 | 6 | 57704110 | 0.31068 | 1.29E-12 | 5.50 | ||
| snp_sb042060867162 | 8 | 5053406 | 0.085761 | 2.56E-06 | 0 | ||
| snp_sb042061028714 | 9 | 56601353 | 0.192557 | 2.76E-07 | 0 | ||
| snp_sb001001045903 | 10 | 5270775 | 0.11165 | 7.46E-06 | 0 | ||
| snp_sb001001049696 | 10 | 7303004 | 0.059871 | 4.02E-08 | 0.96 | ||
| BLINK | snp_sb042060176733 | 1 | 4778713 | 0.404531 | 8.40E-06 | 2.38 | |
| snp_sb042060379940 | 3 | 10836535 | 0.07767 | 2.70E-10 | 10.4 | ||
| snp_sb001000469722 | 4 | 54108806 | 0.18932 | 1.47E-06 | 7.34 | ||
| snp_sb042060589862 | 5 | 59844243 | 0.415858 | 1.89E-06 | 8.19 | ||
| snp_sb001000719254 | 6 | 57704110 | 0.31068 | 1.77E-10 | 5.38 | ||
| snp_sb042060843522 | 7 | 59997937 | 0.119741 | 1.15E-06 | 15.3 | ||
| snp_sb042060850458 | 7 | 63300346 | 0.127832 | 2.46E-06 | 5.29 | ||
| snp_sb042060867162 | 8 | 5053406 | 0.085761 | 5.97E-07 | 12.6 | ||
| 1 MAF, minor allele frequency (>0.05). 2 PVE, phenotypic variation (%).3 PH, plant height (cm).4,5,6,7 Non-significant SNP markers were identified by MLM, MLMM, CMLM, and ECMLM. a SNPs in bold were identified by two GWAS models (farmCPU and BLINK). | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




